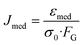Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A monolithic glass chip for active single-cell sorting based on mechanical phenotyping†
Christoph
Faigle
a,
Franziska
Lautenschläger
b,
Graeme
Whyte
 b,
Philip
Homewood
c,
Estela
Martín-Badosa
d and
Jochen
Guck
*ab
b,
Philip
Homewood
c,
Estela
Martín-Badosa
d and
Jochen
Guck
*ab
aBiotechnology Center, Technische Universität Dresden, Dresden, Germany. E-mail: jochen.guck@biotec.tu-dresden.de
bCavendish Laboratory, Department of Physics, University of Cambridge, Cambridge, UK
cDolomite Microfluidics, Royston, UK
dDepartament de Fisica Aplicada i Optica, Universitat de Barcelona, Spain
First published on 16th December 2014
Abstract
The mechanical properties of biological cells have long been considered as inherent markers of biological function and disease. However, the screening and active sorting of heterogeneous populations based on serial single-cell mechanical measurements has not been demonstrated. Here we present a novel monolithic glass chip for combined fluorescence detection and mechanical phenotyping using an optical stretcher. A new design and manufacturing process, involving the bonding of two asymmetrically etched glass plates, combines exact optical fiber alignment, low laser damage threshold and high imaging quality with the possibility of several microfluidic inlet and outlet channels. We show the utility of such a custom-built optical stretcher glass chip by measuring and sorting single cells in a heterogeneous population based on their different mechanical properties and verify sorting accuracy by simultaneous fluorescence detection. This offers new possibilities of exact characterization and sorting of small populations based on rheological properties for biological and biomedical applications.
1. Introduction
Traditionally, interest in the functioning of biological cells has mainly been focused on molecular cell biology. In recent years however, the research focus has increasingly expanded to include biophysical and biomechanical properties of cells. It has been shown that cell mechanics is a sensitive marker to detect functional changes in cells, not only during pathological alterations such as cancer1–4 and metastasis,5 but also during physiological cellular processes such as differentiation,6,7 proliferation8 and motility.9How, then, are mechanical properties measured? There is a variety of experimental possibilities covering a wide range of spatial and temporal scales. Localized stress responses of single cells can be probed using magnetic bead rheometry10,11 or atomic force microscopy.12,13 However, these methods have a low throughput and can only be used on adherent cells. A more global mechanical response of single cells can be measured by optical tweezers14 or microplate manipulation,15 however once again only with a low throughput. On the other end of the temporal spectrum, suspended cells can be rapidly measured by high-throughput methods such as hydrodynamic stretching16,17 or high-frequency modulated viscoelasticity measurements by optical deformation.18 However, with these latter techniques, where mechanics is determined explicitly for each cell, no sorting into mechanical subpopulations has been shown.
Obviously, in heterogeneous cell populations, depending on their type or state, different sub-populations will have different characteristic properties, and the question of how to separate these populations comes to mind. Ideally, this is done without the use of additional markers such as fluorescent tracers or magnetic beads, as they might affect cell behavior. There are a flurry of microfluidic approaches in recent years that can sort cells even at a very high throughput. These include sorting by deterministic lateral displacement,19 inertial microfluidics,20 cell compression in ridges21 or ratched-based microfluidic systems.22 However, these are all passive. Sorting depends on flow geometries which have to be chosen a priori for a certain flow speed and a certain cell property (size and stiffness). They do not offer the possibilities of changing sorting boundaries during an experiment or reusing the same device for different experimental conditions.
To overcome these limitations, the microfluidic optical stretcher (MOS), a dual laser beam trap-based tool that is able to trap cells from a microfluidic flow channel and deform them with optical forces, offers a solution, since it combines several advantageous aspects. The MOS allows cells naturally occurring in the suspension to be measured and allows a reasonably high throughput of around 100 cells h−1, while retaining the capability of detailed viscoelastic measurements. The MOS can measure cell mechanics over a broad optical force and time range, allowing for simultaneous measurement of parameters such as short time elastic responses, long-term creep compliance,6 or cortical tension.23 The non-contact measurement and the microfluidic system open up the possibility of recovery and reuse of cells after measurement. If needed, the setup can easily be upgraded with additional features such as fluorescence microscopy, spectroscopy, or refractive index measurement. Cells that have been measured in the MOS include red blood cells,24,25 white blood cells,9,26 embryonic stem cells,27 fibroblasts28 and cancer cells.29,30 Recently, it was shown that certain cells can be distinguished among different heterogeneous cell populations, based on a variety of viscoelastic parameters.1,6,31 However, it has not yet been possible to separate these characterized populations.
We propose a novel version of the microfluidic optical stretcher based on a design which makes it more simple to set up and handle via the fabrication of an integrated optofluidic chip, consisting of two asymmetrically etched glass pieces that are then permanently bonded. Optimal cell trapping is ensured by plug-and-play like optical fiber insertion, using a three-point alignment technique. The thin and optically flat bottom of the assembled chip allows for high-resolution imaging. In this paper, we report the fabrication and use of this chip, as well as its application for optical sorting of treated cells based solely on viscoelastic properties, confirmed by fluorescence detection.
2. Design principle
Before successfully fabricating a new microfluidic optical stretcher, taking the time to consider design aspects is paramount. These aspects can be broadly categorized into three points: optical manipulation of cells, their observation, and chip-to-world connection and fluidics. Manipulation aspects are those that involve the fundamental principle of the optical stretcher: the trapping and deformation of cells with counterpropagating laser beams. The laser needs to be delivered by a waveguide with a Gaussian beam profile, and the beam path from fiber to cell should be such that the beam profile is not significantly deteriorated, which reflects directly on trapping efficiency. These laser beams need to be aligned on an axis and the height of the axis should be well-adjusted with respect to the microfluidic channel in order to manipulate as many cells as possible. Also, the fabrication material of the chip should offer a high laser damage threshold to ensure a long lifetime.Concerning the observation of measurements, there needs to be optical access to the manipulation region. For high quality measurements, the surface of the microfluidic channel where cells are handled should be as smooth as possible and optically transparent. Chip-to-world connection in its simplest form involves the introduction and extraction of cells via microfluidic connectors and the delivery of the cells to the manipulation region via a microfluidic channel, with a controlled flow. Here it takes the form of a more complex microfluidic design which can be used for sorting cells after measurement. It can also offer other functionalities such as influx of additional reagents for studying the immediate mechanical response of cells. At the same time, cleaved optical fibers have to be brought sufficiently close to the manipulation region inside the chip. In general, the design should be simple to manufacture, use and maintain.
Optical manipulation
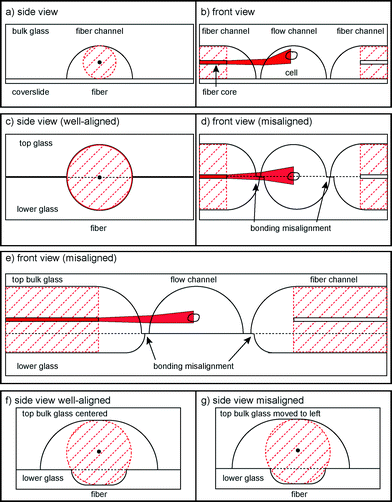
In previous published versions of the MOS, the fundamental requirements concerning optical manipulation were met with varying success. The current state-of-the-art MOS consists of a square glass capillary with microfluidic connectors and perpendicularly aligned optical fibers on an SU-8 structured surface32,33 and fulfils all requirements. Another version is a microstructured PDMS chip with integrated fiber channels.34 However, the height of the laser beam axis cannot be adjusted with respect to the flow channel, so many cells flow underneath the beams and cannot be trapped. Another approach to this problem is building a microstructured glass chip with femtosecond laser micromachining.35,36 Instead of using optical fibers, waveguides are written in bulk glass by the laser at the correct height. However, this process slightly deteriorates beam quality. Although very flexible, this method requires highly specialized equipment for production, a significant cost factor. We decided to use more standard manufacturing processes, by wet etching37 in bulk glass, for our fabrication.One way to do this is to etch flow and fiber channels into one glass piece and cover it with a thin cover slip (Fig. 1a, b). Unfortunately, this does not circumvent the problem that also occurred within the PDMS chip: cells passing beneath the beam axis. Also, the rounded flow channel walls refract the laser beam which reduces the trapping efficiency. It is thus necessary to lower the beam axis with respect to the flow channel. This can be achieved by etching identical and symmetrical channels into two pieces of glass and bonding them (Fig. 1c, d). In this case, the curvature of the channel wall at the point of intersection with the laser beams is much lower. However, it is not possible to align the two glass halves in such a way that there is no offset at exactly the height of the laser axis. This significantly distorts the laser beam profile due to the one-sided phase retardation. We overcame this problem by using two asymmetrically etched glass pieces (Fig. 1e). The upper side contains both the larger part of the optical fiber and the entire flow channel. This way, it is ensured that the beam axis does not intersect with the interface of the glass pieces. By careful choice of the chip layout it is possible to create a design with the dimensions in a similar range as the previously used optical stretcher designs, such as a flow channel with a diameter in the 100 μm range, the distance between the two optical fibers not exceeding a few hundred μm, since stretching efficiency decreases with increasing distance. The most relevant parameter is the height between the trapping location and the bottom of the channel. If the optical trap is placed too high, cells flowing beneath it will not feel enough optical force to be trapped, and if it is placed too low, reflections from the bottom of the channel will cause interference in the trapping field, reducing trapping efficiency and forces.
Once the fibers are in place, the beam path has to be considered. Stretching efficiency is related to the beam width, shape, and laser intensity that the cell encounters in the trap. The complex geometries and material changes that the laser beam encounters after exiting the optical fiber make more detailed consideration necessary. We used FTFD simulations to calculate the optimal geometries (cf. Experimental section).
Furthermore, it is necessary to align the optical fibers exactly along the same axis to ensure stable trapping and symmetric distribution of stresses. In the single capillary setup, this was solved by microstructuring SU-8 channels that held fibers in place and applying pressure from the top. In the PDMS chip the fiber channels had no clearance in their respective channels, so fibers were automatically aligned. In the laser-micromachined chip, waveguides were written in one process and thus along one axis. However, when bonding two glass halves together, imperfect bonding can lead to misalignment, making it impossible to introduce the fibers into the chip. The solution to this problem is achieved by using a 3-point alignment strategy (Fig. 1f, g) to secure the fibers in place, with the roof of the upper channel and the corners of the edge of the bottom channel providing the contact points. In order to reduce the effect of lateral misalignment, the mask used for etching the upper channel is wider than the lower, which gives the top of the upper channel a flat profile. This allows the two glass plates to be laterally misaligned by up to 10 μm without affecting the 3-point alignment and ensures that both fibers are still co-aligned (see the ESI† Fig. S3).
Observation of cells
The preferred bulk material affects the chip both in terms of resistance to high laser power, as well as imaging quality at the trapping region. The materials in the optical stretcher need to be transparent for both infrared lasers and visible light for imaging. PDMS, a clear silicon-based organic polymer, is widely used as a material for microstructuring. However, in combination with the high laser intensity, contaminations in the PDMS chip34 can start to burn, leading to rapid degradation of the chip. Bulk glass does not have these contaminations and can resist the high laser powers needed. With respect to image quality, only distortion-free imaging allows even small cell elongations to be detected, and also offers the possibility of using fluorescence or spectroscopic analysis. One previous version of the optical stretcher written in a monolithic glass chip by femtosecond lasering36 had the problem that the fluidic channel fabrication process led to a rough bottom surface, distorting the imaging. We counteracted the problem by only etching the fluidic channel into the bulk side of the glass and lowering the fiber channels accordingly. This way, the thin glass slide acts as the polished bottom of the fluidic channel, avoiding the negative effects of distortion.Chip-to-world connections
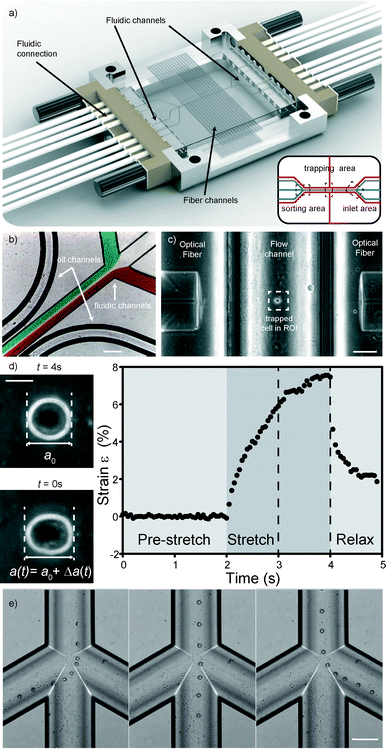
Previous versions of the MOS only involved flowing a cell to and from the manipulation region along a single channel. Usually, standard microfluidic connectors were connected. In this case, it is not possible to have more than one microfluidic channel. Creating more complex channel geometries, such as multiple flow inlets and outlets, or multiple fiber channels, greatly extends the functionality of the optical stretcher. This can only be done by microstructuring, as was done in PDMS34 or in glass, by femtolaser micromachining35 or by wet etching. Two unconnected channel geometries are included in the system: fluidics and optics. The fluidic geometry deals with the transport of cells. To ensure leakage free connection of tubing to the chip, we used an 8-way linear connector (see Fig. 2a) that was tightly pressed to the chip side. The optical-gel channels can be also accessed from the connector.Laminar flow is used to bring cells into the trap. To be able to control the flow speed exactly is necessary since during measurement of the cell, the flow is set to zero. Any flow could lead to rotation of the cell and thus to distortion of the measurement. To ensure laminar flow, the geometries and flow speeds have to be in a low Reynolds regime, and to accurately control the flow within the novel device, differential pressure was applied across the input and output channels. By measuring the fluid flow rate in and out of the chip through various combinations of channels, it is possible to determine the flow resistance of all the internal parts of the chip allowing control of the flow rate in any chosen part of the chip. By controlling the flow out of the chip it is possible to sort the cells into the three different outlets based on any measurable properties (see Fig. 2e). In this case however, we chose not to use this function for our experiments since the sorting region was placed too far away from the trapping region, making it hard to ensure that the correct cell flows into its corresponding channel. Instead, after measurement we use a laser beam from one side to push the cell into streamlines at the trapping area. This takes longer and decreases throughput, but the laminar flow regime ensures that the cell flows into the correct sorting channel. Finally, chip and fluidic connections are held in a custom-designed aluminum frame, making this chip a simple and easy to handle optofluidic add-on for any microscope stage.
3. Experimental
3.1. Simulations
To investigate the consequences of the curved surfaces and different fiber channel heights, we ran FTFD simulations using the software package Meep38 to model the propagation of light through various possible designs of the chip (see the ESI† Fig. S1). The curved walls refract the light while the height of the fiber axis above the bottom of the channel determines if there are reflections on the channel bottom or top surface. By etching the fiber channels into both pieces of glass it is possible to alter the position of the trap, while keeping the fiber channel at a constant height. The trap height above the bottom is chosen by etching the two sides of the fiber channels to different depths, and adding up to the diameter of a standard optical fiber, 125 μm (see the ESI† Fig. S2). The final design consists of a top channel of 86 μm and a bottom channel of 40 μm. This places the trap 27 μm above the floor of the microfluidic channel. Such a design places the fibers 193 μm from the center of the microfluidic channel and results in the trap being 28 μm wide, leaving the edge of the trap 13 μm above the floor, which minimizes its interference effects.3.2. Cell culture
HL60/S4 myeloid precursor cells39–41 were chosen as the model cells for this study, because they naturally grow in the suspension, which means they are measured in their physiological environment in a microfluidic optical stretcher. The cells were incubated at 37 °C with a 5% carbon dioxide level. All optical stretching experiments were performed within 2 h after the cells were taken out of the incubator. For fluorescence, cells were stained with Hoechst 33258 nuclear stain (Sigma-Aldrich, 861405) according to the standard protocol. To increase the deformability of the cells, they were treated with 0.2 μM cytoskeletal drug cytochalasin D (Sigma-Aldrich, C2618) and incubated for 15 minutes at 37 °C. Subsequently, in both cases, cell solutions were removed by centrifugation and cells were resuspended in RPMI 1640 medium.3.3. Microscopy and experimental setup
The principle and set-up of the microfluidic optical stretcher have been described extensively elsewhere.25 The microfluidic chip was fabricated by Dolomite Microfluidics (Royston, UK) according to the design specified above and mounted on an inverted phase contrast microscope (Axio Observer, Carl Zeiss GmbH, Jena, Germany) with a CCD camera. The laser used was a 1060 nm Stretcherlaser Yb-2x33 V 2.0 (Fibotec Fiberoptics GmbH, Meiningen, Germany). After insertion of the optical fibers, the support channels were filled with an index-matching gel (G608N, Thorlabs, Dachau, Germany). Fluidic flow was controlled with a Fluigent MCFS pressure pump (Fluigent, Villejuif, France). The power used for trapping was 0.2 W per fiber, and during stretch 1.2 W per fiber was applied. The microfluidic channel and all connected tubing were filled with phosphate buffered saline. Cells were injected via a 4-way port and care was taken to keep the system air bubble free. Flow speed could be remote-controlled via the microfluidic pressure pump. Every single cell flowed to the trap region, the flow was set to zero, and the cell was trapped by turning on the lasers to trapping power. After waiting until the cell had aligned itself along its major axis and in the center of the channel, the stretching phase was activated. The cell was held at trapping power for two seconds, and then a power rise in step form was applied for two seconds. During another second after the stretch, the cell relaxed at trapping power. During the pre-experiments, the cell was then released by turning off the laser power. During the sorting experiment, power was turned off only on one side for 10 seconds, after which the cell was released. After cell injection, we measured for 30 minutes each batch, allowing 30–50 cells to be measured per experiment. We repeated the experiments three times for each group of cells, leading to a total of around 100 cells per group. We measured the average refractive index of cells of each type using a digital holographic microscope,27 which showed that there was no significant difference in the refractive index (untreated cells = 1.379 ± 0.0036, cytochalasin D-treated cells = 1.380 ± 0.0056).Viability tests were performed using the vital stain trypan blue25 (see the ESI† Fig. S5). While live cells exclude the dye, dead, membrane-compromised and otherwise non-functional cells will allow the dye to permeate the cellular membrane, which is visible under a microscope by a distinct blue tinge inside the cell. Our tests showed that stretched cells were not detectably damaged, which means that cellular viability is not affected by optical stretching.
3.4. Mechanical analysis and sorting
Custom written LABVIEW software was used to control the laser, flow and camera. Edge detection analysis as described by Lincoln et al.33 was performed in real-time. Data for the major axis length a(t) were stored for every time-frame, while a0 was calculated as the mean length of the major axis during the initial trapping period. The time-varying axial strain| ε(t) = (a(t) − a0)/a0, |
The optical stress depends on the geometries of the chip, while the geometric factor takes the size of the cell into account. Calculations with generalized Lorenz–Mie theory42 using custom MATLAB software revealed an average GGF of 0.92.The area of the cell was calculated for each time-frame by triangulation from the center of mass of the cell to two neighboring radial edge points, determined by the program. During the sorting phase, after measurement, the program compared the median compliance value of the cell with a user-defined cut-off value and used a single laser beam to push it to the left or right channel side depending on whether the measured value is larger or smaller than the cut-off.
4. Results and discussion
4.1. Microfluidics and sorting
We developed a microfluidic glass chip consisting of three distinct regions (Fig. 2a inset). The first region consists of three inlet channels which merge into a single channel (Fig. 2b). Measurement and analysis of cells take place in this channel (Fig. 2c and d), the second region, where optical fibers orthogonally inserted from the side into separate channels irradiate the cells. This central channel then splits into a three-channel outlet again, where cells are sorted (Fig. 2e), the third region. To accurately control flow within the device, differential pressure was applied across the input and output channels. More detailed design parameters can be found in the experimental section.At the inlet region, the laminar flow regime allows different fluids to flow in from the side channels without mixing (Fig. 2b). The cells coming in through the middle channel then proceed down the single channel to the measurement region, enveloped by a sheath stream from the side channels. By turning on the laser when a cell is positioned between the two fibers, the cell is trapped in the optical trap (Fig. 2c). Cells can be trapped with as little as 200 mW laser power per fiber. By increasing the laser power, the forces acting on the surface of the cell become large enough to measurably deform cells (Fig. 2d). Although the laser beam passes through a curved channel wall, this is not a problem, since the curvature near the bottom is still low. In this case, the slight refraction is even beneficial since backcoupling into the opposite fiber is avoided (see the ESI† Fig. S1). As a demonstration of the stretching capabilities of the chip, HL60 cells were tested in the chip. HL60 cells were chosen as they are naturally in the suspension, are ideally suited to the optical stretcher and have previously been well characterized in the optical stretcher.6 Also in this new chip, stretching gives rise to characteristic deformation curves (Fig. 2d).
In order to have a choice in fiber-to-cell distances and not be constrained by eventual manufacturing inaccuracies, we placed a range of fiber channels with different fiber end-to-end distances on the channel. We also added two support channels along the side of the microfluidic channel which run perpendicular to the fiber channels. These can be filled with an index matching gel or oil. By matching the refractive index of the otherwise air-filled space between the cleaved fiber end and the hemispherical glass wall of the fluidic channel, re- and deflections of the laser beam are reduced. An added benefit is that the channels allow dust and debris, leftovers from the chip manufacturing or fiber insertion process, to be rinsed away from the core area of the fiber where they can influence the quality of the trap by distorting the beam or even burning. Also, air bubbles in front of the fiber interface can be flushed away. One could also envision optical fibers to be permanently attached to the chip by flowing optical adhesive through the channel and then curing it to firmly secure the fibers in place.
We also added an additional feature that gives users the possibility to easily remove and exchange optical fibers. In the capillary setup,32 exchange of optical fibers was not possible. In the PDMS device,34 it was possible, but due to the soft and sticky material insertion of a glass fiber was tedious and tended to scrape off material which would then contaminate the beam path. In femtosecond chips,35 the fibers have to be placed outside of the chip and their position iteratively optimized for ideal light coupling of fiber to waveguide. They then have to be glued tight. Here, we extended the bottom glass half and fiber grooves of the chip slightly (see the ESI† Fig. S4). Any fiber of the right size can just be placed on the groove and slid forward until it sits tight. There is no contamination from scrape-off or the necessity of attaching fibers permanently. This makes the reuse of the chip for other wavelengths simple, as well as making it safe for transport.
Since the three-way sorting location is quite removed from the trap location and cannot be imaged at the same time, we chose to sort using the trap as a one-sided laser sorter directly at the trap.43 The laminar flow conditions ensure that cells sorted at this location stay on the chosen channel side and the splitting of the streamlines at the junction advects the cell flow into its corresponding channel without any further intervention.
The novel chip presented here has several advantageous aspects over previous microfluidic optical stretcher setups.32,34,36 One is the possibility of using more complex channel geometries. Moreover, low fiber contamination and chip degradation allow for a long lifetime. Low roughness of surfaces permits good imaging, while simple insertion and alignment of optical fibers increases the ease of use. In order to demonstrate its practical use, we proceeded to sort cells in a mixed population by their mechanical phenotype.
4.2. Mechanical phenotyping of populations
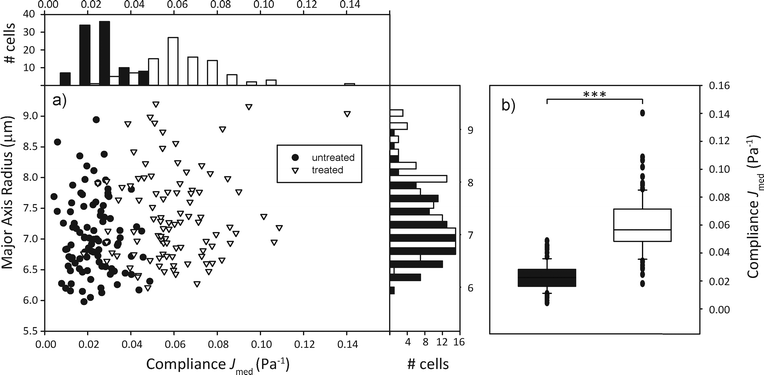
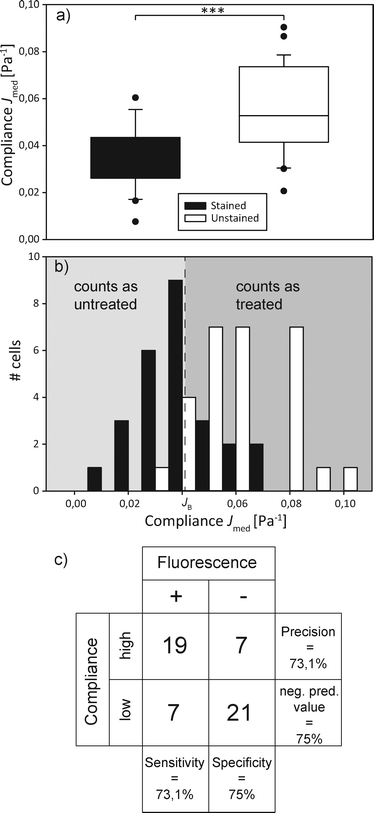
In order to sort a mixed cell population, first the homogeneous populations need to be characterized and relevant parameters extrapolated. We used the human promyelocytic leukemia cells HL-60, in one case treated with the cytoskeletal drug cytochalasin D, in the other case untreated, but with a fluorescent nuclear stain as a control. We expected the cells treated with the drug to be significantly more compliant than the untreated group due to the inhibition of actin polymerization in the cytoskeleton.44 100 cells per group were sent through the microfluidic channel and optically stretched at the location of the optical trap. For each cell, we measured the elongation of the major axis over time and the cell area shortly before the stretch step via real-time analysis of the acquired images. The relevant parameters we extracted from the data were the major axis radius of the cell and the compliance Jmed, which corresponds to the median compliance during the last second of the stretch phase. Fig. 3 shows that the distribution of major axis radii is quite similar in both groups, while the distribution of compliance Jmed clearly shows the mechanical difference between untreated and treated groups.4.3. Sorting of mixed populations based on mechanical phenotyping
In order to demonstrate the proof of concept, i.e. that we are able to distinguish and actively sort single cells according to their mechanical phenotype, we decided on a specific parameter that characterizes a cell randomly selected from a mixed population of treated and untreated cells and sorts it into a group accordingly. The compliance distributions as previously specified were used to calculate a decision boundary JB for the automated sorter based on Bayesian decision theory,45 which minimizes the sorting error to be expected due to the overlap of the compliance distribution functions. Assuming that the cost of each missorted cell is the same, JB = 0.0402 Pa−1 led to the minimized error of 9.37% wrongly sorted cells.We then poured together treated and untreated cells in equal concentrations and proceeded to have them sorted automatically. This entails trapping a cell, turning off the flow, measuring the major axis elongation, comparing the calculated compliance to the boundary value, imaging the cells' nuclear fluorescence for control purposes, turning off one laser for optical sorting of the cells and starting the flow again. This is all controlled by the LabView program. The compliance distribution and a confusion table can be found in Fig. 4. It appears as if the distribution of both types of cells differs from what was found during the preliminary experiments. The distribution of untreated cells appears more smeared out, with higher compliance values than expected, while the distribution of cytochalasin-D treated cells appears slightly shifted towards a lower compliance. This leads to the sorting error of 25.9% being larger than expected. We suspect that this might be due to the fact that cytochalasin-D induced disruption of actin networks is reversible within timescales of half an hour, which leads to the stiffening of the treated cells. During the experiment cytochalasin-D also diffuses from the treated cells into the medium, which also affects untreated cells, making them more compliant. This is the first demonstration of sorting of a mixed cell population by active characterization of the cells' mechanical phenotype.
5. Conclusions
We have demonstrated a significant advancement in the capabilities of cell mechanics measurements using an optical stretcher. By careful choice of the design parameters, a robust, monolithic glass chip has been be produced which is tolerant of misalignment during the fabrication process while still maintaining the critical alignment of the optical fibers required to produce a counterpropagating laser beam trap. The straightforward microfluidic connectors and accurate guiding of the fibres during insertion result in an easily set up optical stretcher which has capabilities beyond the previous state of the art MOS. Lithographic fabrication allows for complex microfluidic channel designs, while a smooth channel surface provides reasonably good imaging quality and allows for the use of fluorescence. The use of this design was demonstrated by successfully sorting cells by their mechanical fingerprint and simultaneous fluorescence detection.Some problems still persist. First, the fabrication process itself is quite time-consuming. Then, due to the fabrication constraints, especially the wet etching, imaging quality is somewhat reduced compared to smooth glass slide channel surfaces, since the semi-circular flow channel distorts the halo of the cell in phase contrast imaging which is beneficial for analysis. Very small deformations cannot be as easily detected as in a rectangular channel. Additionally, the fact that the optical fibers are further away from the trap leads to a lower trapping efficiency, which means that more power is needed for trapping and stretching. Nonetheless, we believe that the advantages this chip design exhibits significantly outweigh the disadvantages. In addition to the experimental possibilities highlighted here, other types of experiment can be envisioned. Making use of the laminar flow, we can create steep chemical gradients that do not mix rapidly. A trapped cell can be pushed in and out of these regions and be subjected to different chemicals in between measurements. A cell can remain trapped in the center while a new solution flows in through the side channel and completely replaces the liquid surrounding the cell. Every single cell could be measured multiple times in different environments, giving new insights into the influence of the surroundings of suspension cells on their viscoelastic behavior. In future generations of this chip, even more accurate measurement of compliance could be done by measuring each cell's refractive index27 and calculating the global geometric factor in real time. Characterization and sorting efficiency of cells might be improved by not limiting oneself to only the median compliance as a possible criterion of differentiation but by devising a range of parameters31 such as the slope of the compliance curve or the relaxation time after stretching, as well as non-mechanical parameters such as the refractive index of a cell, or fluorescence intensity as in fluorescence-actived cell sorting (FACS).
This study has been the first step towards a new device combining detailed viscoelastic characterization of single cells with sorting capabilities, adding marker-free mechanical phenotyping to the usual fluorescence-based flow cytometry. A multitude of exciting new possibilities for biomedical applications can be envisioned.
Acknowledgements
We are grateful to M. Schürmann and J. Scholze for the DHM measurements, to L. Kaderali for discussion of Bayesian decision theory, and to Donald and Ada Olins for the gift of the HL60 subline. This project has received funding from the European Union's Seventh Framework Programme for Research, Technological Development and Demonstration under grant agreement no. 28260 (Starting Grant “LightTouch”). EMB gratefully acknowledges the José Castillejos grant from the Spanish Ministry of Education and Science.References
- H. T. K. Tse, D. R. Gossett, Y. S. Moon, M. Masaeli, M. Sohsman, Y. Ying, K. Mislick, R. P. Adams, J. Rao and D. Di Carlo, Sci. Transl. Med., 2013, 5, 212ra163 CrossRef PubMed.
- M. Lekka, D. Gil, K. Pogoda, J. Dulińska-Litewka, R. Jach, J. Gostek, O. Klymenko, S. Prauzner-Bechcicki, Z. Stachura, J. Wiltowska-Zuber, K. Okoń and P. Laidler, Arch. Biochem. Biophys., 2012, 518, 151–156 CrossRef CAS PubMed.
- S. Suresh, Acta Biomater., 2007, 3, 413–438 CrossRef PubMed.
- J. Guck and E. R. Chilvers, Sci. Transl. Med., 2013, 5, 212fs41 CrossRef PubMed.
- S. Byun, S. Son, D. Amodei, N. Cermak, J. Shaw, J. H. Kang, V. C. Hecht, M. M. Winslow, T. Jacks, P. Mallick and S. R. Manalis, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7580–7585 CrossRef CAS PubMed.
- A. Ekpenyong, G. Whyte, K. Chalut, S. Pagliara, F. Lautenschläger, C. Fiddler, S. Paschke, U. Keyser, E. Chilvers and J. Guck, PLoS One, 2012, 7, e45237 CAS.
- R. D. Gonzalez-Cruz and E. M. Darling, Adipocyte, 2013, 2, 87–91 CrossRef PubMed.
- M. Tsai, R. Waugh and P. Keng, Biophys. J., 1996, 70, 2023–2029 CrossRef CAS.
- J. Guck, F. Lautenschläger, S. Paschke and M. Beil, Integr. Biol., 2010, 2, 575–583 RSC.
- A. Bausch, W. Möller and E. Sackmann, Biophys. J., 1999, 76, 573–579 CrossRef CAS.
- B. Fabry, G. N. Maksym, J. P. Butler, M. Glogauer, D. Navajas and J. J. Fredberg, Phys. Rev. Lett., 2001, 87, 148102 CrossRef CAS.
- K. Franze, M. Francke, K. Gunter, A. F. Christ, N. Korber, A. Reichenbach and J. Guck, Soft Matter, 2011, 7, 3147–3154 RSC.
- R. Mahaffy, S. Park, E. Gerde, J. Käs and C. Shih, Biophys. J., 2004, 86, 1777–1793 CrossRef CAS.
- Y.-Z. Yoon, J. Kotar, G. Yoon and P. Cicuta, Phys. Biol., 2008, 5, 036007 CrossRef PubMed.
- O. Thoumine and A. Ott, J. Cell Sci., 1997, 110, 2109–2116 CAS.
- J. S. Dudani, D. R. Gossett, H. T. K. Tse and D. Di Carlo, Lab Chip, 2013, 13, 3728–3734 RSC.
- D. R. Gossett, H. T. K. Tse, S. A. Lee, Y. Ying, A. G. Lindgren, O. O. Yang, J. Rao, A. T. Clark and D. Di Carlo, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7630–7635 CrossRef CAS PubMed.
- T. Sawetzki, C. Eggleton, S. Desai and D. Marr, Biophys. J., 2013, 105, 2281–2288 CrossRef CAS PubMed.
- J. P. Beech, S. H. Holm, K. Adolfsson and J. O. Tegenfeldt, Lab Chip, 2012, 12, 1048–1051 RSC.
- S. Hur, N. Henderson-MacLennan, E. McCabe and D. Di Carlo, Lab Chip, 2011, 11, 912 RSC.
- G. Wang, W. Mao, R. Byler, K. Patel, C. Henegar, A. Alexeev and T. Sulchek, PLoS One, 2013, 8, e75901 CAS.
- B. K. Lin, S. M. McFaul, C. Jin, P. C. Black and H. Ma, Biomicrofluidics, 2013, 7, 034114 CrossRef PubMed.
- C. J. Chan, G. Whyte, L. Boyde, G. Salbreux and J. Guck, Interface Focus, 2014, 4, 20130069 CrossRef CAS PubMed.
- J. Guck, R. Ananthakrishnan, T. Moon, C. Cunningham and J. Käs, Phys. Rev. Lett., 2000, 84, 5451–5454 CrossRef CAS.
- J. Guck, R. Ananthakrishnan, H. Mahmood, T. Moon, C. Cunningham and J. Käs, Biophys. J., 2001, 81, 767–784 CrossRef CAS.
- F. Lautenschläger, S. Paschke, S. Schinkinger, A. Bruel, M. Beil and J. Guck, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15696–15701 CrossRef PubMed.
- K. J. Chalut, A. E. Ekpenyong, W. L. Clegg, I. C. Melhuish and J. Guck, Integr. Biol., 2012, 4, 280–284 RSC.
- F. Wottawah, S. Schinkinger, B. Lincoln, S. Ebert, K. Müller, F. Sauer, K. Travis and J. Guck, Acta Biomater., 2005, 1, 263–271 CrossRef PubMed.
- J. Guck, S. Schinkinger, B. Lincoln, F. Wottawah, S. Ebert, M. Romeyke, D. Lenz, H. Erickson, R. Ananthakrishnan, D. Mitchell, J. Käs, S. Ulvick and C. Bilby, Biophys. J., 2005, 88, 3689–3698 CrossRef CAS PubMed.
- T. W. Remmerbach, F. Wottawah, J. Dietrich, B. Lincoln, C. Wittekind and J. Guck, Cancer Res., 2009, 69, 1728–1732 CrossRef CAS PubMed.
- T. Kießling, M. Herrera, K. Nnetu, E. Balzer, M. Girvan, A. Fritsch, S. Martin, J. Käs and W. Losert, Eur. Biophys. J., 2013, 42, 383–394 CrossRef PubMed.
- B. Lincoln, S. Schinkinger, K. Travis, F. Wottawah, S. Ebert, F. Sauer and J. Guck, Biomed. Microdevices, 2007, 9, 703–710 CrossRef PubMed.
- B. Lincoln, F. Wottawah, S. Schinkinger, S. Ebert and J. Guck, Cell Mech., 2007, 83, 397–423 CAS.
- F. Gast, P. Dittrich, P. Schwille, M. Weigel, M. Mertig, J. Opitz, U. Queitsch, S. Diez, B. Lincoln, F. Wottawah, S. Schinkinger, J. Guck, J. Käs, J. Smolinski, K. Salchert, C. Werner, C. Duschl, M. Jäger, K. Uhlig, P. Geggier and S. Howitz, Microfluid. Nanofluid., 2006, 2, 21–36 CrossRef CAS.
- N. Bellini, K. C. Vishnubhatla, F. Bragheri, L. Ferrara, P. Minzioni, R. Ramponi, I. Cristiani and R. Osellame, Opt. Express, 2010, 18, 4679–4688 CrossRef CAS PubMed.
- N. Bellini, F. Bragheri, I. Cristiani, J. Guck, R. Osellame and G. Whyte, Biomed. Opt. Express, 2012, 3, 2658–2668 CrossRef PubMed.
- D. C. S. Bien, P. V. Rainey, S. J. N. Mitchell and H. S. Gamble, J. Micromech. Microeng., 2003, 13, 557 CrossRef.
- A. F. Oskooi, D. Roundy, M. Ibanescu, P. Bermel, J. Joannopoulos and S. G. Johnson, Comput. Phys. Commun., 2010, 181, 687–702 CrossRef CAS PubMed.
- Z. Bar-Shavit, S. Teitelbaum, P. Reitsma, A. Hall, L. Pegg, J. Trial and A. Kahn, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 5907–5911 CrossRef CAS.
- A. L. Olins, B. Buendia, H. Herrmann, P. Lichter and D. E. Olins, Exp. Cell Res., 1998, 245, 91–104 CrossRef CAS PubMed.
- A. L. Olins, T. V. Hoang, M. Zwerger, H. Herrmann, H. Zentgraf, A. A. Noegel, I. Karakesisoglou, D. Hodzic and D. E. Olins, Eur. J. Cell Biol., 2009, 88, 203–214 CrossRef CAS PubMed.
- L. Boyde, A. Ekpenyong, G. Whyte and J. Guck, Appl. Opt., 2012, 51, 7934–7944 CrossRef PubMed.
- F. Bragheri, P. Minzioni, R. Martinez Vazquez, N. Bellini, P. Paie, C. Mondello, R. Ramponi, I. Cristiani and R. Osellame, Lab Chip, 2012, 12, 3779–3784 RSC.
- M. Schliwa, J. Cell Biol., 1982, 92, 79–91 CrossRef CAS.
- E. L. Lehmann, Ann. Math. Stat., 1950, 21, 1–26 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4lc01196a |
| This journal is © The Royal Society of Chemistry 2015 |