A droplet-to-digital (D2D) microfluidic device for single cell assays†
Steve C. C.
Shih
ab,
Philip C.
Gach
ab,
Jess
Sustarich
ab,
Blake A.
Simmons
ab,
Paul D.
Adams
bcd,
Seema
Singh
ab and
Anup K.
Singh
*ab
aSandia National Laboratories, 7011 East Ave, Livermore, CA, USA. E-mail: aksingh@sandia.gov; Fax: +1 925 294 3020; Tel: +1 925 294 1260
bJoint Bioenergy Institute (JBEI), 5855 Hollis St., Emeryville, CA, USA
cLawrence Berkeley National Laboratories (LBNL), Building 64R0121, 1 Cyclotron Road, Berkeley, CA 94720, USA
dDepartment of Bioengineering, University of California, Berkeley, CA, USA
First published on 30th October 2014
Abstract
We have developed a new hybrid droplet-to-digital microfluidic platform (D2D) that integrates droplet-in-channel microfluidics with digital microfluidics (DMF) for performing multi-step assays. This D2D platform combines the strengths of the two formats—droplets-in-channel for facile generation of droplets containing single cells, and DMF for on-demand manipulation of droplets including control of different droplet volumes (pL–μL), creation of a dilution series of ionic liquid (IL), and parallel single cell culturing and analysis for IL toxicity screening. This D2D device also allows for automated analysis that includes a feedback-controlled system for merging and splitting of droplets to add reagents, an integrated Peltier element for parallel cell culture at optimum temperature, and an impedance sensing mechanism to control the flow rate for droplet generation and preventing droplet evaporation. Droplet-in-channel is well-suited for encapsulation of single cells as it allows the careful manipulation of flow rates of aqueous phase containing cells and oil to optimize encapsulation. Once single cell containing droplets are generated, they are transferred to a DMF chip via a capillary where they are merged with droplets containing IL and cultured at 30 °C. The DMF chip, in addition to permitting cell culture and reagent (ionic liquid/salt) addition, also allows recovery of individual droplets for off-chip analysis such as further culturing and measurement of ethanol production. The D2D chip was used to evaluate the effect of IL/salt type (four types: NaOAc, NaCl, [C2mim] [OAc], [C2mim] [Cl]) and concentration (four concentrations: 0, 37.5, 75, 150 mM) on the growth kinetics and ethanol production of yeast and as expected, increasing IL concentration led to lower biomass and ethanol production. Specifically, [C2mim] [OAc] had inhibitory effects on yeast growth at concentrations 75 and 150 mM and significantly reduced their ethanol production compared to cells grown in other ILs/salts. The growth curve trends obtained by D2D matched conventional yeast culturing in microtiter wells, validating the D2D platform. We believe that our approach represents a generic platform for multi-step biochemical assays such as drug screening, digital PCR, enzyme assays, immunoassays and cell-based assays.
Introduction
Single cell analysis has become a valuable means to analyze cells since it is able to decipher individual cells within a heterogeneous population. The single cell approach can assist our understanding of complex biological systems on the cellular level that can not be obtained through population averages. This requires meticulous investigations on the growth of single cells in response to a variety of chemical and biological cues. One area of interest is screening single cells for effective biofuel production and microbial toxicity to a variety of chemicals from upstream pretreatment processing of lignocellulosic biomass.Recently, lignocellulosic biofuels have attracted a lot of attention as they provide an attractive alternative to fossil fuels as they are derived from non-food crops and have a lower impact on the environment.1,2 The key steps in cellulosic biofuel production involve the thermochemical pretreatment of biomass, followed with enzymatic saccharification of the pretreated biomass, and fermentation of the resulting sugars into ethanol or other biofuels in a host such as yeast. A promising pretreatment technology is ionic liquid (IL) pretreatment3–5 because of their ability to efficiently dissolve and fractionate biomass and produce higher yield of lignin-free cellulose from both grasses and woody biomass compared to other methods.6 The pretreated cellulose is further broken down into simple sugars for downstream fermentation by cellulases (e.g. β-glucosidases).7,8 At high concentrations, some ILs used for pretreatment are toxic to most enzymes and organisms used for saccharification and fermentation, respectively. However, complete removal of IL from pretreated biomass and sugar is prohibitively expensive. In addition, a consolidated one-pot process with combined pretreatment and saccharification unit operations or consolidated pretreatment–saccharification–fermentation unit operations are highly desirable. To eliminate the burden of complete removal of IL from sugar generated from IL pretreatment and to enable the efforts around one-pot consolidated processes, large numbers of enzyme variants and mutant strains have been generated that need to be screened for tolerance to IL, and selection of promising candidates for further process scale up. Significant efforts have been made to analyze ILs and to determine their effect on hydrolysis rates of pretreated lignocelluloses,9,10 and examine their toxicity in fungi and in bacteria.11 Currently, the screening is done using large populations of cells with microtiter plates and cuvettes.9–12 However, the microtiter plate based screening has a number of drawbacks – it is slow, expensive as it uses a large amount of reagents, and suffers from poor quantification and reproducibility. In addition, in laboratories that lack robot dispensers and aspirators, this process requires a significant amount of manual labor and procedural optimization. Finally, planktonic cells such as yeast and E. coli exhibit erratic culture behavior (e.g. variations in biomass growth), which result in unusual growth patterns.13,14 Hence, an alternative to understand the effects of IL on biofuel-producing cell strains in populations is to isolate and analyze single cells for toxicity towards a specific IL in various concentrations.
Microfluidic platforms offer a potential solution to overcome many of these drawbacks – they can drastically reduce the number of pipetting steps required, lower the cost of reagents by at least an order of magnitude, reduce the time required for screening, and potentially improve the throughput. Droplet-in-channel microfluidics has emerged as a very promising technology where pL–nL monodisperse droplets can be used as an alternative to microtiter wells to conduct high-throughput assays.15,16 This is a two-phase system where aqueous droplets are formed with a surrounding oil phase (there are also multiple phase systems17–19). The droplet-in-channel format has been used for many applications including culturing and analyzing of single cells,20–22 multistep enzyme assays,23–25 and clinical diagnostics.26,27 A challenge with this format is the serial addition of reagents – akin to pipetting. Many different schemes have been proposed to add one reagent at a time to the aqueous droplet: droplet fusion,28,29 electrocoalescence,30 picoinjection,31,32 and specialized microfabricated structures to induce droplet merging.33,34 These are innovative methods that require exquisite control over flow rates, timing, fluidic resistance, rate of droplet generation, and pressure fluctuation. Here, we present an alternative form of droplet microfluidics which has been shown to be an elegant platform for reagent addition via droplet merging – digital microfluidics (DMF).35,36 Digital microfluidics is well-suited for adding reagents in parallel and reagents can be mixed on demand without the requirement of optimal and precise flow rates. A DMF device typically comprises of an open array of electrodes covered with a dielectric and a hydrophobic layer. To move droplets, electric potentials are applied to the electrode such that electrostatic forces (i.e. qE) are generated which allow droplets to perform a number of different operations (e.g. move, merge, mix, split, and dispense).36 The low reagent consumption and facile connection to analytical instruments has made DMF a popular platform for cell culture and assays.37–42 Typically, a density of cells is cultured in a droplet on the DMF platform and contains a surfactant, Pluronics,43,44 to minimize bio-fouling on the digital microfluidic surface. Although Pluronics has no adverse effects in the gene expression of mammalian cells45 (and no change in their proliferation42), the inhomogeneous nature of the Pluronics is not suitable for post-processing techniques such as mass spectrometry (MS) and/or HPLC.46
In this report, we discuss a novel proof-of-principle architecture that combines the strengths of the two droplet microfluidic platforms (which we call droplet-to-digital microfluidics – D2D microfluidics) while overcoming the weaknesses of the individual devices. This device allows us to analyze cells without the use of Pluronics (e.g. F68 and F127) which permits compatibility with post-processing techniques such as HPLC and MS. Furthermore, it may also enhance the lifetime of device use since Pluronics is not a permanent solution to prevent biofouling.43,44 The most important advantage is the device configuration, which enables droplet generation by droplets-in-channels with parallel addition of reagents (on demand dispensing from reservoirs and splitting of droplets) and single cell analysis by digital microfluidics. Furthermore, this new technique allows manipulation of wide range of volumes (pL volumes for droplet-in-channels and pL–μL volumes for DMF) and is easily adaptable for automation. Here, we have applied our D2D device to analyzing ILs and determining their effects on downstream processes related to biofuel production (e.g. microbial growth and ethanol production). We propose that this system can be applied to a wide range of applications that requires screening of single cells that involves addition of multiple reagents with a variety of volumes, automation, and post-processing analysis.
Materials and methods
Reagents and materials
Unless otherwise specified, general-use reagents were purchased from Sigma Aldrich. Sacchromyces cervisiae strain BY4742 was obtained from the JBEI Registry (https://registry.jbei.org). SYTO-9, a stain for nucleic acid in yeast was purchased from Life Technologies (Grand Island, NY).D2D microfluidic device fabrication
Droplet-to-digital (D2D) microfluidic devices were fabricated in the University of California Biomolecular Nanotechnology Center (UC Berkeley BNC) fabrication facility, using a transparent photomask printed at Fineline Imaging (Colorado Springs, CO). Fabrication reagents and supplies included SU-8-5, SU-8-2025, SU-8-2075, S-1811 and SU-8 Developer from Microchem (Newton, MA), gold- and chromium-coated glass slides from Telic (Valencia, CA), indium tin oxide (ITO) coated glass slides (Delta Technologies, Stillwater, MN), Aquapel from TCP Global (San Diego, CA), MF-321 positive photoresist developer from Rohm and Haas (Marlborough, MA), Standard KI/I2 gold etchant from Sigma, CR-4 chromium etchant was from OM Group (Cleveland, OH), and AZ-300T photoresist stripper from AZ Electronic Materials (Somerville, NJ).To fabricate a DMF device, it involved three steps: 1) electrode patterning on the bottom plate, 2) dielectric deposition layer, and 3) channel/spacer features. The first step involving DMF fabrication bearing patterned electrodes and contact pads were formed by photolithography and etching as described previously41 with an exposure time of 5 s (40mW cm−2) using an OAI Series 200 Aligner (San Jose, CA USA). For the second step, these devices were plasma treated under 20% O2 and RF power of 20 W for 3 min and were then coated with a 7 μm layer dielectric using SU-8-5 following Microchem's instructions for spin speed and bake times. For the channel/spacer features (step 3), the devices were then plasma treated (using the same conditions as above) and coated with a layer of SU-8-2075 to pattern channels and a spacer with a height of 140 μm. Spin speeds, soft and hard bake times, and development times were following Microchem's instructions. After development, these were rinsed and dried with isopropanol and diH2O and were hard-baked for 15 min at 200 °C. To create a hydrophobic layer, DMF devices and ITO top-plates were coated with 0.2 μm filtered Aquapel. After 15 min, the Aquapel was removed with a kimwipe and rinsed with diH2O. This was then dried with N2 and left at room temperature for 1 h.
The droplet microfluidic devices were fabricated using soft lithography techniques. Briefly, we first fabricated the microfluidic channels using SU-8-2025 photoresist on a 4" silicon wafer (i.e. a master mold). Following Microchem's guidelines, the structure of the master has a height of 140 μm, generating the same dimension height in the PDMS mold. The master was silanized with trichloro(1H,1H,2H,2H-perfluorooctyl) silane (97%, Aldrich) overnight. PDMS (Sylgard 184, Dow Corning, Midland, MI) mixed at a ratio of 10 to 1 (base to curing agent) was poured onto the master, placed in a vacuum to remove bubbles for 12 h, cured at 80 °C for 1 h, and peeled away. The inlet and outlet holes were punched into the layer of channels using a Harris Uni-Core with a tip diameter of 0.75 mm. We exposed the PDMS and a glass substrate to oxygen plasma (Harrick Plasma, Ithaca, NY) for 3 min at max RF power and bonded the PDMS and substrate in contact to form a permanent seal.
To assemble the capillary interface (which was purchased from IDEX – item no. 1572, PEEK tubing 360 μm O.D, 150 μm I.D. × 5 ft), one end of the capillary was inserted into the outlet of the droplet microfluidic device and the other end was inserted into the channel on the DMF device (a channel directed to the 300 μm electrodes). After insertion, the capillary was adhered to the device using 40 μm adhesive plate seal (PlateMax Ultraclear Sealing Film, Axygen, Union City, CA USA) and epoxy by UV curing for 1–2 min (Fig. 1a). An exploded view is shown in ESI† (Fig. S1).
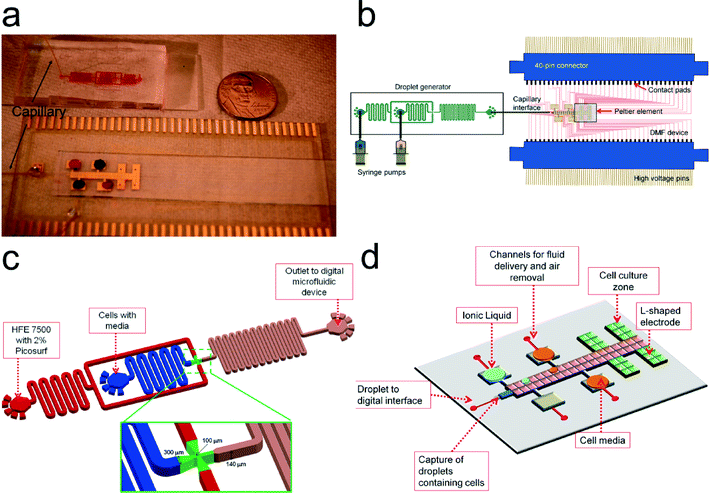 | ||
| Fig. 1 Droplet to Digital (D2D) microfluidic device. (a) Image of device with capillary connecting the droplet-in-channel to DMF device. Colored dye is used to outline the channel droplet device and to show droplets in reservoirs of digital microfluidic device. (b) Schematic of the D2D device. Fluid is pressure-driven by syringe pumps into the droplet generator. Droplets from the generator are driven to the DMF device (via capillary) and are actuated by high voltage (HV) signals. The DMF device is connected to two 40-pin connectors which guide HV signals to the contact pads of the DMF device. A Peltier element (to control temperature) is situated below the four cell culture regions on the DMF device. (c) Schematic of the droplet-in-channel device used to generate ~20 nL droplets containing a single cell. In (d), the digital microfluidic design includes four culture regions with L-shaped electrodes, four reservoir electrodes (used for ionic liquid, cell media, and waste), a channel layer (shown in red) that is used for 1) fluid delivery, 2) air bubble removal, and 3) interfacing the capillary from the droplet device to the digital microfluidic device. A dielectric layer is not shown for clarity. An exploded view of the DMF device is shown in ESI† Fig. S1. | ||
D2D microfluidic automated control
The droplets on the digital microfluidic device were controlled by an in-house C++ program which was used to control the application of driving potentials using an automated feedback control system for high-fidelity droplet movement. The feedback system is described elsewhere41,47,48 and was used to monitor droplet movement on the device and droplet evaporation. Briefly, to move a droplet on a destination electrode, a 200 ms pulse of driving potential is applied to the destination electrode relative to the top-plate electrode. The output from the feedback system is connected to an analog input of an RBBB Arduino microcontroller (Modern Device, Providence, RI), which controls the control board containing solid-state switches (responsible for applying potential to the DMF device). The Arduino microcontroller was also used to control the Peltier cooler, which is used to maintain constant surface temperature on the DMF during cell culture. The neMESYS syringe pumps was controlled by an in-house C++ program that enables control of the flow rate of the liquids/droplets in the droplet generator and the oil injection between the top and bottom layer of the DMF device to prevent evaporation of droplets on the DMF during cell culture. A figure showing the automated feedback system with the signal flow is shown in Fig. 3. The automation code includes Arduino and C++ code to run the microfluidic experiments (see ESI†).Macroscale ionic liquid screening experiments
Macroscale growth assays were performed using the Saccharomyces cerevisiae strain BY4742. A cryopreserved stock of the strain was streaked on agar plates of SD medium and supplemented with 20 g L−1 of glucose and incubated at 30 °C. All media components were of cell culture or molecular biology grade. For well-plate screening assays, 5 mL starter cultures were grown at 30 °C to saturation in SD medium supplemented with 20 g L−1 glucose in 30 mL test tubes with shaking at 200 rpm for 16–20 h. The optical density of the starter cultures was determined at 600 nm (OD 600) with the Tecan Infinite F200 Pro (Tecan Ltd, San Jose, CA) well-plate reader. The starter cultures were diluted to a final OD of 0.05 (measured with a spectrophotometer using a 1 cm cuvette) with SD medium, supplemented with either 20 g L−1 of glucose spiked with 1-ethyl-3-methylimidazolium acetate (abbreviated as [C2mim] [OAc]), 1-ethyl-3-methylimidazolium chloride ([C2mim] [Cl]), sodium chloride (NaCl), or sodium acetate (NaOAc) to give final concentrations of 0 (control), 37.5, 75, and 150 mM. Cultures were distributed into 24-well plates (800 μL per well). Plates were incubated at 30 °C with shaking for 24 h in a Tecan Infinite F200 Pro plate reader and OD600 was acquired every 20 min. To generate growth curves, the measured OD was corrected by subtracting the blank OD (i.e. only SD media) for each ionic liquid concentration and were plotted in the natural log scale. All experiments were repeated at least three times.To quantitatively determine the impact of the ionic liquids on yeast growth, the growth kinetics were calculated using the following equation:
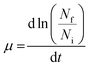 | (1) |
Microfluidic ionic liquid screening experiments
To ensure monodisperse droplet generation and prevention of merging of droplets in the generator, channels were coated with Picoglide-1™ (0.5% w/w FC-40) (Dolomite Microfluidics, Charlestown, MA) using a neMESYS (Cetoni, Korbussen, Germany) syringe pump with a flow rate of 0.2 μL s−1 for 5 min and further incubated with the Picoglide for 1 h. The channels were then rinsed at the same flow rate with only HFE 7500 for 5 min and then dried in room temperature for 30 min.To prepare cells for microfluidic analysis, 5 mL starter cultures were grown at 30 °C to saturation in SD medium supplemented with 20 g L−1 glucose in 30 mL test tubes with shaking at 200 rpm for 16–20 h. After incubation, traces of cell debris and clumping can occur (which can cause clogging in the microfluidic device) and therefore the cells were transferred to fresh media. To minimize clumping of cells, the culture was sonicated in a heated water bath (30 °C) for 5 min. 80 μL of cells were then diluted in 1 mL of fresh media and then transferred to a 2.5 mL High-Precision Glass Syringe with Tubing connector 1/4-28 UNF thread (Cetoni) containing 500 μL of HFE 7500. This dilution was performed such that the majority of droplets generated contained no more than one cell (i.e. we are using Poisson's statistics).15 Although this protocol results in the formation of some empty droplets, these droplets were manually actuated with droplets with a cell. Droplets containing two or more cells (<10% of the droplets) were analyzed as a droplet containing single cell. To generate the droplets containing a single cell, the oil (i.e. HFE 7500 with 0.5% v/v Picosurf) flow rate was set to 0.1 μL s−1 and then it was set to 0.05 μL s−1 when the oil reaches the T-junction. At the same time, the aqueous (i.e. cells in media) flow rate was set to 0.005 μL s−1 which generated ~20 nL droplets containing a single cell. To image the cells inside the channel, cells were stained with SYTO-9 following the manufacturer's instructions. We used the same prepared cells for replicates and re-prepared fresh cells and media for other IL/salt conditions.
To prepare the digital microfluidic device for single cell screening, the capillary was injected with HFE 7500 using a 3 mL syringe before connecting it to the droplet microfluidic device. This ensures there are no air bubbles trapped in the capillary during operation. To inject liquids (cell media and 300 mM ionic liquid/salt) into the reservoirs, we used two techniques. The first technique consisted of pipetting 3.0 μL liquids directly onto the reservoirs, filling the surrounding area with biocompatible oil (i.e. HFE 7500), and then placing the ITO top-plate. The second technique consisted of placing a capillary into the channel that is connected to the reservoir (see red channels in Fig. 1d) and injecting the liquid into reservoirs with the syringe pump. The advantage of using technique two is the continuous filling of the reservoirs as the liquid in the reservoir is depleted using our automated control with feedback. The filling of the reservoir with liquid is performed with the ITO top plate and in biocompatible oil. After injection of the liquids, a serial dilution is conducted to create four concentrations: 0 mM (control), 37.5 mM, 75 mM, and 150 mM of ionic liquid using the automated system by generating a script using an in-house C++ program.
To generate growth curves, images of the cells were captured at different time intervals (0, 2, 6, and 24 h) using an inverted Olympus IX71 microscope connected to a scientific grade CCD camera (iXon EMCCD, Andor Technology, South Windsor, CT) at a 40× magnification. The cells from the images were counted manually or using Image J and these values were plotted vs. time. Growth kinetics were calculated using eqn (1) (above) and were tested for significance using a one-way ANOVA test and a Tukey's post hoc test using a P-value < 0.05.
Ethanol production in yeast
To quantify ethanol production in yeast, the cultures (after 24 h) on the D2D device were pipetted into 0.6 mL centrifuge tubes containing 0.3 mL of fresh media to ensure anaerobic growth. The tubes were sealed with parafilm and incubated at 30 °C with shaking at 200 rpm. After 24 h, the cultures were filtered using a 0.2 μm membrane filter to collect the supernatant (and to remove yeast cells) before HPLC analysis. The quantification of ethanol was conducted using an Agilent 1200 HPLC equipped with a refractive index detector (Agilent, Santa Clara CA). Separations were achieved using an Aminex HXP-87H (300 × 7.8 mm, 9 μm) analytical column (Bio-Rad, Hercules, CA USA) with a Micro-Guard Cation H guard column (30 × 4.6 mm) at 50 °C. A mobile phase of 4 mM H2SO4 at 0.6 mL min−1 was used. A calibration curve was generated through the use of external standards.Results and discussion
Droplet-to-digital (D2D) microfluidic device and operation
The goal of this work was to develop a microfluidic system to screen and to analyze cells that are tolerant to different types and concentrations of IL. Our microfluidic system joins a small group of studies that have used a combination (“hybrid”) of microfluidic platforms. Although in these studies these “hybrid” systems are presented as digital-to-channel systems (which is different from our droplet-to-digital system), they do present interesting innovations that helped guide our work.49,50 Abdelgawad et al.49 presents a preliminary single-plate DMF configuration with a PDMS channel. However, it suffers a key limitation in which single plate DMF devices are not capable of dispensing droplets from reservoirs – a requirement if we are to create a library of samples with different concentrations. Watson et al. presented a second generation hybrid microfluidic device that uses a digital microfluidic device formed on a top substrate which is mated to a network of microchannels. Here, the fabrication is more complex (e.g. drilling holes for transferring reagents) and requires precise alignment procedures. Therefore, in this work, we chose instead to create two devices and mate them by using a capillary interface without specialized fabrication or precise alignment (as shown by some groups,51–54 a capillary can be easily interfaced to a digital microfluidic device). Here, we describe the first implementation of a droplet-to-digital microfluidic device that is capable of parallel addition of multiple reagents on demand and single cell analysis.To accurately compare the effect of various parameters on cell growth and ethanol production, it is important to start with a single cells in each droplet. In designing the D2D system shown in Fig. 1, we evaluated a number of alternative strategies to obtain a single cell in a droplet, including pipetting a low density of cells into a reservoir on the DMF and use actuation to dispense a single cell from the reservoir. Dispensing on DMF can be accomplished by breaking a larger droplet into smaller droplets with reproducible volumes.55 We attempted to generate a droplet with a single cell by starting with a larger droplet (reservoir) containing a low density of cells. However, cells in the reservoir tend to move away from dispensing electrode during actuation making it hard to dispense them. This behavior was attributed to the low-conductance of the cell media, which results in moving cells away from the high field.56 Furthermore, dispensing a volume from a larger droplet is a random process on the digital microfluidic device and it is very difficult to obtain consecutive successful dispensed droplets containing only a single cell and to predict when a dispensed droplet has a single cell. On the other hand, encapsulation of single cells is easily accomplished in droplet-in-channel microfluidics by careful balancing of aqueous flow (containing cells) and oil flow and has been demonstrated by many groups.57,58 Hence, we used a droplet generator to create droplets encapsulating a single cell (with some droplets generated with no cells or two or more cells) (Fig. 1c). This device made out of PDMS was stable over the course of many droplet generations, and could be used months later without loss of operation.
As shown in Fig. 1, a key feature of our design is mating of two droplet devices via a capillary. Droplets with a single cell are generated in the droplet-in-channel chip and are pressure-driven through the capillary and translated onto the digital microfluidic device (Fig. 1d; see exploded view in ESI† Fig. S1). The step-by-step process is shown in Fig. 2. Here, a droplet is generated in a biocompatible oil phase (i.e. HFE 7500) with 0.5% Picosurf surfactant (Fig. 2a). We used this surfactant and oil as the combination has been shown to be biocompatible (and we believe that the surfactant and oil aided our prevention for biofouling on the DMF surface).16,22 The concentration of the surfactant was optimized to minimize merging of droplets inside the channel but at the same time allow merger with droplets containing the cell media and ionic liquid on the digital microfluidic device. As the droplet reaches the capillary inlet, it is transferred to the DMF device where it reaches a section containing ten (a 5 × 2 array) 300 μm × 300 μm inter-digitated electrodes. This droplet is immediately actuated and merged with a IL/salt droplet on the digital microfluidic device (shown in frames 1–5 in Fig. 2b). Serial dilutions of the IL/salt were formed by merging a droplet (0.1 μL) containing cell media and a 300 mM IL/salt droplet. By mixing this droplet in a circular pattern,59 it produced a 2× dilution of the ionic liquid (frames 1–5 in Fig. 2c). This 150 mM droplet was then split into two droplets. One droplet was actuated to the cell culture zone and the other was used for subsequent mixing with a dispensed cell media droplet resulting in droplets containing 75 and 37.5 mM concentrations (frame 6 – Fig. 2c). Such operations would be very difficult to employ on a microchannel device as it would require valving or injection/coalescence techniques demanding optimal timing and control.
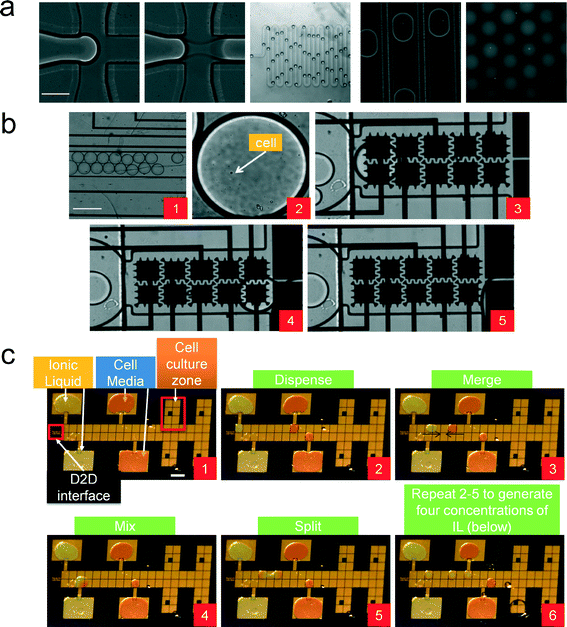 | ||
| Fig. 2 Device operation of the D2D microfluidic device. (a) A series of images showing droplet encapsulation of a single cell. Fluorescent dye (SYTO-9) was used to image the yeast cell. Scale bar is 200 μm. (b) Droplets from the generator are pressure-driven into the DMF device (frame 1). Frame 2 shows a droplet encapsulating a single cell and actuated on DMF electrodes (frames 3 and 4). The 20 nL droplet from the generator is then mixed with a larger droplet (100 nL) (frame 5). Scale bar is 300 μm. (c) Frames 1–5 show the actuation scheme to generate a 150 mM IL droplet. Frames 2–5 are repeated for the other concentrations of IL, which results in four droplets having a 0, 37.5, 75, or 150 mM IL concentration (frame 6). These droplets capture a single cell (from the generator) and are actuated to their own separate culture region. Scale bar is 2 mm. (See ESI† Video S1) | ||
D2D automation system
An attractive feature of microfluidic platforms is easier automation (via control of voltages and fluid flow) than conventional fluid-handling robots. Here, we designed and built an integrated automated microfluidic system that is capable of culturing cell in different conditions (Fig. 3). The platform comprises four core components: a feedback system for droplet sensing and detection, temperature control for culturing cells, flow rate control for droplet generation, and automated droplet movement on the DMF device. Automating these components together is challenging, as sophisticated control is required to ensure proper droplet movement and continuous cell culturing operation. Hence, a primary goal for the work reported here was the development of an automation system that is capable to automate single cell droplet generation and movement and to culture cells with unattended operation for extended periods of time. Shih et al.41,47,48 recently reported an impedance sensing and feedback control system for high-fidelity droplet movement and sensing on digital microfluidic devices. The system is very useful for manipulating “sticky” droplets such as those containing proteins. We adapt this system for droplet actuation and sensing droplets on the D2D microfluidic system and incorporate additional features (e.g. controlling flow rates) to ensure compatibility with other microfluidic paradigms (e.g. droplet-in-channels) and continuous cell culturing and analysis.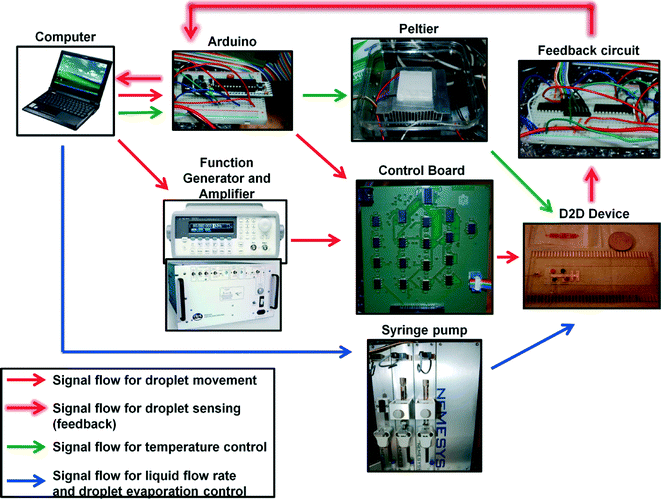 | ||
| Fig. 3 Automation system controlling the D2D microfluidic device. This schematic shows the components and the signal flow that control droplet movement and sensing on the device, temperature control by the Peltier element, and flow rates from the syringe pump. The Arduino is used to control the Peltier and to maintain constant temperature in the culture regions on the DMF. An in-house C++ program is used to control neMESYS syringe pumps to maintain steady flow rates and to prevent droplet evaporation. Scripts are written in C++ using Codeblocks 10.05 (http://www.codeblocks.org/home) using their default GCC compiler which is included as ESI.† | ||
The automation workflow consists of executing five procedures: 1) flow rate control, 2) droplet sensing, 3) IL dilutions, 4) mixing and cell culture, and 5) evaporation prevention. The flow control consists of controlling a high-precision neMESYS syringe pump and flow is initially applied to start generation of the droplets. Once the droplets are created and contain a single cell, they are pressure driven to the inter-digitated 300 μm electrodes on the DMF device via capillary. During the droplet generation phase, the feedback system continually (every 200 ms) applies 200 V (operating at 8 kHz) pulses to the 300 μm electrodes to check if a droplet from the generator is on the electrode. When a droplet is “sensed” on the electrode (i.e., detecting the difference in impedance when the electrode is not bearing a droplet and when there is a droplet on the electrode), it immediately stops the flow and the droplet containing a single cell is mixed with a droplet containing IL (IL dilutions were generated on the DMF previous to the flow rate control – see Fig. 2c and ESI† video). These four droplets are actuated to the cell culture zone and the Peltier heater was immediately ramped to its desired temperature (30 °C). To culture the cells on DMF, we started a mixing sequence where we would circulate the droplet with cells around the 2 × 3 electrode array (incl. L-shaped electrode). Each actuation consists of 100 ms pulses and with a 500 ms break step (i.e. no electrodes are actuated). In initial studies, we observed that without this break step, dielectric breakdown would occur during the cycling of the droplet (~60% of the devices; N = 15). However, with an added break step to the mixing sequence, it eliminated the breakdown and we were able to achieve high-fidelity droplet movement. To prevent evaporation of the droplets, the device was initially flooded with oil (~1–2 mL of HFE 7500). The automation system injects oil between the top and bottom layers of the device through a capillary using the channels fabricated on the DMF device during cell culture. To start the oil infusion process, an electric potential of 200 V at 8 kHz is applied to a reference electrode (i.e. any electrode that does not hold a droplet) every 5 s. Two parameters are used for the infusion process: Voil and Vmeasured. Voil represents the measured voltage across the reference electrode not bearing a droplet – in other words, with only oil between the top and bottom plates (we measured Voil = 0.174 ± 0.014 V; N = 12 on our devices). Vmeasured is the voltage of the reference electrode during operation. If Vmeasured < Voil (which represents the reference electrode being exposed to air – oil layer is evaporating), then oil is injected into the device at a flow rate of 10 μL s−1. The flow would stop when the reference electrode reaches the threshold value (i.e. Vmeasured > Voil). Two syringes were used for this process and were each equipped with a 2-way valve that allows refill of the syringes and injection of oil into the device. Only one was used during operation and when it becomes empty, the other filled syringe is activated while the empty syringe is set to refill. This new automation system was capable of implementing flow control of droplets, temperature control, droplet manipulation and movement, and preventing droplet evaporation. We speculate that this approach will be useful for applications that require long incubation periods and automation (e.g. cell culturing in different conditions, screening clinical samples, or enzymatic chemical reactions).
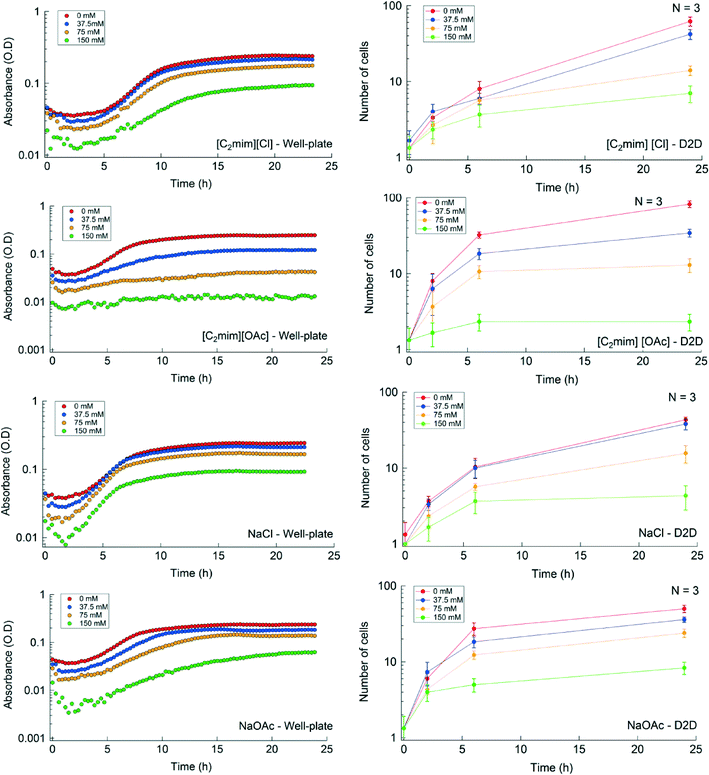
Impact of IL on growth and ethanol production
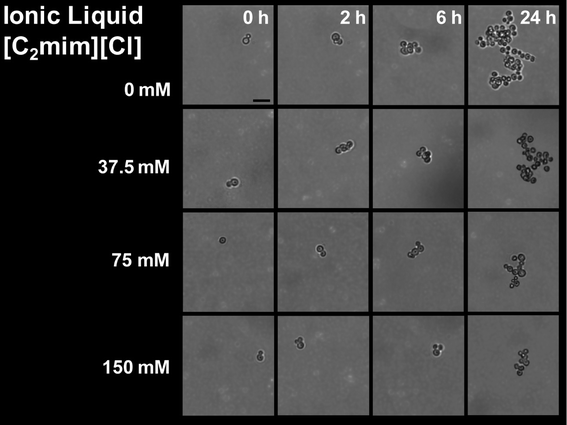
We used the automated D2D microfluidic device to determine the effect of IL/salt type and concentration on the morphological structure and growth of a single yeast cell and compared it to conventional well-plate experiments. To conduct a microfluidic screen, the cultures were interrogated with four concentrations of each IL or salt (the automated screening workflow is shown in ESI† Fig. S2). For both platforms, the culture conditions (e.g. Peltier heater temperature and IL concentration) were maintained the same and a total of 16 IL/salt conditions were evaluated. To minimize clumping of yeast, we circulated the droplet of yeast cells in a circular pattern (as described in the methods section) during culturing on the D2D device using the automation system. The single cell proliferation images cultured in [C2mim] [Cl] are shown in Fig. 4. The morphology of the yeast for [C2mim] [Cl], NaCl, and NaOAcs were unchanged during the 24 h at all tested concentrations. [C2mim] [OAc] at 75 mM and at 150 mM concentrations led to lysing of cells indicating that the ionic liquid is toxic to the yeast and will inhibit growth at these concentrations (see ESI† Fig. S3). Fig. 5 and Table S3† depict the growth curves and the growth rates respectively for the IL conditions tested in the well-plate and in the D2D microfluidic device. The growth curves shown in Fig. 5 are in qualitative agreement with respect to their growth profile (i.e. no IL shows largest growth at ~24 h) for both platforms. The replicates of the control experiments (i.e. intra-experiments) showed good reproducibility whereas the inter-experiments (experiments between different IL/salt) showed some variation at 2 h and 6 h on the D2D device (p = 0.004 and p = 0.001, respectively) but not in well-plates (see ESI† Fig. S4). We speculate the cells are exposed to fluctuating temperatures during preparation, in the syringe pumps, and in the droplet generator. These fluctuating conditions can directly affect their growth rates as the yeast travels through these various zones. Another cause for heterogeneity could be that the encapsulated single cell is at a different stage of its life cycle. In addition, growth was low or inhibited at concentrations greater than 37.5 mM for [C2mim] [OAc] and at 150 mM for NaOAc. Some studies have shown that acetic acid in its dissociated form is known to have an inhibitory impact on microbial growth60 which could explain the differences in morphology at 0 h and at 24 h and the decrease in their growth at these IL concentrations. For the other ionic liquids/salts tested, it showed no inhibitory effect for concentrations below 37.5 mM (where growth was similar in trend as the control).We calculated the growth rates for yeast grown in both well-plates and D2D platforms and determined that some calculated growth rates (in well-plates) were very similar in values. For example, yeast cultured in NaOAc have growth rates of 0.228 h−1, 0.268 h−1, and 0.241 h−1 for 0, 37.5, and 75 mM concentrations respectively; whereas for yeast cultured in D2D, we obtained 0.510 h−1, 0.444 h−1, and 0.379 h−1 for the same concentrations respectively. These disparities in the growth rates of the D2D device were of statistical difference (p = 0.015) while the well-plate methods were not statistically significant (p = 0.62). Dewan et al.58 and Wilson et al.61 recently reported that the heterogeneity of population of cells is less prominent compared to single cells due to “averaging out” the population heterogeneity58 or cell-to-cell variation in the single cell analysis.61 Therefore, we hypothesize that these effects may explain the differences in growth rates described here.
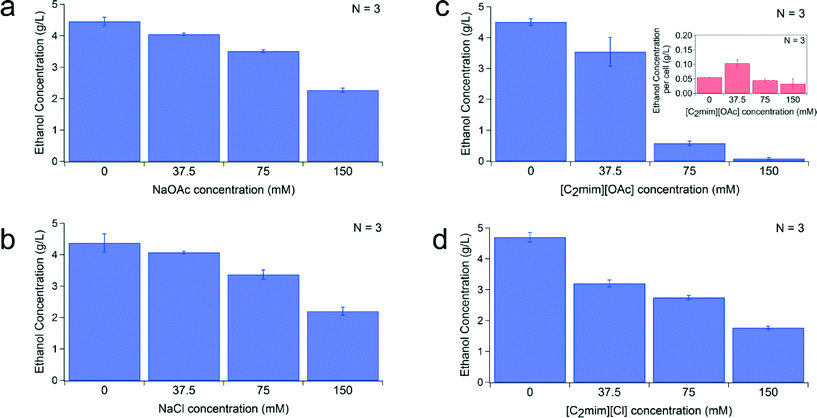
We also studied the impact of IL type and concentration on ethanol production in yeast (see description in Fig. S2†). We evaluated the influence of IL/salt on ethanol production at 16 conditions (same as above). Cultures were grown on the D2D microfluidic device for 24 h and then cultured in anaerobic chamber for another 24 h. Ethanol concentration was evaluated by HPLC. Ethanol eluted at ~20 min and was well separated from the other constituents (as shown in the chromatogram Fig. S5†). The ethanol production for each ionic liquid at four different concentrations is shown in Fig. 6. The total ethanol production for each ionic liquid decreased with increasing IL concentration (Fig. 6a–d). At higher concentrations of ionic liquid (75 and 150 mM), NaCl, NaOAc, and [C2mim] [Cl] exhibited a slight decrease (1.2–2×) in their ethanol production, while ionic liquid containing 1-ethyl-3-methylimidazolium acetate (i.e. [C2mim] [OAc]) showed a significant decrease (150 mM resulted in 58× decrease) in their ethanol production compared to the control (0 mM). We plotted the ethanol produced per cell for this ionic liquid (Fig. 6c inset) and it shows at 75 and 150 mM only produces 0.03–0.04 g L−1 of ethanol per cell. This is much lower compared to the other ionic liquid and salts which produce 0.15–0.55 g L−1 of ethanol per cell at these high concentrations. This observation is expected since [C2mim] [OAc] at these high concentrations is known to be toxic and to affect the fermentative metabolism.62 Development of an engineered IL-tolerant host can potentially eliminate or reduce the adverse effects of these solvents.
Conclusion
We have developed a novel automated droplet-to-digital (D2D) microfluidic platform for screening the effect of ionic liquids on cell growth and ethanol production. We also developed an automation system for conducting serial dilutions without pipetting, control temperature, and vary pressure flow (for preventing droplet evaporation and for delivering droplets with a single cell). In addition, this platform allows for the addition of reagents in parallel and single cell analysis. We tested our platform to screen multiple IL conditions that showed [C2mim] [OAc] had inhibitory effects on yeast growth and therefore significantly reduced their ethanol production. We also showed from statistical analysis that conducting single cell analysis using microfluidics resulted in observing differences in growth kinetics, which was not pronounced in the well-plate analysis. Here we demonstrate a 4-plex format but much higher level of multiplexing can be achieved using higher number of pixels in the array63 or using bussed electrode configurations.64 Furthermore, we have shown that this method allows a 600-fold reduction in volume compared to well-plate formats (which require from 200–800 μL) while our assay only requires a total of ~120 nL. Although the single cell screening demonstrated here is focused on studying ionic liquid with biofuel applications, the automated microfluidic device demonstrated are applicable to many different organisms and may prove useful for a diverse range of applications in single cell analyses that require frequent reagent addition and mixing steps.Acknowledgements
The authors thank Marijke Frederix and Dr. Aindrila Mukhopadhyay for the yeast isolate; Susan Yilmaz for initial help with microscopy; Jennifer Gin, Edward Baidoo, Helcio Burd for their assistance with operating the HPLC and chromatogram analysis; This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org), supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. Sandia is a multi-program laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the United States Department of Energy's Nuclear Security Administration under contract DE-AC04-94AL85000.References
- B. A. Simmons, D. Loque and H. W. Blanch, Genome Biol., 2008, 9, 242 CrossRef PubMed
.
- P. P. Peralta-Yahya and J. D. Keasling, Biotechnol. J., 2010, 5, 147–162 CrossRef CAS PubMed
.
- A. P. Dadi, S. Varanasi and C. A. Schall, Biotechnol. Bioeng., 2006, 95, 904–910 CrossRef CAS PubMed
.
- R. P. Swatloski, S. K. Spear, J. D. Holbrey and R. D. Rogers, J. Am. Chem. Soc., 2002, 124, 4974–4975 CrossRef CAS PubMed
.
- S. Singh, B. A. Simmons and K. P. Vogel, Biotechnol. Bioeng., 2009, 104, 68–75 CrossRef CAS PubMed
.
- C. Li, B. Knierim, C. Manisseri, R. Arora, H. V. Scheller, M. Auer, K. P. Vogel, B. A. Simmons and S. Singh, Bioresour. Technol., 2010, 101, 4900–4906 CrossRef CAS PubMed
.
- D. Klein-Marcuschamer, P. Oleskowicz-Popiel, B. A. Simmons and H. W. Blanch, Biomass Bioenergy, 2010, 34, 1914–1921 CrossRef CAS
.
- D. Klein-Marcuschamer, P. Oleskowicz-Popiel, B. A. Simmons and H. W. Blanch, Biotechnol. Bioeng., 2012, 109, 1083–1087 CrossRef CAS PubMed
.
- Q. Li, Y. C. He, M. Xian, G. Jun, X. Xu, J. M. Yang and L. Z. Li, Bioresour. Technol., 2009, 100, 3570–3575 CrossRef CAS PubMed
.
- M. Zavrel, D. Bross, M. Funke, J. Buchs and A. C. Spiess, Bioresour. Technol., 2009, 100, 2580–2587 CrossRef CAS PubMed
.
- H. Pfruender, R. Jones and D. Weuster-Botz, J. Biotechnol., 2006, 124, 182–190 CrossRef CAS PubMed
.
- Q. Huang, Q. Wang, Z. Gong, G. Jin, H. Shen, S. Xiao, H. Xie, S. Ye, J. Wang and Z. K. Zhao, Bioresour. Technol., 2013, 130, 339–344 CrossRef CAS PubMed
.
- G. B. Fogel and C. F. Brunk, Anal. Biochem., 1998, 260, 80–84 CrossRef CAS PubMed
.
- W. A. Duetz, L. Ruedi, R. Hermann, K. O'Connor, J. Buchs and B. Witholt, Appl. Environ. Microbiol., 2000, 66, 2641–2646 CrossRef CAS PubMed
.
- T. Schneider, J. Kreutz and D. T. Chiu, Anal. Chem., 2013, 85, 3476–3482 CrossRef CAS PubMed
.
- L. Mazutis, J. Gilbert, W. L. Ung, D. A. Weitz, A. D. Griffiths and J. A. Heyman, Nat. Protoc., 2013, 8, 870–891 CrossRef CAS PubMed
.
- T. Kong, J. Wu, K. W. Yeung, M. K. To, H. C. Shum and L. Wang, Biomicrofluidics, 2013, 7, 44128 CrossRef PubMed
.
- Z. Liu and H. C. Shum, Biomicrofluidics, 2013, 7, 44117 CrossRef PubMed
.
- D. Saeki, S. Sugiura, T. Kanamori, S. Sato and S. Ichikawa, Lab Chip, 2010, 10, 357–362 RSC
.
- S. Koster, F. E. Angile, H. Duan, J. J. Agresti, A. Wintner, C. Schmitz, A. C. Rowat, C. A. Merten, D. Pisignano, A. D. Griffiths and D. A. Weitz, Lab Chip, 2008, 8, 1110–1115 RSC
.
- J. Clausell-Tormos, D. Lieber, J. C. Baret, A. El-Harrak, O. J. Miller, L. Frenz, J. Blouwolff, K. J. Humphry, S. Koster, H. Duan, C. Holtze, D. A. Weitz, A. D. Griffiths and C. A. Merten, Chem. Biol., 2008, 15, 427–437 CrossRef CAS PubMed
.
- E. Brouzes, M. Medkova, N. Savenelli, D. Marran, M. Twardowski, J. B. Hutchison, J. M. Rothberg, D. R. Link, N. Perrimon and M. L. Samuels, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14195–14200 CrossRef CAS PubMed
.
- C. Chang, J. Sustarich, R. Bharadwaj, A. Chandrasekaran, P. D. Adams and A. K. Singh, Lab Chip, 2013, 13, 1817–1822 RSC
.
- L. Mazutis, J. C. Baret, P. Treacy, Y. Skhiri, A. F. Araghi, M. Ryckelynck, V. Taly and A. D. Griffiths, Lab Chip, 2009, 9, 2902–2908 RSC
.
- S. L. Sjostrom, Y. Bai, M. Huang, Z. Liu, J. Nielsen, H. N. Joensson and H. Andersson Svahn, Lab Chip, 2014, 14, 806–813 RSC
.
- L. Yu, M. C. Chen and K. C. Cheung, Lab Chip, 2010, 10, 2424–2432 RSC
.
- D. Pekin, Y. Skhiri, J. C. Baret, D. Le Corre, L. Mazutis, C. B. Salem, F. Millot, A. El Harrak, J. B. Hutchison, J. W. Larson, D. R. Link, P. Laurent-Puig, A. D. Griffiths and V. Taly, Lab Chip, 2011, 11, 2156–2166 RSC
.
- L. Mazutis, J. C. Baret and A. D. Griffiths, Lab Chip, 2009, 9, 2665–2672 RSC
.
- H. Song, M. R. Bringer, J. D. Tice, C. J. Gerdts and R. F. Ismagilov, Appl. Phys. Lett., 2003, 83, 4664–4666 CrossRef CAS PubMed
.
- B. Ahn, K. Lee, R. Panchapakesan and K. W. Oh, Biomicrofluidics, 2011, 5, 24113 CrossRef PubMed
.
- B. O'Donovan, D. J. Eastburn and A. R. Abate, Lab Chip, 2012, 12, 4029–4032 RSC
.
- D. J. Eastburn, A. Sciambi and A. R. Abate, PLoS One, 2013, 8, e62961 CAS
.
- L. Xu, H. Lee, R. Panchapakesan and K. W. Oh, Lab Chip, 2012, 12, 3936–3942 RSC
.
- X. Niu, S. Gulati, J. B. Edel and A. J. deMello, Lab Chip, 2008, 8, 1837–1841 RSC
.
- A. R. Wheeler, Science, 2008, 322, 539–540 CrossRef CAS PubMed
.
- K. Choi, A. H. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef CAS PubMed
.
- N. Vergauwe, D. Witters, F. Ceyssens, S. Vermeir, B. Verbruggen, R. Puers and J. Lammertyn, J. Micromech. Microeng., 2011, 21, 054026 CrossRef
.
- D. Witters, N. Vergauwe, S. Vermeir, F. Ceyssens, S. Liekens, R. Puers and J. Lammertyn, Lab Chip, 2011, 11, 2790–2794 RSC
.
- P. T. Kumar, F. Toffalini, D. Witters, S. Vermeir, F. Rolland, M. Hertog, B. M. Nicolai, R. Puers, A. Geeraerd and J. Lammertyn, Sens. Actuators, B, 2014, 199, 479–487 CrossRef CAS
.
- S. H. Au, S. C. C. Shih and A. R. Wheeler, Biomed. Microdevices, 2011, 13, 41–50 CrossRef CAS PubMed
.
- S. C. C. Shih, I. Barbulovic-Nad, X. Yang, R. Fobel and A. R. Wheeler, Biosens. Bioelectron., 2013, 42, 314–320 CrossRef CAS PubMed
.
- I. Barbulovic-Nad, H. Yang, P. S. Park and A. R. Wheeler, Lab Chip, 2008, 8, 519–526 RSC
.
- V. N. Luk, G. Mo and A. R. Wheeler, Langmuir, 2008, 24, 6382–6389 CrossRef CAS PubMed
.
- S. H. Au, P. Kumar and A. R. Wheeler, Langmuir, 2011, 27, 8586–8594 CrossRef CAS PubMed
.
- S. H. Au, R. Fobel, S. P. Desai, J. Voldman and A. R. Wheeler, Integr. Biol., 2013, 5, 1014–1025 RSC
.
-
R. Rodríguez-Díaz, T. Wehr and S. Tuck, Analytical techniques for biopharmaceutical development, M, Dekker, Boca Raton, 2005 Search PubMed
.
- S. C. C. Shih, R. Fobel, P. Kumar and A. R. Wheeler, Lab Chip, 2011, 11, 535–540 RSC
.
- S. C. C. Shih, H. Yang, M. J. Jebrail, R. Fobel, N. McIntosh, O. Y. Al-Dirbashi, P. Chakraborty and A. R. Wheeler, Anal. Chem., 2012, 84, 3731–3738 CrossRef CAS PubMed
.
- M. Abdelgawad, M. W. Watson and A. R. Wheeler, Lab Chip, 2009, 9, 1046–1051 RSC
.
- M. W. Watson, M. J. Jebrail and A. R. Wheeler, Anal. Chem., 2010, 82, 6680–6686 CrossRef CAS PubMed
.
- J. Gorbatsova, M. Jaanus and M. Kaljurand, Anal. Chem., 2009, 81, 8590–8595 CrossRef CAS PubMed
.
- H. Kim, M. S. Bartsch, R. F. Renzi, J. He, J. L. Van de Vreugde, M. R. Claudnic and K. D. Patel, J. Lab. Autom., 2011, 16, 405–414 CrossRef CAS PubMed
.
- H. Kim, M. J. Jebrail, A. Sinha, Z. W. Bent, O. D. Solberg, K. P. Williams, S. A. Langevin, R. F. Renzi, J. L. Van De Vreugde, R. J. Meagher, J. S. Schoeniger, T. W. Lane, S. S. Branda, M. S. Bartsch and K. D. Patel, PLoS One, 2013, 8, e68988 CAS
.
- N. Thaitrong, H. Kim, R. F. Renzi, M. S. Bartsch, R. J. Meagher and K. D. Patel, Electrophoresis, 2012, 33, 3506–3513 CrossRef CAS PubMed
.
- R. B. Fair, Microfluid. Nanofluid., 2007, 3, 245–281 CrossRef CAS
.
- G. H. Markx, Organogenesis, 2008, 4, 11–17 CrossRef PubMed
.
- B. L. Wang, A. Ghaderi, H. Zhou, J. Agresti, D. A. Weitz, G. R. Fink and G. Stephanopoulos, Nat. Biotechnol., 2014, 32, 473–478 CrossRef CAS PubMed
.
- A. Dewan, J. Kim, R. H. McLean, S. A. Vanapalli and M. N. Karim, Biotechnol. Bioeng., 2012, 109, 2987–2996 CrossRef CAS PubMed
.
- P. Paik, V. K. Pamula and R. B. Fair, Lab Chip, 2003, 3, 253–259 RSC
.
- B. Maiorella, H. W. Blanch and C. R. Wilke, Biotechnol. Bioeng., 1983, 25, 103–121 CrossRef CAS PubMed
.
- J. L. Wilson, S. Suri, A. Singh, C. A. Rivet, H. Lu and T. C. McDevitt, Biomed. Microdevices, 2014, 16, 79–90 CrossRef PubMed
.
- Y. V. Nancharaiah and A. J. Francis, Bioresour. Technol., 2011, 102, 6573–6578 CrossRef PubMed
.
- B. Hadwen, G. R. Broder, D. Morganti, A. Jacobs, C. Brown, J. R. Hector, Y. Kubota and H. Morgan, Lab Chip, 2012, 12, 3305–3313 RSC
.
- S. C. C. Shih, N. S. Mufti, M. D. Chamberlain, J. Kim and A. R. Wheeler, Energy Environ. Sci., 2014, 7, 2366–2375 CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4lc00794h |
| This journal is © The Royal Society of Chemistry 2015 |