Elemental and molecular mass spectrometry for integrated selenosugar speciation in liver and kidney tissues of maternal feeding and supplemented rats
Héctor
González-Iglesias
a,
María Luisa
Fernández-Sánchez
a,
Ying
Lu
b,
Sonia
Fernández Menéndez
a,
Spiros A.
Pergantis
b and
Alfredo
Sanz-Medel
*a
aDepartment of Physical and Analytical Chemistry, Faculty of Chemistry, University of Oviedo, Julián Clavería, 8, 33006 Oviedo, Spain. E-mail: asm@uniovi.es
bDepartment of Chemistry, Environmental Chemical Processes Laboratory, University of Crete, Voutes, Heraklion, 71003 Crete, Greece
First published on 9th October 2014
Abstract
The development of methods assessing the nutritional value and metabolism of selenium are of growing interest. In this work, the integrated used of a methodology based on HPLC-isotope pattern deconvolution (IPD)-ICP-MS and a molecular tandem mass spectrometric technique, such as HPLC-APCI-MS/MS, in the selected reaction monitoring (SRM) mode, was applied to quantify and identify the selenosugar SeGalNAc in liver and kidney tissues of lactating rats fed with formula milk supplemented with 77selenite. The SeGalNAc levels found in liver and kidney of maternal feeding rats (kidney 23 ± 3 ng g−1; liver 26 ± 3 ng g−1) were much higher than those found in supplemented (kidney 9.9 ± 0.3 ng ng−1; liver 10 ± 4 ng g−1) and non-supplemented rats (kidney 3.4 ± 0.5 ng g−1; liver 4 ± 1 ng g−1). The percentage of exogenous SeGalNAc for the supplemented group in kidney and liver reached 32 ± 1% and 30 ± 10%, respectively. Conversely, the percentage of exogenous selenium in high molecular weight selenospecies reached values higher than 58%. Thereby, most exogenous selenium seems to be incorporated into the synthesis of selenoproteins, indicating that the turnover rates are different for the different species and their synthesis might occur in different tissue compartments. Finally, the identification of SeGalNAc was confirmed in liver and, for the first time to our knowledge, in the kidney cytosol of maternal feeding and supplemented rats. Overall, we expect that the judicious use of elemental and molecular mass spectrometry tools to obtain integrated quantitative Se speciation information might help to expand our knowledge of selenium metabolism.
Introduction
Selenium is an essential trace element required for the correct development and wellbeing of plants, animals, and humans.1 The multiple roles of this element are related to its incorporation into proteins in the form of selenocysteine, the 21st amino acid used for protein synthesis. Twenty-five genes encoding selenoproteins have been already identified in humans,2,3 while twenty-four have been found in the mouse and rat genome, although only a few of them have been functionally characterized.4 Selenoproteins including glutathione peroxidases, thioredoxin reductases and iodothyronine deiodinases have biological functions in oxido-reduction processes, redox signaling, antioxidant defense, thyroid hormone metabolism, and immune responses.5The main source of trace elements is food, milk being the primary supply of nutrients for the newborn during the first months of life. Nevertheless, when breastfeeding is not enough or not possible, formula milk appears as an alternative, approved by regulatory committees. Selenium levels in formula milk are similar to or higher (as a result of supplementation) than those found in human milk, although the physicochemical form in which the element occurs is different.6 There is growing interest in the production of selenium-enriched milk and nutritional supplements. Selenium supplementation in formula milk is usually carried out in the form of selenite (SeO32−) salt. As already mentioned here, the nutritional value of selenium is critically dependent on the chemical form in which it occurs in a given food,7,8 and so the development of methods assessing the speciation, metabolism and nutritional status of selenium is currently in high demand.
Several compartmental models, based on the chemical form in which selenium is supplemented, have been proposed to establish selenium fate and distribution in the body.9–11 In the “selenite model”, once this element is absorbed through the gastrointestinal tract, it is reduced to selenide in the enterocyte and transferred to the liver in a form bound to albumin. In the liver it is used for the synthesis of selenoproteins, cellular glutathione peroxidase (cGHSPx), and metabolites (i.e., selenosugars), and later is re-excreted to the bloodstream and transferred to the kidneys, where it is degraded and used for the synthesis of extra-cellular glutathione peroxidase and selenosugars.10,12 However, current literature only contains sparse data on Se species in the liver and kidneys, two vital multifunctional organs. In fact, the urine content of Se metabolite methyl 2-acetamido-2-deoxy-1-seleno-β-D-galactopyranoside (SeGalNAc), identified in rat and human urine13–15 and in porcine liver,16 could be a better selenium nutritional biomarker than the currently used total selenium concentrations.17
Recently, a new methodology based on the use of stable isotopes and isotope pattern deconvolution (IPD) in connection with HPLC-ICP-MS, has been developed to study selenium metabolism in lactating rats fed with formula milk supplemented with 77selenite.18–20 This methodology provides unique quantitative information about the tracer, tracee, and their elemental species in biological tissues and fluids. In these studies, the discrimination of the fate of endogenous (natural) and supplemented (enriched 77Se) selenium and their catabolised selenospecies was carried out in rat urine,18 feces,19 serum20 and erythrocytes.20
The identification and characterization of SeGalNAc in urine has been previously reported by using HPLC coupled to atmospheric pressure chemical ionization (APCI) tandem mass spectrometry (MS/MS).21 However, there is a lack of detection of SeGalNAc in liver and kidney due to its low concentration. Suzuki et al. have reported the presence of the selenosugar in rat liver and urine, with detection based on the retention times of the standard materials used for reference, although they did not conduct any conclusive identification by means of molecular mass spectrometry techniques.22,23 To overcome this, the use of HPLC-APCI-MS/MS in the selected reaction monitoring (SRM) mode has been proven to be very useful in the detection of small quantities of a given compound in a mixture as long as the mass of the compound is known (targeted analysis). Thereby, Lu et al.16 have proposed the conclusive identification of SeGalNAc in porcine liver based on the monitoring of characteristic SeGalNAc SRM transitions, the SRM intensity ratios and HPLC retention times in comparison with those of a SeGalNAc standard. Recently, HPLC-ICP-MS and molecular mass spectrometry (HPLC – electrospray – MS/MS in SRM mode) were also used in a complementary fashion to monitor small selenium species over time in both serum and urine of volunteers treated with different selenium supplements, confirming the presence of selenosugars and the trimethylselenonium ion.24
In this work, an array of elemental and molecular mass spectrometry tools are used for integrated quantitative Se speciation to expand our knowledge of selenium metabolism. Hence, we further explore the use of HPLC-IPD-ICP-MS for the quantification of total selenium and of selenium-containing biomolecules in tissues in an attempt to gain further insight into the metabolism of this element in mammals. Furthermore, identification of the selenosugar SeGalNAc in liver and kidney of lactating rats (after Se supplementation by the enriched stable isotope compound, 77selenite) is demonstrated using HPLC-APCI-MS/MS in the SRM mode.
Experimental
Instrumentation
An ICP-MS model Agilent 7500ce (Agilent Technologies, Tokyo, Japan) equipped with an octapole reaction cell was used for total selenium determination and quantitative selenium speciation. A flow of 4 mL min−1 of H2 was used for interference suppression. Plasma operating conditions are given in Table 1 for time resolved and isotope dilution modes. An HPLC system hyphenated to ICP-MS included a Shimadzu pump (LC-20AD, Kyoto, Japan), a Rheodyne six-port injector (CA, USA) with a 50 μL sample loop, and a column.| Plasma parameters | |
|---|---|
| RF power/W | 1500 |
| Plasma gas flow rate/L min−1 | 15 |
| Auxiliary gas flow rate/L min−1 | 1.1 |
| Sampling depth/mm | 5.8 |
| Ion lens setting | Daily optimized for best sensitivity of 10 μg L−1 Li, Co, Y and Tl |
| Reaction cell parameters | |
|---|---|
| H2 gas/mL min−1 | 4 |
| *Octapole bias/V | −13 |
| *QP bias/V | −12 |
| Data acquisition parameters (IPD analysis) | |
|---|---|
| Acquisition mode | ID analysis |
| Monitored isotopes | 74, 76, 77,78, 79, 80, 81, 82, 83 |
| Points per peak | 3 |
| Acquisition time per point/s | 4 |
| Replicates | 5 |
| Data acquisition parameters (IPD-post column) | |
|---|---|
| Acquisition mode | Time resolved analysis |
| Monitored isotopes | 74, 76, 77,78, 79, 80, 81, 82 |
| Points per peak | 1 |
| Integration time (per peak)/s | 0.3 |
| Chromatographic conditions (HPLC-IPD-ICP-MS) | |
|---|---|
| Multimode size exclusion and anionic exchange | Shodex Asahipak GS-520 HQ (300 mm × 7.5 mm i.d., 7 μm particle size) |
| Mobile phase | 40 mM NH4Ac, 3% MeOH, pH = 7.4 |
| Flow rate | 0.7 mL min−1 |
| Injection volume | 50 μL |
| APCI-MS/MS parameters | |
|---|---|
| Discharge current | 4.0 μA |
| Vaporizer temperature | 400 °C |
| Capillary temperature | 300 °C |
| Collision cell pressure | 1.0 m Torr |
| Chromatographic conditions (HPLC-APCI-MS/MS) | |
|---|---|
| Reversed-phase (RP) | Waters Atlantis C18 (100 mm × 3 mm i.d., 3.5 μm particle size) |
| Mobile phase | 40 mM NH4Ac, 3% MeOH, pH = 7.4 |
| Flow rate | 0.8 mL min−1 |
| Injection volume | 50 μL |
A triple quadrupole mass spectrometer (TSQ Quantum, Thermo Electron, San Jose, CA, USA) with an atmospheric pressure chemical ionization (APCI) source was operated in the positive ion mode to identify the SeGalNAc compound. A Thermo Finnigan Surveyor HPLC system with a manual injector fitter with a 50 μL loop was coupled on-line to the APCI-MS/MS according to the conditions given in Table 1.
A combined size exclusion and ion exchange separation column (Shodex Asahi Pack GS-520 HPLC) was used for the quantitative speciation of Se in liver and kidney and the preconcentration of the selenosugar (SeGalNAc) from the cytosolic fractions. A reversed-phase (RP) column (Atlantis C18, Waters Corporation, USA) was used for the selenosugar identification in liver and kidney of lactating rats by HPLC-APCI-MS/MS.
An Ultra-turrax T-25 (IKA Labortechnik, Staufen, Germany) instrument was employed for liver and kidney homogenizations.
Two centrifuges were used for the preparation of liver and kidney cytosolic fractions: a Biofuge Stratos Heraeus (Thermo Fisher Scientific, Germany) centrifuge, and an Avanti J-26xp (Beckman Coulter, USA) ultracentrifuge.
A LYOLAB 3000 (Heto-Holten A7S, Allerod, Denmark) lyophilizer was also employed for selenosugar preconcentration.
Reagents and materials
Enriched 77Se (94.75% abundance) and 74Se (98.84% abundance) were obtained from Cambridge Isotope Laboratories (Andover, MA, USA). These selenium standards have been previously characterized both in isotopic composition and selenium concentration, as indicated by González-Iglesias et al.18,19A non-supplemented commercial formula milk, provided by Laboratorios Ordesa (Barcelona, Spain), and containing low amounts of essential elements was used for rat feeding. The level of selenium was determined by ICP-MS analysis, obtaining 50 ± 5 ng Se g−1 powder. The product contained (g per 100 g): protein (12.5), lactose (50), fat (22), carbohydrate (70), and minerals (3.5).
Synthetically prepared SeGalNAc standard25 was provided by Professor K. A. Francesconi (Institute of Chemistry-Analytical Chemistry, Karl-Franzens University Graz, Austria).
Standard bovine liver reference material SRM 1577a was purchased from the National Institute of Standards and Technology (NIST, USA).
Ammonium acetate, methanol, sodium chloride, tris(hydroxymetyl) aminomethane hydrochloride (TRIS), and acetonitrile of analytical grade were also used.
Distilled de-ionized water (18 MΩ cm) was obtained by means of a Milli-Q system (Millipore).
Animals
The animal experiments were carried out in the Animal Unit Laboratory of the University of Oviedo following the guidelines established by The Local Ethics Committee (R. D. 223/1988) and The Animal Experiments Directive (86/609/EEC) on the protection of animals used for experimental and other scientific purposes. The experimental treatments performed were previously described by González-Iglesias et al.18,19,26 In brief, two-week-old Wistar rats were maintained in metabolic cages (three rats/cage) at 22 °C with a light/dark cycle of 12/12 h for two weeks. Rats were then randomized into 3 groups (3 rats per group). Afterwards, a group of three of those rats, labeled as non-supplemented group, was fed with reconstituted formula milk ad libitum for another two weeks. Another group of three rats, labeled as supplemented group, was fed with the same reconstituted formula milk but supplemented with 77Se in the chemical form of selenite and at a concentration of 0.15 μg Se g−1 powder milk, for two weeks. Finally, a rat reference group was fed with maternal milk for two weeks also, and labeled as maternal-feeding group.The animals were sacrificed at the end of the study, and after whole body perfusion and central longitudinal incision into the abdominal wall, liver and kidney organs were harvested.
Liver and kidney samples preparation
Liver and kidneys were cleaned with cold ultrapure water and homogenized with an Ultra-turrax T-25 system with 4 volumes (w/v) of cold 50 mM Tris–HCl buffer solution at pH = 7.4, under a nitrogen atmosphere and in an ice-water bath. All the samples were stored at −20 °C until use.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 minutes to sediment non-suspended material. The supernatants were subjected to a second ultracentrifugation at 105
000 × g for 30 minutes to sediment non-suspended material. The supernatants were subjected to a second ultracentrifugation at 105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 90 minutes at 4 °C, to obtain the cytosolic fractions.27
000 × g for 90 minutes at 4 °C, to obtain the cytosolic fractions.27
Determination of total Se in liver and kidney by IPD-ICP-MS
Isotope pattern deconvolution (IPD) is a chemometric technique based on multiple least squares, which has been applied in connection with ICP-MS and stable isotopes to study selenium metabolism in lactating rats after milk supplementation with enriched 77SeO32−.18–20 This mathematical tool enables the selenium quantification of both endogenous (natural) and exogenous (supplemented) origin in the animals with very good accuracy and with a time-saving experimental setting, providing quantitative information about the tracer and the tracee in biological tissues. Liver and kidney-mineralized samples, containing selenium of natural abundance (natSe, endogenous), the isotopically enriched metabolic tracer (77Se, exogenous), and the appropriate amount of an enriched 74Se standard for the quantifications, were analyzed by ICP-MS, under the conditions showed in Table 1 and following the procedure described by González-Iglesias et al.18 The IPD methodology was used to calculate the concentration of endogenous and exogenous selenium present in liver and kidney tissues. Bovine liver reference material was used for validating the utilized selenium quantitation procedure in biological samples, after its mineralization.Quantitative speciation of Se in liver and kidney by HPLC-IPD-ICP-MS
Chromatographic separation of Se-containing chemical species in liver and kidney cytosolic fractions was carried out in a Shodex Asahi Pack GS-520 HPLC column, coupled on-line to the ICP-MS (see operating conditions in Table 1). Fifty microliters of liver or kidney cytosolic fraction from each rat were injected, and the Se-chemical-species eluting from the column were quantitated by post-column IDA following previous reports.19,20,28 Briefly, a 74Se-enriched standard solution of the appropriate concentration was continuously introduced (at 0.1 mL min−1) using a peristaltic pump at the end of the column through a T-piece. The emerging equilibrated mixture of isotopes was nebulized into the plasma. The intensities in the chromatogram (counts s−1) were converted into two independent natural and exogenous (supplemented) Se mass flow chromatograms (ng min−1), by applying the IPD procedure described earlier. Finally, the Se amount of each selenium chemical species, containing natural or exogenous selenium, was determined in each chromatographic peak by area integration.Selenosugar identification in liver and kidney by APCI-MS/MS
The identification of SeGalNAc in liver and kidney preconcentrated cytosolic fractions was carried out by means of a molecular tandem mass spectrometry technique, i.e. HPLC-APCI-MS/MS, using the optimized tuning conditions shown in Table 1. Selective and sensitive monitoring for SeGalNAc was carried out using the selected reaction monitoring (SRM) mode, with the specific transitions previously described elsewhere.16,21Results and discussion
Total natural/exogenous Se determinations in liver and kidney by IPD-ICP-MS
IPD-ICP-MS methodology was used to study natural and/or supplemented selenium distribution in liver and kidney of lactating rats. In order to validate the sample digestion procedure and the IPD approach, previously developed for quantitation of total natural and exogenous selenium,18,19 a certified reference material (bovine liver, NIST, SRM 1577a) was analyzed. Sample containing a certified amount of Se with natural isotopic abundance, was spiked with the two selected enriched Se isotopes (74Se and 77Se), before acidic microwave digestion. The selenium isotopes were measured by ICP-MS in the digested samples and the concentrations of natural selenium (reference value) and exogenous Se (spiked 77Se) were determined from the selenium isotope composition measured in the sample by applying the IPD methodology.18,19Table 2 shows the observed quantitative determination values of selenium in the bovine liver reference material for both natural Se (certified value) and for 77Se (tracer) obtained, which agreed well with the certified Se (natural Se) and the added 77Se (tracer).| Reference material | Natural selenium (ng g−1) | 77Se-enriched (ng g−1) | ||
|---|---|---|---|---|
| Certified | Found | Added | Found | |
| NIST SRM 1577a bovine liver | 710 ± 70 | 690 ± 40 | 650 | 630 ± 40 |
Next, the IPD approach was conducted to investigate natural and supplemented total selenium quantification in rat liver and kidneys, as follows: nine livers and eighteen kidneys from nine rats (classified as maternal feeding, non-supplemented and supplemented groups, 3 rats per group) were treated as indicated in the Experimental section. Samples were spiked with a known amount of 74Se before their mineralization and Se isotope abundance determination was carried out by ICP-MS. From those experimental abundance values, IPD was used to quantify the amount of endogenous (natSe) and supplemented (77Se) selenium present in every sample under study. Samples were analyzed in triplicate and the obtained results for the three rat groups are shown in Table 3.
| Supplemented | Non supplemented | Maternal feeding | |||||
|---|---|---|---|---|---|---|---|
| natSe | 77Se | totalSe | % 77Se | total Se | total Se | ||
| Kidney (ng g−1) | Rat 1 | 105 ± 5 | 165 ± 5 | 165 ± 10 | 61 | 74 ± 3 | 317 ± 6 |
| Rat 2 | 115 ± 6 | 170 ± 7 | 285 ± 13 | 60 | 88 ± 5 | 288 ± 3 | |
| Rat 3 | 124 ± 6 | 155 ± 4 | 309 ± 10 | 56 | 91 ± 4 | 415 ± 8 | |
| Average | 115 ± 10 | 163 ± 8 | 278 ± 17 | 59 ± 3 | 84 ± 9 | 340 ± 67 | |
| Liver (ng g−1) | Rat 1 | 150 ± 4 | 248 ± 3 | 398 ± 7 | 62 | 110 ± 5 | 602 ± 4 |
| Rat 2 | 110 ± 5 | 250 ± 4 | 360 ± 9 | 69 | 125 ± 4 | 572 ± 7 | |
| Rat 3 | 140 ± 3 | 275 ± 8 | 415 ± 4 | 60 | 140 ± 6 | 707 ± 6 | |
| Average | 133 ± 21 | 258 ± 15 | 391 ± 36 | 66 ± 5 | 125 ± 15 | 627 ± 71 | |
The observed total levels of selenium in liver and kidneys were highly related to the total amount of selenium ingested. Indeed, under physiological conditions, selenium is mainly stored in liver and kidney, but with deficient intake, its amount is markedly reduced in liver, while in kidney it is maintained.29 This observation is consistent with the above data, since the ratio between total selenium for the maternal feeding group versus the supplement group obtained for liver reached 1.6 times, while in kidney this ratio reached 1.2. Moreover, the hepatic Se concentration decrease in rats fed with the Se deficient diet (non supplemented) reflects an insufficient supply of selenium and is coherent with the decreased levels of selenium circulating in the body. Furthermore, at the end of the supplementation period the % of exogenous selenium in the supplemented group (58–60) is very similar to those values previously found in urine19 and serum,20 suggesting the slow turnover (metabolism and catabolism) of selenium-containing biomolecules between body tissues and the bio-fluids.
Selenium quantitative speciation in rat liver and kidney cytosolic fractions by HPLC-IPD-ICP-MS
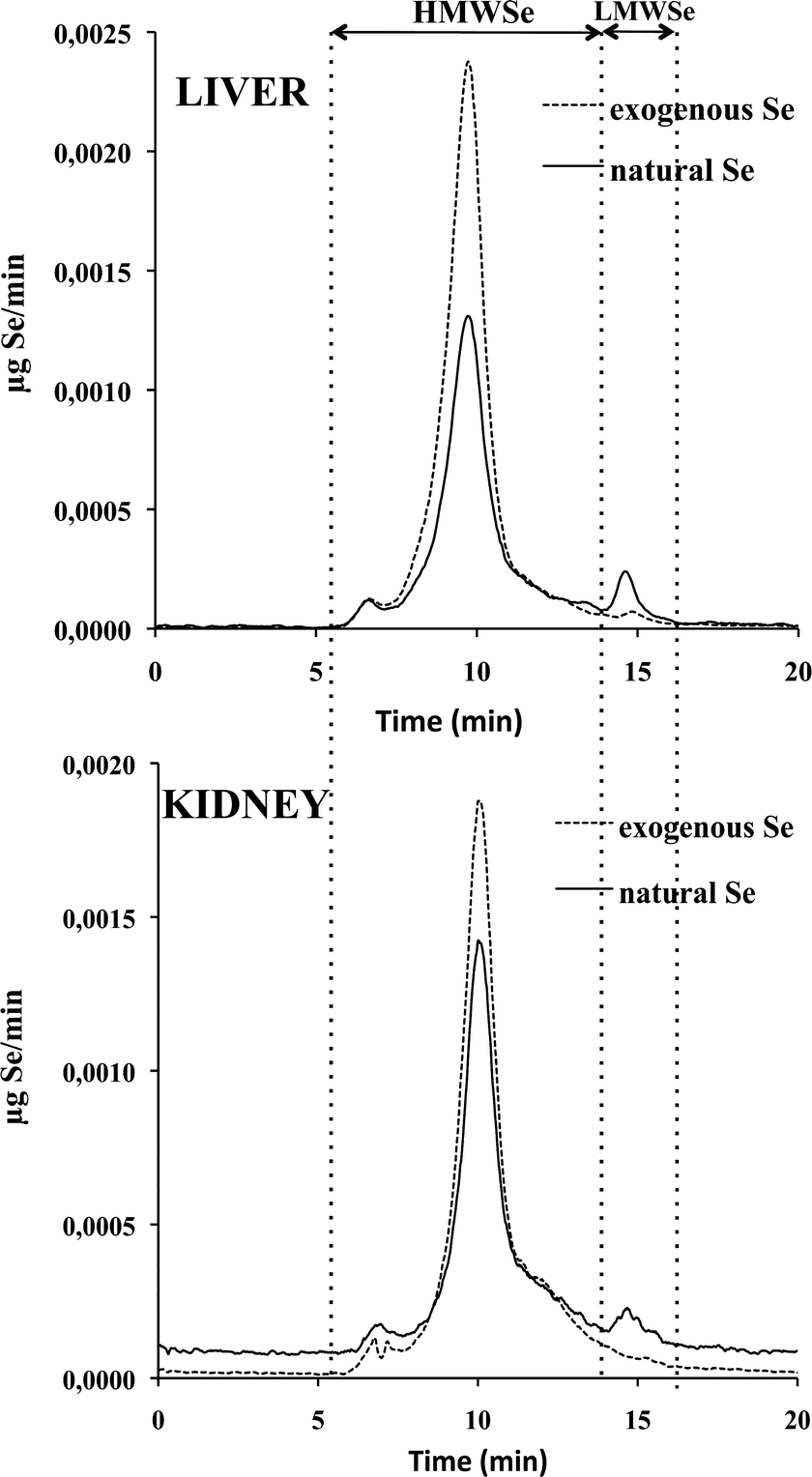
The HPLC-IPD-ICP-MS methodology was applied to determine the distribution of selenium (natural and exogenous) chemical species in the rat liver and kidney cytosolic fractions of maternal, non-supplemented and supplemented groups. Quantitative Se speciation was performed by post-column isotope dilution analysis by continuous mixing of the sample flow with an enriched 74Se solution. All the Se isotopes were monitored and the corresponding chromatograms obtained. Applying the IPD model at every point in the chromatogram, the two mass flow chromatograms for natural and exogenous (77Se) selenium were obtained for the liver and kidney cytosolic fractions, and the amount of the selenium-chemical species calculated by integration of the chromatographic peaks (see Procedures).Fig. 1 illustrates the chromatographic separation of selenospecies obtained in the cytosolic fractions of liver and kidney of supplemented rats, as indicated in the Experimental procedures. The speciation analysis, both in liver and kidney (according to the column calibration based on the retention time observed) revealed the presence of two main regions: a predominant region composed of high molecular weight selenium biomolecules (HMWSe, from 5 to 14 min) and a less abundant region of low molecular weight selenospecies (LMWSe, at 14 min).
In the HMWSe region, two main selenoproteins have been identified to date in rat liver and kidney, the cellular glutathione peroxidases (cGSHPx) and phospholipid hydroperoxide glutathione peroxidases (PHGSHPx).30,31 The major peak observed may correspond to the retention time of cGSHPx standard (tr ∼ 10 min), although its further identification was not carried out in this work which is focused on the selenosugar species.
In the LMWSe region, two selenosugars have been identified in rat liver: the SeGalNAc and its precursor.12,16,32 Conversely, in rat kidney the presence of SeGalNAc has not been reported so far. The small natural Se peak detected in LMWSe region matched with the standard compound SeGalNAc (tr = 14.4–15.2 min), both in liver and kidney samples. In any case, the chemical identity of this metabolite was further confirmed by HPLC-APCI-MS/MS.
Similar elemental profiles to those shown in Fig. 1, for the supplemented group, were obtained for liver and kidney of the maternal feeding and non-supplemented groups. The amounts of the selenium-chemical species for the HMWSe and the SeGalNAc metabolite (tr = 14.8 min) were determined by IPD. Obtained results are summarized in Table 4.
| Selenospecies (ng Se g−1) | Supplemented | Non supplemented | Maternal feeding | ||||
|---|---|---|---|---|---|---|---|
| natSe | 77Se | totalSe | % 77Se | total Se | total Se | ||
| Kidney | HMWSe | 93 ± 16 | 129 ± 22 | 222 ± 38 | 58 ± 9 | 64 ± 12 | 272 ± 20 |
| SeGalNAc | 6.7 ± 0.1 | 3.2 ± 0.2 | 9.9 ± 0.3 | 32 ± 1 | 3.4 ± 0.5 | 23 ± 3 | |
| Liver | HMWSe | 112 ± 40 | 185 ± 54 | 297 ± 94 | 62 ± 2 | 91 ± 16 | 561 ± 34 |
| SeGalNAc | 7 ± 3 | 3 ± 1 | 10 ± 4 | 30 ± 10 | 4 ± 1 | 26 ± 3 | |
It is well known that total selenium concentration is not representative of the real functional activity of selenoproteins, because the element is incorporated into a large variety of proteins and metabolites, with different biological functions and activities.1 Liver and kidney are the foremost organs responsible for selenium metabolism, since most Se-proteins of the body are synthesized in the liver and their excretion as Se-metabolites is regulated by kidney. Interestingly, the levels of HMWSe in liver are much higher than those found in kidney for all the three groups under study, while SeGalNAc levels are very similar in liver and kidney within each group. However, the obtained ratio between SeGalNAc levels for the maternal feeding group versus supplement group reached 2.6 times for liver, and 2.3 for kidney. These ratios are higher than those observed for total selenium comparisons (see below), indicating that the selenosugar variations between groups could be a better potential biomarker than total selenium.
Selenosugars have been recognized as predominant excretory metabolites of selenium. In both liver and kidneys analyzed in the supplemented group, it should be noted that the HMWSe region contains mainly exogenous selenium (77Se), while the LMWSe region (where the SeGalNAc is present) contains almost entirely endogenous selenium (natSe). Thereby, most exogenous selenium seems to be incorporated into the synthesis of selenoproteins as GHSPx. As expected, these results suggest that turnover rates are different for HMWSe and LMWSe (SeGalNAc), indicating that the synthesis of Se proteins is preferential at the supplementation levels used in our experiments (virtually only natural Se is present in the form of Se metabolite, coming from protein catabolism).
Identification of SeGalNAc in liver and kidney by HPLC-APCI-MS/MS
As stated above, SeGalNAc could be a better selenium nutritional biomarker rather than the total selenium measurement, hence the identification of this metabolite must be further confirmed. The nature of the minor peak eluting at LMWSe (see Fig. 1) was investigated by HPLC-APCI-MS/MS in the rat liver and kidney preconcentrated cytosolic fractions. The use of APCI source has been previously described for SeGalNAc identification in human urine and porcine liver, since it provides improved sensitivity and reduced matrix effects.16,21 The SeGalNAc detection was performed using the selected reaction monitoring (SRM) mode which is based on the observed collision-induced dissociation (CID) of this Se species.21The best SRM transitions and their corresponding collision energy were optimized using the SeGalNAc standard (50 μL injected at a concentration of 50 μg Se L−1). For the molecular ion [SeGalNAc + H+] (m/z = 300) these values were: m/z 300 → 204 (10 eV); m/z 300 → 186 (10 eV); m/z 300 → 144 (20 eV); m/z 300 → 138 (20 eV). Fig. 2 shows the four SRM transitions monitored for the SeGalNAc standard (tr = 6.2 min), by reverse phase HPLC-APCI-MS/MS. The chromatogram peak areas for each of the transitions allowed for determining the ratios of the four SRM transitions for SeGalNac (i.e., 138/144, 138/204, 144/204, 186/144, and 186/204). Comparing the ratios for the SeGalNAc standard with those ratios obtained for liver and kidney cytosolic preconcentrated fractions is used to identify the presence of this selenosugar in real samples.16
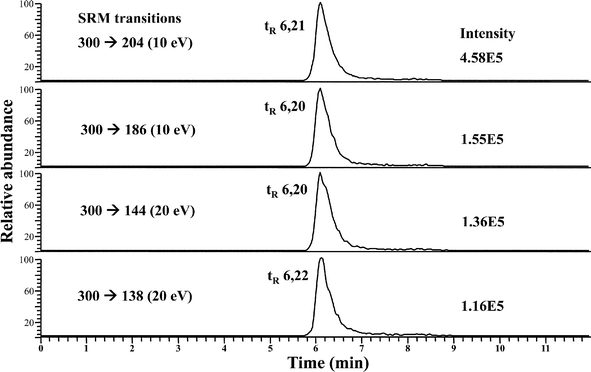 | ||
| Fig. 2 RP HPLC-APCI-MS/MS chromatograms from a SeGalNAc standard (after 50 μL injection at a concentration of 50 μg Se L−1), in which four SRM transitions were monitored. | ||
To minimize matrix effects, we carried out a preconcentration of the selenosugars from the cytosolic fractions of liver and kidney as described in the Experimental procedures. Liver and kidney fractions from maternal feeding and supplemented rats were analyzed. Fifty microliters of each fraction was injected into the HPLC-APCI-MS/MS system and SRM transitions were monitored. It should be noted that in maternal feeding rats, the main molecular ion resulting from [SeGalNAc + H+] corresponds to m/z 300, while the main ion for the supplemented group corresponds to m/z 297, since liver and kidneys are enriched in the 77Se stable isotope. Therefore, SRM transitions from [77SeGalNAc + H+] (m/z 277) → 204, 186, 144, and 138, were also monitored and their intensities used to calculate SRM ratios. It should be mentioned at this point that the product ions produced upon CID from either m/z 300 (80SeGalNAc) or m/z 297 (77SeGalNAc) have the same m/z values (i.e. 204, 186, 144 and 138). This is because in both cases their initial CID step is the loss of CH3SeH.21
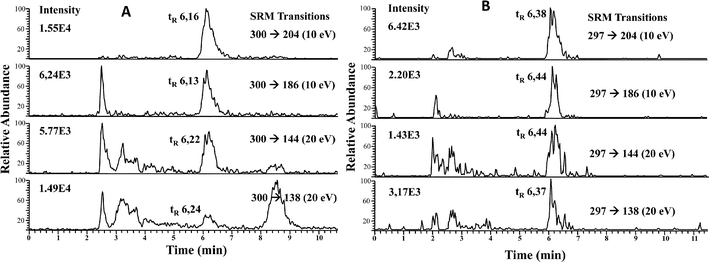 | ||
| Fig. 3 RP HPLC-APCI-MS/MS chromatograms of LMWSe preconcentrated liver cytosolic fraction from maternal feeding (A) and supplemented rats (B). | ||
| Liver | SRM ratio (for m/z 300 precursor ion) | |||||
|---|---|---|---|---|---|---|
| Samples analysed | m/z 138/144 | m/z 138/204 | m/z 138/186 | m/z 144/204 | m/z 186/144 | m/z 186/204 |
| SeGalNAc standard (n = 10) | 0.86 ± 0.01 | 0.26 ± 0.01 | 0.80 ± 0.02 | 0.30 ± 0.01 | 1.08 ± 0.01 | 0.32 ± 0.02 |
| Liver fraction (maternal) (n = 3) | 0.83 ± 0.08 | 0.25 ± 0.03 | 0.80 ± 0.12 | 0.29 ± 0.01 | 1.11 ± 0.01 | 0.33 ± 0.01 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| SRM ratio (for m/z 297 precursor ion) | ||||||
|---|---|---|---|---|---|---|
| Samples analysed | m/z 138/144 | m/z 138/204 | m/z 138/186 | m/z 144/204 | m/z 186/144 | m/z 186/204 |
| SeGalNAc standard (n = 10) | 0.86 ± 0.04 | 0.26 ± 0.01 | 0.79 ± 0.08 | 0.29 ± 0.01 | 1.10 ± 0.05 | 0.32 ± 0.02 |
| Liver fraction (supplemented) (n = 3) | 1.06 ± 0.25 | 0.27 ± 0.04 | 0.83 ± 0.02 | 0.30 ± 0.03 | 1.06 ± 0.06 | 0.29 ± 0.02 |
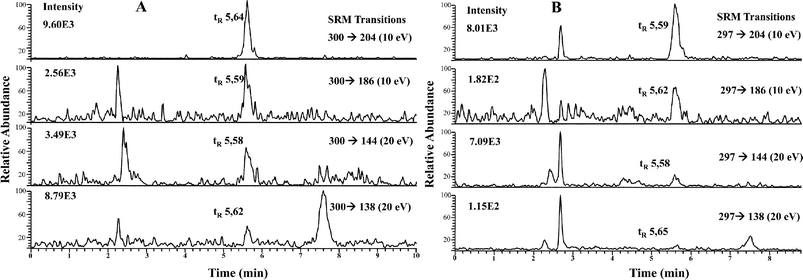 | ||
| Fig. 4 RP HPLC-APCI-MS/MS chromatograms of LMWSe preconcentrated kidney cytosolic fraction from maternal feeding (A) and supplemented rats (B). | ||
| Kidney | SRM ratio (for m/z 300 precursor ion) | |||||
|---|---|---|---|---|---|---|
| Samples analyzed | m/z 138/144 | m/z 138/204 | m/z 138/186 | m/z 144/204 | m/z 186/144 | m/z 186/204 |
| SeGalNAc standard (n = 10) | 0.85 ± 0.02 | 0.25 ± 0.02 | 0.80 ± 0.03 | 0.29 ± 0.02 | 1.06 ± 0.02 | 0.30 ± 0.02 |
| Kidney fraction (maternal) (n = 3) | 0.95 ± 0.09 | 0.28 ± 0.03 | 0.85 ± 0.04 | 0.29 ± 0.03 | 1.07 ± 0.08 | 0.33 ± 0.01 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| SRM ratio (for m/z 297 precursor ion) | ||||||
|---|---|---|---|---|---|---|
| Samples analyzed | m/z 138/144 | m/z 138/204 | m/z 138/186 | m/z 144/204 | m/z 186/144 | m/z 186/204 |
| SeGalNAc standard (n = 10) | 0.79 ± 0.04 | 0.23 ± 0.01 | 0.76 ± 0.03 | 0.29 ± 0.01 | 1.05 ± 0.01 | 0.31 ± 0.01 |
| Kidney fraction (supplemented) (n = 3) | 0.78 ± 0.02 | 0.22 ± 0.02 | 0.89 ± 0.05 | 0.28 ± 0.01 | 0.88 ± 0.08 | 0.25 ± 0.04 |
Conclusions
The use of enriched stable isotopes in connection with ICP-MS detection, HPLC separation and mathematical calculations based on IPD, provides unique quantitative information about the tracer, the tracee, and their existing selenospecies in biological tissues (i.e., liver and kidneys). The quantitative metabolic data obtained may be used to further the understanding of compartmental modeling of essential and toxic trace elements and to develop new kinetic studies related to their species metabolism and nutritional value.Unfortunately, ICP-MS information is elemental and therefore unsuitable for the conclusive identification of selenium-containing proteins or metabolites. Thus, molecular mass spectrometry techniques (e.g., HPLC-APCI-MS/MS, in the SRM mode) have to be used for the eventual selenium-chemical species identification in complex biological samples. That is, “integrated” chemical speciation seems mandatory these days to investigate nutritional value of formulas and supplements of Se (e.g., those used for baby nutrition).
Regarding total Se levels in liver and kidneys, our results show that under low selenium dietary conditions, its amount is markedly reduced in the liver while this reduction in kidney is lower, as shown in supplemented and non-supplemented groups of this study. Thus, selenium supplementation (as selenite) increases selenium levels in the body of lactating rats. However, the observed values were well below the levels found in breast fed rats (indicating a higher bioavailability of selenium coming from the selenospecies present in maternal milk). In that vein, Rodriguez de la Flor et al.33 investigated the selenium speciation in human breast milk, and they compared the results obtained with those for infant formulae. Total selenium levels in formula milk are similar to or higher than those found in human milk, although the physicochemical form in which the element occurs is different. Indeed, as we previously stated, in breast milk selenium was mainly distributed in the high-medium molecular weight region while in the formula milk selenium is mainly distributed in the low molecular weight region.
That is, total selenium concentration alone is not representative of the metabolic and nutritional activity of selenium. It is well known that this element is incorporated in multiple proteins through different metabolic pathways.1 The synthesis of most of the selenoproteins and metabolites occurs in liver preferentially and then in the kidneys.1,2 The integrated Se speciation in such tissues here demonstrates that high molecular weight selenium-chemical species levels are much higher in liver than those found in kidney. Interestingly, the selenosugar SeGalNAc levels (low molecular weight species) are quite similar in liver and kidney, within each rat group. These findings indicate that the turnover rates are different for the different species and their synthesis might occur in different tissue compartments.
Finally, the low concentration of this selenium metabolite in liver and kidney makes it very difficult to carry out its identification by classical molecular mass spectrometry. However, sample pretreatment followed by the use of HPLC-APCI-MS/MS in the SRM mode allowed the identification of SeGalNAc in rat liver and, for the first time to our knowledge, in the kidney cytosol of maternal feeding and supplemented rats.
Acknowledgements
Authors are grateful for the financial support from the “Fundación para el Fomento en Asturias de la Investigación Científica Aplicada y la Tecnología” (FICYT) and “Fondo Europeo de Desarrollo Regional” (FEDER) through the project referenced FC-11-PC10-46, and the “Plan Nacional de I+D+I” (Spanish Ministry of Science and Innovation or MICINN, and FEDER Program) through MICINN CTQ2010-16636. “Laboratorios Ordesa” (Barcelona, Spain) and “Fundación Grupo Castrillo” (Spain) are gratefully acknowledged as well. S.A.P. wishes to acknowledge co-funding of this research by the European Union-European Regional Development Fund and Greek Ministry of Education/EYDE-ETAK through program ESPA 2007–2013/EPAN II/Action “SYNERGASIA” (09ΣYN-13-832).References
- M. Roman, P. Jitaru and C. Barbante, Metallomics, 2014, 6, 25–54 RSC.
- L. V. Papp, J. Lu, A. Holmgren and K. K. Khanna, Antioxid. Redox Signaling, 2007, 9(7), 775–806 CrossRef CAS PubMed.
- G. V. Kryuko, S. Castellano, S. V. Novoselov, A. V. Lobanov, O. Zehtab, R. Guigó and V. N. Gladyshev, Science, 2003, 300(5624), 1439–1443 CrossRef PubMed.
- K. M. Barnes, J. K. Evenson, A. M. Raines and R. A. Sunde, J. Nutr., 2009, 139, 199–206 CrossRef CAS PubMed.
- B. Moghadaszadeh and A. H. Beggs, Physiology, 2006, 21, 307–315 CrossRef CAS PubMed.
- R. R. de la Flor St. Remy, M. L. Fernández-Sánchez, J. López Sastre and A. Sanz-Medel, J. Anal. At. Spectrom., 2004, 19, 616–622 RSC.
- A. Sanz-Medel, Spectrochim. Acta, Part B, 1998, 53, 197–211 CrossRef.
- Z. Pedrero and Y. Madrid, Anal. Chim. Acta, 2009, 634, 135–152 CrossRef CAS PubMed.
- M. L. Janghorbani, L. J. Kasper and V. R. Young, Am. J. Clin. Nutr., 1984, 40, 208–218 CAS.
- B. H. Patterson, O. A. Levander, K. Helzlsouer, P. A. McAdam, S. A. Lewis, P. R. Taylor, C. Veillon and L. A. Zech, Am. J. Physiol., 1989, 257, R556–R567 CAS.
- C. A. Swanson, B. H. Patterson, O. A. Levander, C. Veillon, P. R. Taylor, K. Helzlsouer, P. A. McAdam and L. Zech, Am. J. Clin. Nutr., 1991, 54, 917–926 CAS.
- K. T. Suzuki and Y. Ogra, Food Addit. Contam., 2002, 19, 974–983 CrossRef CAS PubMed.
- B. Gammelgaard and L. Bendhal, J. Anal. At. Spectrom., 2004, 19, 135–142 RSC.
- Y. Ogra, K. Ishiwata, H. Takayama, N. Aimi and K. T. Suzuki, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2002, 767, 301–312 CrossRef CAS.
- V. Díaz Huerta, J. Szpunar, R. Lobinski, M. L. Fernández Sánchez and A. Sanz-Medel, J. Anal. At. Spectrom., 2003, 18, 1471–1476 RSC.
- Y. Lu and S. A. Pergantis, Metallomics, 2009, 1(4), 346–352 RSC.
- Y. Kobayashi, Y. Ogra, K. Ishiwata, H. Takayama, N. Aimi and K. T. Suzuki, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(25), 15932–15936 CrossRef CAS PubMed.
- H. González-Iglesias, M. L. Fernández Sánchez, J. I. García Alonso and A. Sanz-Medel, Anal. Bioanal. Chem., 2007, 387, 707–713 CrossRef PubMed.
- H. González-Iglesias, M. L. Fernández Sánchez, J. A. Rodríguez-Castrillón, J. I. García Alonso and A. Sanz-Medel, J. Anal. At. Spectrom., 2009, 24, 460–468 RSC.
- H. González-Iglesias, M. L. Fernandez-Sanchez and A. Sanz-Medel, in Analytical Techniques for Clinical Chemistry: Methods and Application, ed. S. Caroli and G. Záray, Wiley, 2012, ch. 22, pp. 627–649 Search PubMed.
- S. Letsiou, V. Nischwitz, P. Traar, K. A. Francesconi and S. A. Pergantis, Rapid Commun. Mass Spectrom., 2007, 21(3), 343–351 CrossRef CAS PubMed.
- K. T. Suzuki, C. Doi and N. Suzuki, Toxicol. Appl. Pharmacol., 2006, 217, 185–195 CrossRef CAS PubMed.
- K. T. Suzuki, L. Somekawa, K. Kurasaki and N. Suzuki, Toxicol. Appl. Pharmacol., 2006, 217, 43–50 CrossRef CAS PubMed.
- S. Kokarnig, A. Tsirigotaki, T. Wiesenhofer, V. Lackner, K. A. Francesconi, S. A. Pergantis and D. Kuehneltet, J. Trace Elem. Med. Biol., 2014 DOI:10.1016/j.jtemb.2014.06.012 , pii: S0946-672X(14)00117-5. [Epub ahead of print].
- P. Traar, F. Belaj and K. A. Francesconi, Aust. J. Chem., 2004, 57, 1051–1053 CrossRef CAS.
- H. González-Iglesias, M. L. Fernández-Sánchez, J. López-Sastre and A. Sanz-Medel, Electrophoresis, 2012, 33(15), 2407–2415 CrossRef PubMed.
- K. T. Suzuki, L. Somekawa, K. Kurasaki and N. Suzuki, J. Health Sci., 2006, 52(5), 590–597 CrossRef CAS.
- A. Sanz-Medel, M. L. Fernández-Sánchez, H. González-Iglesias and J. B. López-Sastre, Pure Appl. Chem., 2010, 82(2), 447–460 CrossRef CAS.
- Y. Thomassen and J. Aaseth, Occurrence and distribution of selenium, CRC Press, Boca Raton, FL, 1989, ch. 8, pp. 169–212 Search PubMed.
- A. M. Michelson, J. Environ. Pathol., Toxicol. Oncol., 1998, 17(3–4), 233–239 CAS.
- M. Maiorino, J. B. Wissing, R. Brigelius-Flohe, F. Calabrese, A. Roveri, P. Steinert, F. Ursini and L. Flohe, FASEB J., 1998, 12, 1359–1370 CAS.
- Y. Kobayashi, Y. Ogra and K. T. Suzuki, J. Chromatogr. B: Biomed. Sci. Appl., 2001, 760, 73–81 CrossRef CAS.
- R. R. de la Flor St. Remy, M. L. Fernández Sánchez, J. B. López Sastre and A. Sanz-Medel, J. Anal. At. Spectrom., 2004, 19, 1104–1110 RSC.
| This journal is © The Royal Society of Chemistry 2015 |