Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic devices for imaging neurological response of Drosophila melanogaster larva to auditory stimulus†
Reza
Ghaemi
a,
Pouya
Rezai
ab,
Balaji G.
Iyengar
ac and
Ponnambalam Ravi
Selvaganapathy
*a
aDepartment of Mechanical Engineering, McMaster University, Hamilton, ON, Canada. E-mail: selvaga@mcmaster.ca
bDepartment of Mechanical Engineering, York University, Toronto, ON, Canada
cQiptera Solutions Inc. McMaster Innovation Park, Hamilton, ON, Canada
First published on 11th December 2014
Abstract
Two microfluidic devices (pneumatic chip and FlexiChip) have been developed for immobilization and live-intact fluorescence functional imaging of Drosophila larva's Central Nervous System (CNS) in response to controlled acoustic stimulation. The pneumatic chip is suited for automated loading/unloading and potentially allows high throughput operation for studies with a large number of larvae while the FlexiChip provides a simple and quick manual option for animal loading and is suited for smaller studies. Both chips were capable of significantly reducing the endogenous CNS movement while still allowing the study of sound-stimulated CNS activities of Drosophila 3rd instar larvae using genetically encoded calcium indicator GCaMP5. Temporal effects of sound frequency (50–5000 Hz) and intensity (95–115 dB) on CNS activities were investigated and a peak neuronal response of 200 Hz was identified. Our lab-on-chip devices can not only aid further studies of Drosophila larva's auditory responses but can be also adopted for functional imaging of CNS activities in response to other sensory cues. Auditory stimuli and the corresponding response of the CNS can potentially be used as a tool to study the effect of chemicals on the neurophysiology of this model organism.
1. Introduction
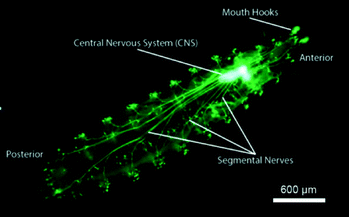
Drosophila melanogaster is a widely used model organism for studying human biology and diseases at the molecular-genetic level.1–3 This is due to its many advantages such as molecular-genetic, developmental, cellular/neuronal simplicity, genetic tractability and the increasingly incisive application of advanced optical methods for live imaging of biological processes. At its larval stages, Drosophila contains different types of sensory neurons that are patterned in a segmental configuration. They sense various environmental cues (e.g. mechanical, visual and chemical) and relay information to the Central Nervous System (CNS) to help elicit stereotypic motor behaviors. This simple architecture continues to be exploited for studying numerous developmental-genetic and neurobiological problems primarily through deploying surgical, histological, transgenic and behavioral methods.4,5Studying the behavioral responses in the larval stage of Drosophila using neuroimaging methods is challenging because the larva exhibits robust digging and burrowing behavior. This behavior is carried out by a cylindrical body wall that contains segmentally iterated sets of skeletal muscles and a specialized structure at the anterior end called the cephalopharyngeal skeleton (CPS). The latter is operated by specialized muscles to enable digging into food substrates.6 This digging movement is an impediment to temporal imaging of fluorescence activities in the larval sensory neurons and the CNS (see the ESI† S2). Conventional immobilization methodologies involving the use of anesthetic drugs will affect animals' neurophysiological status.7 Other methods, such as the use of tissue glue, used to immobilize embryos are irreversible, use solvents that could affect the physiological state and do not completely immobilize the CNS. Ideally, immobilization has to be performed in a simple and reversible manner while still allowing sensory stimulus to affect the larva. Miniaturized microfluidic devices are best suited for this purpose.
Microfabricated lab-on-chip devices are increasingly being used in the study of various model organisms such as Caenorhabditis elegans8,9 and Drosophila10–12 as they enable automated immobilization of these small organisms. After immobilization, visualization and tracking of cellular and physiological responses in vivo can be performed through their transparent body wall without motion artifacts. Microfluidic-based immobilization techniques for C. elegans have been developed using chemical (CO2) or mechanical (tapering microchannels or encapsulation using deflectable PDMS membranes) approaches.13–16 In the case of Drosophila, microfluidic devices have also been recently developed mostly to automate the embryo assays17,18 (self-assembly of eggs and morphogenesis,19,20 developmental studies21 and injection19,22) and less attention has been given to on-chip larval studies. Immobilization of Drosophila larvae is more difficult than that of C. elegans as it exerts stronger force. Complicating matters further, the internal organs of interest such as the CNS capsule that needs to be visualized can loosely move inside the hemolymph-filled body cavity even if the outer body is completely immobilized by encapsulation. Recently, mechanical encapsulation10 and CO2 anesthetic exposure11 approaches have been used to immobilize Drosophila larva. They allow whole-larval body compression inside the chip so that neuronal transport processes10 and sensory neuron regeneration upon injury11 can be visualized. Both these devices reduce the movement artifacts as compared to the freely moving larva but do not eliminate them. The use of anesthetic leads to spurious neurobehavioral responses and the use of encapsulation prevents the exposure of the larva to external sensory stimulus.
Very recently, behavioral responses of Drosophila larva in reaction to mechanical stimuli (vibration and sound) have been studied.23,24 Zhang et al.23 used Ca2+ imaging and electrophysiological methods and found that Drosophila larvae's Cho and class IV DA neurons responded optimally to sound waves at 500 Hz frequency. In these studies, the immobilization for Ca2+ imaging was achieved by compressing the larvae (in saline) between coverslips. This is a manual process and the degree of compression, the access of the larva to the external stimuli and the orientation of the animal are all variable and dependent on the operator. It is also conceivable that the overt whole-body mechanical compression could be disruptive for the full range of endogenous sensory-motor activities to take place.
In this paper, we demonstrate two lab-on-chip designs that standardize the immobilization of the larva while still allowing access to external stimuli and for the first time, conduct an on-chip study of the CNS activity of Drosophila larvae evoked by acoustic signals. The use of acoustic signals enabled fast, automated, remote and tunable stimulation of specimens. Specifically we investigated these responses fluorescently at the ventral cord of the CNS, an anatomical structure where a large majority of sensory afferents synapse with interneurons. We expressed a new genetically encoded calcium sensor GCaMP5 (ref. 25) in all sensory and cholinergic interneurons. Our devices are engineered to stabilize the CNS specifically from ongoing motor movements and the resulting internal hemolymph displacements. We demonstrate that localized anchoring of the larval CPS permits functional imaging of CNS in response to auditory stimuli. Our larval-lab-on-a-chip platforms will also be useful for studying CNS responses to other sensory cues (light, chemosensory, tactile, electric/magnetic fields).
2. Methods
The neurological response of Drosophila larvae to auditory stimuli was studied using two different lab-on-chip designs. Device design and fabrication, experimental procedures, data acquisition and processing as well as animal preparation are described in this section.2.1. Design of the microfluidic chips
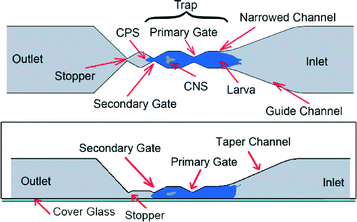
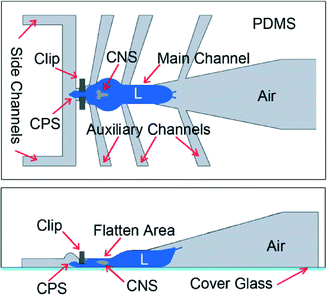
Drosophila larva's burrowing and locomotory behaviors make the CNS capsule move inside the hemolymph-filled body cavity because it is loosely anchored (Fig. 1). In order to immobilize the CNS of the larva and to subsequently study its neurological responses to auditory stimulus, two chips, named the pneumatic chip and the FlexiChip, were designed.2.2. Device fabrication
Devices were fabricated by 3D printing of two plastic master molds that dimensionally corresponded to the design discussed in section 2.1 for the pneumatic chip and the FlexiChip. Following master mold fabrication, soft lithography26 was used for conventional PDMS (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio base
1 ratio base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) agent, SYLGARD® 184) casting, curing (70 °C, 2 h), bonding to glass slides (80 s, 50 W, plasma oxygen) and installation of inlet/outlet tubes (for pneumatic chip, silicone tubing, 3/16" ID × 5/16" OD, Cole-Parmer Canada Inc.). The glass wire in the FlexiChip was placed into the 3D mold at the location of the clip before casting the PDMS into the mold.
agent, SYLGARD® 184) casting, curing (70 °C, 2 h), bonding to glass slides (80 s, 50 W, plasma oxygen) and installation of inlet/outlet tubes (for pneumatic chip, silicone tubing, 3/16" ID × 5/16" OD, Cole-Parmer Canada Inc.). The glass wire in the FlexiChip was placed into the 3D mold at the location of the clip before casting the PDMS into the mold.
2.3. Experimental setup
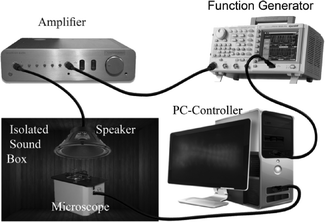
The experimental setup (Fig. 4) consisted of a sound-insulated box (custom made to isolate environmental noises using acoustic sound damping foam UL 94, Parts-Express, USA), an acoustic signal generation system (function generator (AFG3022B, Tektronix, CA), amplifier (RAMSA WO-1200, Panasonic, CA) and speaker (Eminence Beta-12CX coaxial 12", Parts-express, USA)), an optical/fluorescence imaging system (Lumascope 500, single color 488 nm Ex. fluorescence, 40× magnification, Etaluma, CA), the microfluidic device and a software control system (LabVIEW©, flyCapture2© and ImageJ© software).The function generator connected to the amplifier was controlled through a custom LabVIEW© code and was used to generate the desired pure-tone sinusoidal voltage signal inputs to the speaker. The speaker was installed on the roof of the sound insulating box. Various voltage frequency and intensity levels were generated and amplified to the speaker and the corresponding sound frequency (50–5000 Hz) and intensity (95–115 dB) produced in the box were measured using a mini sound level meter (DT-85A, CEM). This was done to calibrate the speaker and the sound insulating box. Frequency ranges were selected to cover the hearing range of the response of Drosophila23 and intensity levels were selected using preliminary experiments that produced a response in the CNS.
The microscope was positioned inside the box right beneath the speaker with a 15 cm distance between its focal plane and the speaker. The microscope was controlled by software and used in the optical mode for loading the animal and in the fluorescence mode for imaging GCaMP5 activities in the CNS of an immobilized larva.
2.4. Animal loading
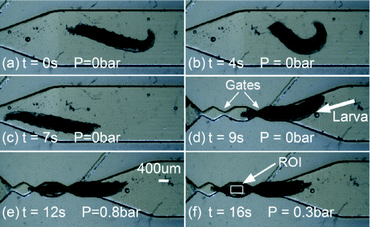 | ||
| Fig. 5 Steps to load the larva using the pneumatic chip. (a–d) The larva swam freely into the trap, and (e–f) the larva was pneumatically moved into the trap and immobilized. Time-lapsed fluorescence imaging was then conducted on the CNS located inside the Region of Interest (ROI). Scale bar = 400 μm for all figures is shown in Fig. 5e. | ||
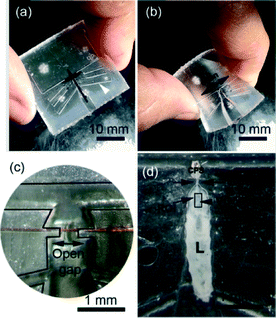 | ||
| Fig. 6 Steps to load the larva using the FlexiChip. The chip (a) is bent (b) so that the clip (c) opens. Then, the animal is inserted into the gap and the chip is released and sealed by a coverslip (d). Time-lapsed fluorescence imaging was then conducted on the CNS located inside the Region of Interest (ROI).31 | ||
Afterwards, a cover-glass was placed on top of the larva (Fig. 6d) before visualization of fluorescence activities in the ventral cord aspect of the CNS (see the ESI† S4) where a large majority of afferent sensory inputs from the body wall arrive. The larval posterior-end protruded into a funnel shaped outer chamber that was open to ambient air. This allowed respiration to continue through posterior spiracles during live imaging. The procedure for loading the larva into the chip takes approximately 5 min.
2.5. Automated animal testing
After the animal was properly loaded into the trap and immobilized, as shown in Fig. 5 and 6, the auditory response of the larva was examined at the abdominal ganglia region of the ventral cord. A custom-made LabVIEW© code controlling the function generator was used to generate a step-like periodic series of acoustic waves (5 s on and 5 s off) while the animal's CNS fluorescence signal activities were recorded in a movie format using the microscope. Each on-portion cycle of the applied wave corresponded to one frequency (50, 100, 200, 500, 1000, 2000 or 5000 Hz) and one intensity (95, 105 or 115 dB) level. The experiment was continued automatically until the entire 21 frequency–intensity combinations were tested. The animal was then washed off the chip and another one was loaded to repeat the experiments. The movies were then analysed as discussed below for quantification of neuronal activities.2.6. Data acquisition and processing
The movies recorded for each animal were analyzed by ImageJ© software (National Institutes of Health, USA) to quantify the fluorescence intensity variations in the CNS in response to the applied acoustic signals. The RGB image sequences for each video were converted to 8-bit black and white images (256 shades of gray). After subtracting the background with a rolling ball radius of 100 pixels, a Region of Interest (ROI in Fig. 5f and 6d) covering the CNS was selected and the mean gray value for the entire image stack was measured inside the ROI and recorded in an Excel file. The intensity variation in each condition was calculated by taking the ratio of the increase in the mean gray value in the ROI during the stimuli to the mean gray value 2 seconds before sound was applied in each experiment.It is important to note that the inherent movement of the animal also results in an increase in CNS activity that may lead to elevated baseline reading. Movement was measured as the change in the center of mass of the CNS and experiments that had high CNS movement were not included in the analysis.
2.7. Animal preparation
Larvae of the genotype w, Cha-Gal4/CyO; UAS-GCaMP5/TM3, Sb were used for imaging CNS activity in response to auditory stimulations. Heterozygotes and homozygotes were not separated before testing. Expression of the GCaMP5 GECI was conducted using the Gal4/UAS system.27 Through standard fly crosses, a stable fly stock was created containing two transgenes: (1) Cha-Gal4 is a promoter sequence of choline acetyltransferase (Cha) driving the expression of the Gal4 transcription factor,28 and (2) UAS-GCaMP5 transgene contains the binding sites for the Gal4 transcription factor.25 Thus, in the Cha-Gal4/CyO; UAS-GCaMP5/TM3 strain, all sensory and central neurons that express the choline acetyltransferase gene express the GCaMP5 calcium sensor. The GCaMP calcium sensor is a circularly permuted protein containing the Green Fluorescent Protein (GFP), calcium binding protein called calmodulin, and the M13 (calmodulin binding) peptide.29 Influx of Ca2+ during neuronal activity triggers a conformational change of GCaMP so that solvent access to the chromophore is prevented, thus resulting in a higher level of fluorescence.30 GCaMP5 is a recently developed high signal-to-noise ratio calcium sensor.25 This genotype was generated through a standard genetic crossing scheme. Third instar stage larvae were isolated using a fine brush, washed with distilled water and dried on a tissue paper before loading into the chips.3. Results and discussion
3.1 Acoustic response of Drosophila 3rd instar larva
After immobilizing the larva (3rd instar) inside each of the PDMS devices, as shown in Fig. 5 and 6, neuronal activities in the CNS in response to a sound wave (5 s duration, 200 Hz frequency and 105 dB intensity) were measured using the experimental setup described before (Fig. 4). We measured the frequency and intensity of the sound inside both of the devices and found them to be the same as those outside. As shown in Fig. 7 (response of the immobilized larva in the pneumatic chip), the animal's CNS activity increased by 21% (reported by an increase in average fluorescence intensity) upon exposure to the sound signal and returned to its original state within 0.5 s after the signal was turned off (image not shown due to similarity to Fig. 7a). This type of response was observed consistently for 5 animals tested under the same acoustic conditions and each time it was tested.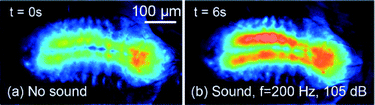 | ||
| Fig. 7 Snapshots of the fluorescence activities in the CNS of a larva (a) before and (b) while it was exposed to a 5 s duration sound wave (200 Hz and 105 dB) in the pneumatic chip. | ||
3.2 Investigation of the effect of sound frequency and intensity on Drosophila larva CNS activities
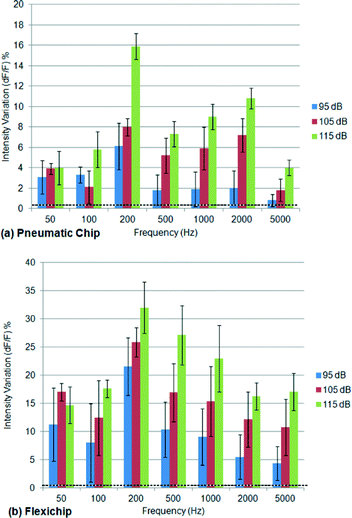
The interesting observation of a significant CNS activity in response to a sound signal (Fig. 7) encouraged us to investigate this phenomenon further in detail. Hence, we recorded the CNS activities of Drosophila 3rd instar larvae in response to sound signals of various intensity (95–115 dB) and frequency (50–5000 Hz) levels using both the pneumatic chip and the FlexiChip (Fig. 8, averaged for n = 5 animals).As shown in Fig. 8, the animals tested in both chips demonstrated a statistically significant increase in CNS activities when the frequency of the sound signal was increased from 50 Hz to 200 Hz. A further increase in frequency resulted in reduced CNS activities as compared to the 200 Hz condition. In addition to a peak in response at 200 Hz, a secondary but less significant peak in CNS response was also observed at 2000 Hz and only inside the pneumatic chip (Fig. 8a). This peak was more significantly pronounced at higher intensity levels (105 and 115 dB). In contrast, at 95 dB the difference between the peak frequency and the rest can be observed but is not statistically significant due to the lower signal to noise ratio (see the ESI† S3).
The level of mechanical vibrations induced by the sound waves at frequencies less than 50 Hz on the chip did not allow clear imaging of the CNS. The CNS response continued to decrease further beyond 5000 Hz (data not shown). The increase in sound intensity from 95 dB to 115 dB resulted in the corresponding increase in CNS activity. The increase in the sound level resulted in the reduction in the signal to noise ratio, indicating that the auditory response of the fly at a higher sound level was clearer.
In order to compare this method with other immobilization methods such as anesthetization, the response of 3rd instar larvae to pure tone sounds was measured before and after exposure to ether. The results indicate that the response to auditory stimulus was quite noticeably reduced in anesthetized larvae compared to the control sample, as shown in the ESI† (S1).
The peak observed in the CNS response at 200 Hz sound waves is in contrast to the recently reported observation that the optimal neuronal response to auditory stimulus in the larva occurs at 500 Hz. We noted two major differences between our experimental design and that used by Zhang et al.23 First, in Zhang et al.,23 the behavioral, calcium imaging and electrophysiological measurements involved the placement of the animals (or semi-intact preparations) directly on top of a speaker that delivered the auditory stimulus which may have coupled some of the vibrations to the larva as tactile stimulations. The use of whole body compression provides sufficient contact of the substrate to activate Cho sensory neurons that are spread throughout the body. Second, our devices exhibit physical separation of the speaker from the device, ensuring that it is the sound waves that cause the response while also providing a better simulation of sound cues that occur in nature. Since the larva is a burrowing animal, it is likely that the sensory neurons in the posterior abdominal segments are better tuned to a sound frequency that matches the wing beat frequency of the predatory wasp. We also test two different designs to separate any possible tactile stimulation effects.
4. Conclusions
With the availability of a new generation of reagents for monitoring neuronal activity through imaging and electrophysiological methods, it is necessary to develop custom engineered microdevices to facilitate experimental manipulations in intact-living specimens. We designed and evaluated two devices to anchor the Drosophila larval CNS so that stable optical recording of its neuronal activities could be conducted. The reduction in CNS movement was achieved through an on-chip mechanism that isolated the larval segments within which the CNS capsule is suspended. These microfluidic chips allowed us to stabilize the CNS specifically from ongoing motor movements and the resulting internal hemolymph displacements while the immobilization technique did not use any anaesthetic drugs which would affect animals' neurophysiological status. The pneumatic chip allowed automated animal loading, immobilization and unloading and it held the larva under positive fluid pressure to reduce the CNS movement entirely. However, since this is a closed-configuration chip, access of the larva to sound stimulation was indirect. The FlexiChip allowed for manual loading, unloading and immobilization. The posterior end of the larva inside the FlexiChip was open, thus allowing the larva to respire, and also for the acoustic vibrations to reach the larval body directly. The stability of the CNS inside both chips enabled the visualization of neuronal activities using a Genetically Encoded Calcium Indicator (GECI) probe, called GCaMP5, in response to auditory stimuli. Both chip designs allowed the stable recording of GCaMP5 fluorescence activity in the CNS. We report an optimal GCaMP5 response at 200 Hz. In conclusion, our customized larval lab-on-chip platforms allow the integration of functional imaging with a sensory-motor response. We anticipate that our intact larva-on-a-chip will also be useful for other studies that involve calcium imaging as well as optogenetic and electrophysiological approaches.Acknowledgements
The authors gratefully acknowledge financial support from the Natural Science and Engineering Research Council of Canada (NSERC) through their Discovery Program, The Canada Research Chairs Program and the Ontario Ministry of Research and Innovation through their Early Researchers Award.Notes and references
- S. Tickoo and S. Russell, Curr. Opin. Pharmacol., 2002, 2, 555–560 CrossRef CAS.
- S. Lenz, P. Karsten, J. B. Schulz and A. Voigt, J. Neurochem., 2013, 127, 453–460 CrossRef CAS PubMed.
- E. Bier, Nat. Rev. Genet., 2005, 6, 9–23 CrossRef CAS PubMed.
- J. B. Duffy, Genesis, 2002, 34, 1–15 CrossRef CAS PubMed.
- H. Kohsaka, S. Okusawa, Y. Itakura, A. Fushiki and A. Nose, Dev., Growth Differ., 2012, 54, 408–419 CAS.
- A. Schoofs, S. Niederegger, A. van Ooyen, H. Heinzel and R. Spiess, J. Insect Physiol., 2010, 56, 695–705 CrossRef CAS PubMed.
- S. Mondal, S. Ahlawat and S. P. Koushika, J. Visualized Exp., 2012, 12, 372–385 Search PubMed.
- P. Rezai, S. Salam, P. Selvaganapathy and B. P. Gupta, in Integrated Microsystems, ed. K. Iniewski, CRC Press, 2011, pp. 581–608 Search PubMed.
- S. E. Hulme and G. M. Whitesides, Angew. Chem., Int. Ed., 2011, 50, 4774–4807 CrossRef CAS PubMed.
- S. Mondal, S. Ahlawat, K. Rau, V. Venkataraman and S. P. Koushika, Traffic, 2011, 12, 372–385 CrossRef CAS PubMed.
- M. Ghannad-Rezaie, X. Wang, B. Mishra, C. Collins and N. Chronis, PLoS One, 2012, 7, e29869 CAS.
- E. S. Heckscher, S. R. Lockery and C. Q. Doe, J. Neurosci., 2012, 32, 12460–12471 CrossRef CAS PubMed.
- T. V. Chokshi, A. Ben-Yakar and N. Chronis, Lab Chip, 2009, 9, 151–157 RSC.
- K. Chung, M. M. Crane and H. Lu, Nat. Methods, 2008, 5, 637–643 CrossRef CAS PubMed.
- C. L. Gilleland, C. B. Rohde, F. Zeng and M. F. Yanik, Nat. Protoc., 2010, 5, 1888–1902 CrossRef CAS PubMed.
- F. Zeng, C. B. Rohde and M. F. Yanik, Lab Chip, 2008, 8, 653–656 RSC.
- J. R. Fakhoury, J. C. Sisson and X. J. Zhang, Microfluid. Nanofluid., 2009, 6, 299–313 CrossRef CAS.
- K. I. Wang, Z. Salcic, J. Yeh, J. Akagi, F. Zhu, C. J. Hall, K. E. Crosier, P. S. Crosier and D. Wlodkowic, Biosens. Bioelectron., 2013, 48, 188–196 CrossRef CAS PubMed.
- G. T. Dagani, K. Monzo, J. R. Fakhoury, C. C. Chen, J. C. Sisson and X. Zhang, Biomed. Microdevices, 2007, 9, 681–694 CrossRef PubMed.
- T. J. Levario, M. Zhan, B. Lim, S. Y. Shvartsman and H. Lu, Nat. Protoc., 2013, 8, 721–736 CrossRef PubMed.
- E. M. Lucchetta, M. S. Munson and R. F. Ismagilov, Lab Chip, 2006, 6, 185–190 RSC.
- S. Zappe, M. Fish, M. P. Scott and O. Solgaard, Lab Chip, 2006, 6, 1012–1019 RSC.
- W. Zhang, Z. Yan, L. Y. Jan and Y. N. Jan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 13612–13617 CrossRef CAS PubMed.
- T. Ohyama, T. Jovanic, G. Denisov, T. C. Dang, D. Hoffmann, R. A. Kerr and M. Zlatic, PLoS One, 2013, 8, e71706 CAS.
- J. Akerboom, T. W. Chen, T. J. Wardill, L. Tian, J. S. Marvin, S. Mutlu, N. C. Calderon, F. Esposti, B. G. Borghuis, X. R. Sun, A. Gordus, M. B. Orger, R. Portugues, F. Engert, J. J. Macklin, A. Filosa, A. Aggarwal, R. A. Kerr, R. Takagi, S. Kracun, E. Shigetomi, B. S. Khakh, H. Baier, L. Lagnado, S. S. Wang, C. I. Bargmann, B. E. Kimmel, V. Jayaraman, K. Svoboda, D. S. Kim, E. R. Schreiter and L. L. Looger, J. Neurosci., 2012, 32, 13819–13840 CrossRef CAS PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- A. H. Brand and N. Perrimon, Development, 1993, 118(2), 401–415 CAS.
- P. M. Salvaterra and T. Kitamoto, Gene Expression Patterns, 2001, 1(1), 73–82 CrossRef CAS.
- J. Nakai, M. Ohkura and K. Imoto, Nat. Biotechnol., 2001, 19(2), 137–141 CrossRef CAS PubMed.
- J. Akerboom, J. D. Rivera, M. M. Guilbe, E. C. Malavé, H. H. Hernandez, L. Tian, S. Hires, J. S. Marvin, L. L. Looger and E. R. Schreite, J. Biol. Chem., 2009, 284(10), 6455–6464 CrossRef CAS PubMed.
- http://figshare.com/articles/FlexiChip1_0_A_simple_device_for_in_vivo_imaging_of_sensory_motor_activities_in_the_Drosophila_larval_CNS/644498 .
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4lc01245c |
| This journal is © The Royal Society of Chemistry 2015 |