Multicolored silver nanoparticles for multiplexed disease diagnostics: distinguishing dengue, yellow fever, and Ebola viruses†
Abstract
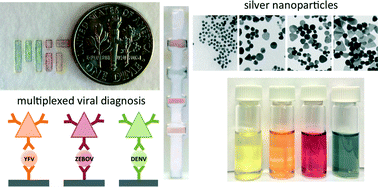
Rapid point-of-care (POC) diagnostic devices are needed for field-forward screening of severe acute systemic febrile illnesses. Multiplexed rapid lateral flow diagnostics have the potential to distinguish among multiple pathogens, thereby facilitating diagnosis and improving patient care. Here, we present a platform for multiplexed pathogen detection using multi-colored silver nanoplates. This design requires no external excitation source and permits multiplexed analysis in a single channel, facilitating integration and manufacturing.
- This article is part of the themed collection: Coronavirus articles - free to access collection

 Please wait while we load your content...
Please wait while we load your content...