Open Access Article
Open Access ArticleThermal stability, antioxidant, and anti-inflammatory activity of curcumin and its degradation product 4-vinyl guaiacol
Tuba
Esatbeyoglu
*a,
Katrin
Ulbrich
b,
Clemens
Rehberg
b,
Sascha
Rohn
b and
Gerald
Rimbach
a
aInstitute of Human Nutrition and Food Science, University of Kiel, Germany. E-mail: esatbeyoglu@foodsci.uni-kiel.de; Fax: +49 431 880 2628; Tel: +49 431 880 5333
bInstitute of Food Chemistry, Hamburg School of Food Science, University of Hamburg, Germany
First published on 14th January 2015
Abstract
Curcumin is a secondary plant metabolite present in Curcuma longa L. Since curcumin is widely used as a food colorant in thermally processed food it may undergo substantial chemical changes which in turn could affect its biological activity. In the current study, curcumin was roasted at 180 °C up to 70 minutes and its kinetic of degradation was analyzed by means of HPLC-PDA and LC-MS, respectively. Roasting of curcumin resulted in the formation of the degradation products vanillin, ferulic acid, and 4-vinyl guaiacol. In cultured hepatocytes roasted curcumin as well as 4-vinyl guaiacol enhanced the transactivation of the redox-regulated transcription factor Nrf2, known to be centrally involved in cellular stress response and antioxidant defense mechanisms. The antioxidant enzyme paraoxonase 1 was induced by roasted curcumin and 4-vinyl guaiacol. Furthermore, roasted curcumin and 4-vinyl guaiacol decreased interleukin-6 gene expression in lipopolysaccharide stimulated murine macrophages. Current data suggest that curcumin undergoes degradation due to roasting and its degradation product exhibit significant biological activity in cultured cells.
Introduction
The curcuminoid curcumin (1,7-bis-(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione or diferuloyl-methane) occurs in the rhizomes of turmeric (Curcuma longa) and gives turmeric its characteristic yellow color.1 Chemically, curcumin is an unsaturated diketone of two ferulic acid units which are connected through a methylene group.2 Recently, potential health benefits of curcumin including anti-oxidative, anti-inflammatory, anti-carcinogenic, and even neuroprotective properties have been described.2–6 Previously, it has been shown that curcumin exhibits cell signaling activity in cultured cells as well as in vivo.7 In fact, curcumin induces the transcription factor Nrf22 and the anti-atherogenic enzyme paraoxonase 1 (PON1).8Nrf2 is a redox-regulated transcription factor which plays a central role in antioxidant defense and stress response mechanisms. Under basal conditions, Nrf2 is bound to its inhibitor Keap1 (Kelch-like ECH associated protein 1) in the cytoplasm. The modification of cysteine residues of Keap1 by electrophiles may disrupt the binding between Nrf2 and Keap1. Nrf2 then migrates into the nucleus where it interacts with small Maf proteins and binds as a heterodimer to the so-called antioxidant-response element (ARE), thereby driving the expression of genes encoding phase II and antioxidant proteins.9,10
PON1 is mainly expressed in the liver and circulates in the blood bound to high-density lipoprotein (HDL) and prevents or delays the oxidation of low-density lipoprotein (LDL).2,8,11,12
Dietary plant bioactives such as curcumin may mediate anti-inflammatory activity by down-regulating the gene expression of pro-inflammatory cytokines including interleukin-6 (IL-6).
Although curcumin per se has been identified as a potent inducer of Nrf2 and PON1 and a potent suppressor of inflammation,2,13,14 little is known regarding the biological activity of curcumin degradation products. Given that curcumin is widely used as a food ingredient in thermally processed food, it may undergo substantial chemical changes.15,16 However, the biological activity of curcumin degradation products due to roasting is largely unknown. In many recipes, applied worldwide, curcuma and curry powders are thermally processed due to cooking, baking and roasting. In fact, curcuma and curry powder may be roasted without further ingredients in a pan or pot prior the addition of liquid ingredients such as water or fat.
Therefore, in the present study curcumin was roasted under defined experimental conditions and degradation was monitored by means of HPLC-PDA and LC-MS, respectively. The biological activity of a curcumin degradation product, regarding the induction of Nrf2 and PON1 and the down-regulation of IL-6, was then studied in cultured cells.
Materials and methods
Chemicals
Curcumin was purchased from Fisher Scientific (Schwerte, Germany). Ferulic acid and 4-vinyl guaiacol were obtained from Sigma-Aldrich (Taufkirchen, Germany). The purity was checked by HPLC analysis, and was >97% for each compound. HPLC solvents were purchased from Roth (Karlsruhe, Germany) and were of HPLC grade quality.High glucose (4.5 g L−1) Dulbecco's modified Eagle's medium with sodium pyruvate and L-glutamine, high glucose (4.5 g L−1) Dulbecco's modified Eagle's medium without L-glutamine, Dulbecco's PBS (DPBS) without Ca and Mg, fetal bovine serum, L-glutamine 200 mM (100×), RPMI 1640 with 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM Hepes, 4.5 g L−1 glucose and 1.5 g L−1 NaHCO3; penicillin/streptomycin (100×; 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1 penicillin; 10 mg mL−1 streptomycin) and G-418 sulfate (50 mg mL−1) were obtained from PAA (Coelbe, Germany) and PAN-Biotech (Aidenbach, Germany). Fetal bovine serum was purchased from Gibco® Life Technologies (Carlsbad, California, USA). Dimethyl sulfoxide (DMSO; ROTIDRY® ≥99.5%) and Neutral Red were obtained from Carl Roth (Karlsruhe, Germany) and curcumin (CAS no. 458-37-7) from Alexis Biochemicals (Lausen, Switzerland) for the cell culture experiments. The dual-luciferase® reporter assay system, luciferase assay system, phRL-TK and passive lysis buffer were purchased from Promega (Mannheim, Germany). JetPEI and peqGOLD TriFast™ were purchased from Peqlab Biotechnologie (Erlangen, Germany) and Pierce™ BCA Protein Assay Kit from Thermo Scientific (Rockford, IL, USA). pARE_GIGPx_Luc was kindly provided by A. Banning and R. Brigelius-Flohé (DIfE, Potsdam, Germany). LPS from Salmonella enterica serotype enteritidis (Sigma) was dissolved in phosphate buffered saline (PBS; stock solution 1 mg mL−1) and stored at −20 °C. SensiFAST™ SYBR® No-ROX One-Step Kit was obtained from Bioline (Luckenwalde, Germany). Primers for interleukin-6 (IL-6) and elongation factor 2 (EF-2) were obtained from Eurofins Genomics (Ebersberg, Germany).
000 U mL−1 penicillin; 10 mg mL−1 streptomycin) and G-418 sulfate (50 mg mL−1) were obtained from PAA (Coelbe, Germany) and PAN-Biotech (Aidenbach, Germany). Fetal bovine serum was purchased from Gibco® Life Technologies (Carlsbad, California, USA). Dimethyl sulfoxide (DMSO; ROTIDRY® ≥99.5%) and Neutral Red were obtained from Carl Roth (Karlsruhe, Germany) and curcumin (CAS no. 458-37-7) from Alexis Biochemicals (Lausen, Switzerland) for the cell culture experiments. The dual-luciferase® reporter assay system, luciferase assay system, phRL-TK and passive lysis buffer were purchased from Promega (Mannheim, Germany). JetPEI and peqGOLD TriFast™ were purchased from Peqlab Biotechnologie (Erlangen, Germany) and Pierce™ BCA Protein Assay Kit from Thermo Scientific (Rockford, IL, USA). pARE_GIGPx_Luc was kindly provided by A. Banning and R. Brigelius-Flohé (DIfE, Potsdam, Germany). LPS from Salmonella enterica serotype enteritidis (Sigma) was dissolved in phosphate buffered saline (PBS; stock solution 1 mg mL−1) and stored at −20 °C. SensiFAST™ SYBR® No-ROX One-Step Kit was obtained from Bioline (Luckenwalde, Germany). Primers for interleukin-6 (IL-6) and elongation factor 2 (EF-2) were obtained from Eurofins Genomics (Ebersberg, Germany).
Roasting of curcumin and ferulic acid
For the roasting experiments 1 mL of a 5 mM methanolic curcumin or ferulic acid solution was dried under nitrogen. The roasting was carried out in glass ampoules at 180 °C in a behrotest ET1 thermoblock (Behr Labortechnik, Germany). Roasting was conducted for 70 minutes and samples taken every 10 minutes by cooling down the corresponding ampoule in an ice bath. After the roasting process samples were re-dissolved in aqueous methanol (70%) for HPLC analysis and re-dissolved in DMSO for cell culture experiments.Analysis of the curcumin and its degradation products using HPLC-PDA and LC-PDA/ESI-MSn
The Knauer HPLC-System consisted of a quaternary pump (S1000), autosampler (S3950) and a photodiode array detector (S2600). A gradient elution using acetonitrile (B) and 0.1% formic acid (A) was carried out on a 150 × 3.0 mm i.d., 5 μm, Luna C18 column (Phenomenex Ltd, Aschaffenburg, Germany) connected to a 4.0 × 3.0 mm i.d. guard column of the same material. Gradient elution was performed as follows: 20% B (1 min); 20–50% B in 59 min; 50–90% B in 5 min; 90% B (5 min); 90–20% B in 2 min; 20% B (8 min); all at a flow rate of 0.7 mL min−1 and a column temperature of 25 °C. The detection was performed simultaneously at 325 nm, 350 nm and 280 nm. Spectra were recorded from 200 to 500 nm. All compounds were identified by their retention times and UV-spectra in comparison to reference substances. Calibration curves were used for quantification.To additionally prove the identity of the phenolic structures due to curcumin roasting, LC-PDA/ESI-MSn was carried out on a system consisting of an UHPLC Ultimate 3000 RS with Chromeleon 6.8 software (Dionex GmbH, Idstein, Germany), and an ESI-MSn amaZon ETD with software trapControl 7.0, HyStar 3.2 and DataAnalysis 4.0 (all Bruker Daltonik, Bremen, Germany). The HPLC consisted of a binary pump, an autosampler, a column compartment, and a photodiode array detector. The LC method was similar to the LC-PDA method as described above, with the exception of a reduced flow rate of 0.5 mL min−1. ESI-MS/MS experiments were recorded in negative ion mode and a scanning range between 50–1500 m/z. The capillary voltage was set to 4.5 kV and the capillary temperature was at 350 °C. N2 was used as dry gas and helium as collision gas. A dry gas flow of 10 L min−1 and a pressure of 55 psi for the nebulizer were set. The automatic MS/MS mode was chosen for the experiments.
Cell lines
Cells of the human liver hepatoma derived cell line Huh7 (Institute of Applied Cell Culture, Munich, Germany) were cultured in high glucose (4.5 g L−1) Dulbecco's modified Eagle's medium (with sodium pyruvate, with L-glutamine) supplemented with 10% (v/v) fetal bovine serum, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin in a humidified atmosphere of 5% CO2 at 37 °C.Huh7 liver hepatoma cells of human origin stably transfected with a 1009 bp [−1013, −4] fragment of the human PON1 promoter (kindly provided by X. Coumoul, INSERM, France) were cultivated high glucose (4.5 g L−1) Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum, 2 mmol L−1L-glutamine, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin and 100 μg mL−1 G418 sulfate at 37 °C (5% CO2).
RAW 264.7 murine macrophages (Institute of Applied Cell Culture, Munich, Germany) were cultured in high glucose (4.5 g L−1) RPMI 1640 medium (with 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM Hepes and 1.5 g L−1 NaHCO3) supplemented with 10% (v/v) fetal bovine serum, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin in a humidified atmosphere of 5% CO2 at 37 °C.
Curcumin, which was used as positive control, pure 4-vinyl guaiacol, and samples of roasted curcumin were dissolved in DMSO. Stock solutions (100 mmol L−1) were stored at −80 °C until further use.
Neutral red cell viability assay
The Neutral Red Assay17,18 was used to determine cell viability. Confluent cells were harvested and seeded at 1.5 × 105 cells for PON1-Huh7 as well as Huh7 and 0.8 × 105 cells for RAW264.7 per well in 24-well plates. PON1-Huh7, Huh7 and RAW264.7 cells were treated with 1–100 μmol L−1 roasted curcumin at 180 °C and 4-vinyl guaiacol for 48 (PON1-Huh7) or 24 h (Huh7 and RAW264.7). The culture medium containing the test substances was replaced with fresh serum-containing medium including 50 μg mL−1 of Neutral Red. After incubation for 2 h, the medium was removed. Cells were extracted with ethanol–water–glacial acetic acid (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 49
49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v). After 15 min incubation at room temperature on a shaking platform the absorbance was measured in a plate reader (Labsystems iEMS Reader, Helsinki, Finland) at 540 nm. Non cytotoxic concentrations of the test compounds were used for further analyses. Two independent experiments were performed at least in triplicate.
1, v/v/v). After 15 min incubation at room temperature on a shaking platform the absorbance was measured in a plate reader (Labsystems iEMS Reader, Helsinki, Finland) at 540 nm. Non cytotoxic concentrations of the test compounds were used for further analyses. Two independent experiments were performed at least in triplicate.
Reporter gene assay for measuring Nrf2 transactivation
Huh7 cells (0.15 × 106 cells per well) were incubated in 24-well plates for 24 h. Huh7 cells were transiently transfected with an expression vector containing a 25 bp oligonucleotide derived from the promoter region of the human gastrointestinal glutathione peroxidase 2 (GIGPx)19 comprising the conserved ARE-motif and the reporter gene firefly luciferase (pARE_GIGPx). The Renilla reniformis luciferase gene phRL-TK was applied for normalization. Transfection was performed using jetPEI transfection reagent according to manufacturer's protocol. After 24 h transfection, Huh7 cells were incubated with 1, 5, 10 and 15 μmol L−1 roasted curcumin or 1, 5, 10 and 25 μmol L−1 4-vinyl guaiacol as well as 20 μmol L−1 curcumin as positive control for further 24 h. Subsequently, cells were lysed with passive lysis buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) and firefly and Renilla luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega, Mannheim, Germany) according to manufacturer's description (Infinite 200 microplate reader, Tecan Group Ltd, Crailsheim, Germany). Two independent experiments were performed at least in triplicate.
5, v/v) and firefly and Renilla luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega, Mannheim, Germany) according to manufacturer's description (Infinite 200 microplate reader, Tecan Group Ltd, Crailsheim, Germany). Two independent experiments were performed at least in triplicate.
Reporter gene assay for measuring PON1 transactivation
PON1-Huh7 cells were seeded at a density of 0.15 × 106 cells per well in 24-well plates and incubated for 24 h. Cells were treated with 1, 5, 10 and 25 μmol L−1 roasted curcumin at 180 °C or 1, 5, 10, 25 and 50 μmol L−1 4-vinyl guaiacol as well as 20 μmol L−1 curcumin for 48 h. Subsequently, cells were washed with PBS, lysed with cell culture lysis reagent (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) and subjected to luciferase activity measurement (Luciferase Assay System) by luminescence reading (Infinite 200 microplate reader, Tecan, Crailsheim, Germany). Results were normalized to total cell protein which was determined by the BCA protein assay according to manufacturer's description. Two independent experiments were performed at least in triplicate.
5, v/v) and subjected to luciferase activity measurement (Luciferase Assay System) by luminescence reading (Infinite 200 microplate reader, Tecan, Crailsheim, Germany). Results were normalized to total cell protein which was determined by the BCA protein assay according to manufacturer's description. Two independent experiments were performed at least in triplicate.
RNA isolation and real time PCR for measuring anti-inflammatory activity
RAW264.7 macrophages were seeded at a density of 0.5 × 106 cells per well in 6-well plates for 24 h. Cells were treated with 15 and 20 μmol L−1 roasted curcumin at 180 °C or 25, 50 and 100 μmol L−1 4-vinyl guaiacol for 24 h. LPS (10 ng mL−1) was added for 6 h, respectively. RNA was isolated using peqGOLD TriFast™ according to manufacturer's instructions.Primers for murine genes IL-6 and EF-2 were designed by Primer3 software. IL-6: F: 5′-TAGTCCTTCCTACCCCAATTTCC and R: 5′-TTGGTCCTTAGCCACTCCTTC; EF-2: F: 5′-GCGTGCCAAGAAAGTAGAGG and R: 5′-AAGATGGGGCCAGGATGAG.
Real time PCR was performed with a SensiFAST™ SYBR® No-ROX One-Step Kit (Bioline, Luckenwalde, Germany). Gene expression was normalized to the housekeeping gene EF-2.
Statistical analyses
Statistical calculations were conducted using IBM SPSS software Version 22. Data were analyzed for normality of distribution by Shapiro-Wilk. Normally distributed data were compared by Student's t-test and not normally distributed data by Mann–Whitney U-test. Data are expressed as means ± SD. Significance was accepted at p < 0.05.Results
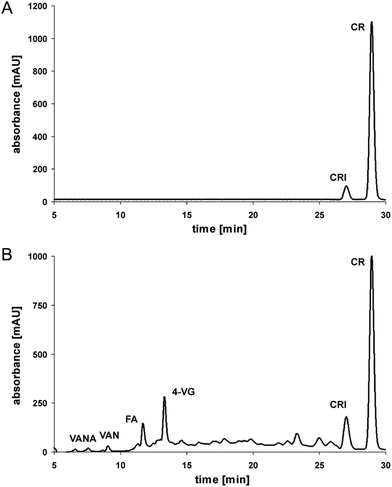
Degradation of curcumin during roasting at 180 °C
Under model roasting conditions, curcumin is strongly degraded. In the present experiment only 30% of the initial curcumin concentration was left after 5 minutes. Whereas ferulic acid did not accumulate significantly during curcumin roasting, its further degradation to 4-vinyl guaiacol is detectable as a dominant product, although formed in relatively low concentrations (Fig. 1).This was also evident when roasting pure ferulic acid: 4-vinyl guaiacol is formed and further degraded after an accumulation in low concentrations of 20% from the initial ferulic acid concentration with a maximum at 50 minutes of roasting. It has been reported, that the degradation of curcumin leads finally to volatile phenolic compounds such as vanillin, guaiacol, and isoeugenol.16 However, in the present experiment only trace levels of vanillin have been detected.
Taking all the degradation data into account, then step 1 (curcumin to ferulic acid) is comparatively faster than step 2 (ferulic acid to 4-vinyl guaiacol) (Fig. 2), as only small amounts of intact curcumin are still present, with only small amounts of ferulic acid, but comparatively higher amounts of 4-vinyl guaiacol. Step 3 (4-vinyl guaiacol to volatile compounds) is slower than steps 1 and 2. Only traces of vanillin have been identified and we detected an accumulation of 4-vinyl guaiacol (Fig. 1).
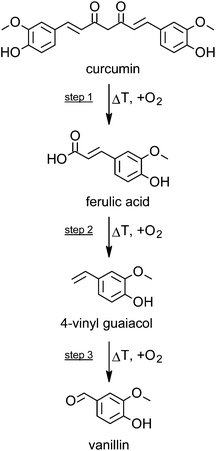 | ||
| Fig. 2 Scheme of the degradation of curcumin during roasting at 180 °C and corresponding formation of selected breakdown products. | ||
It has to be considered that all the volatile compounds formed having different boiling points and therefore affecting the steady states of these reactions. Further, they have different odor threshold levels and therefore contributing very specifically to the flavor. Only a few minutes of thermal treatment, which is in accordance with homelike heat treatment of curcuma, significantly affected the concentrations of curcumin, ferulic acid, 4-vinyl guaiacol, and the other volatile compounds. With regard to the bioactivity of curcumin this might be of importance.
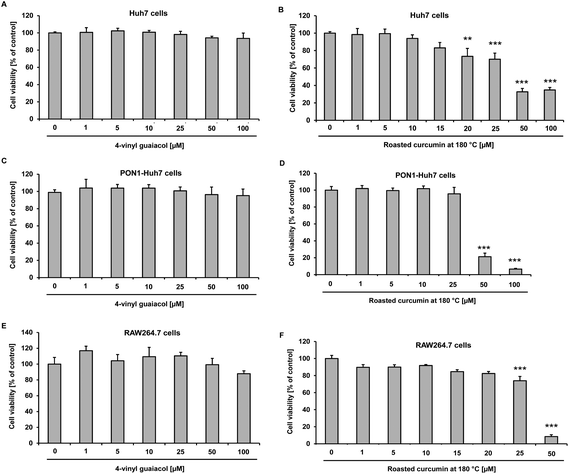
Effects of 4-vinyl guaiacol and roasted curcumin on cell viability in Huh7, PON1-Huh7 and RAW264.7 cells
In order to determine the cytotoxicity of roasted curcumin and 4-vinyl guaiacol, Huh7, PON1-Huh7 and RAW264.7 cells were incubated with increasing concentrations of the test compounds for 24 h (Huh7 and RAW264.7) and 48 h (PON1-Huh7), respectively. Cell viability was determined spectrophotometrically by the Neutral Red assay at 540 nm. Under the conditions investigated, in all three cell lines 4-vinyl guaiacol did not exhibit any cytotoxicity up to 100 μmol L−1 (Fig. 3A, C and E). However, the incubation of Huh7 and PON1-Huh7 with increasing concentrations of roasted curcumin led to dose-dependent cytotoxicity (Fig. 3B and D). Roasted curcumin did not exhibit any cytotoxicity up to a concentration of 20 μmol L−1 in RAW264.7 cells (Fig. 3F). In Huh7 and PON1-Huh7 cells curcumin per se was not cytotoxic at concentrations of up to 20 μmol L−2,8 and in RAW264.7 cells up to 10 μmol L−1.13Nrf2 transactivation of 4-vinyl guaiacol and roasted curcumin
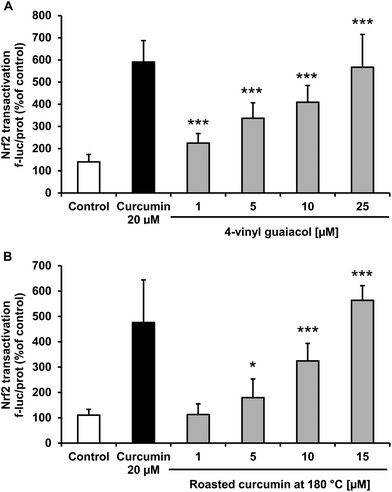
The transcription factor Nrf2 plays a central role in antioxidant and phase II defense mechanisms. Nrf2 activation may be associated with decreased cellular oxidative stress. Treatment of Huh7 hepatocytes with 4-vinyl guaiacol and roasted curcumin significantly enhanced, in a dose-dependent manner, Nrf2 transactivation as monitored by a reporter gene assay (Fig. 4).PON1 transactivation of 4-vinyl guaiacol and roasted curcumin
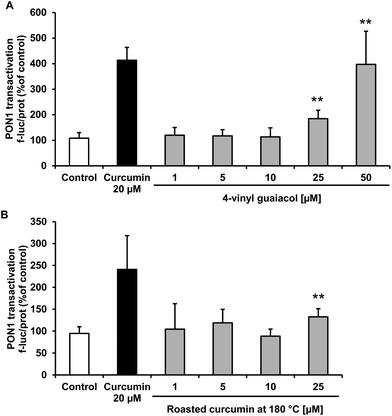
Treatment of HuH7 cells with pure 4-vinyl guaiacol resulted in a dose-dependent significant increase in PON1 transactivation (Fig. 5A) and roasted curcumin slightly induced PON1 at 25 μmol L−1 (Fig. 5B).Effects of 4-vinyl guaiacol and roasted curcumin on interleukin-6 gene expression
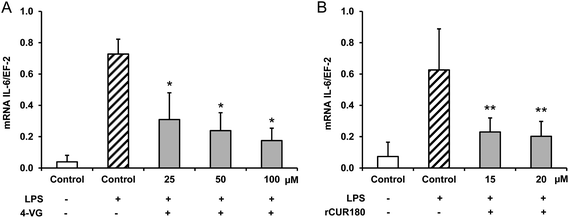
The treatment of murine LPS stimulated RAW264.7 macrophages with 4-vinyl guaiacol and roasted curcumin resulted in a significant down-regulation of IL-6 mRNA levels (Fig. 6A and B).Discussion
According to literature data, curcumin is highly susceptible in terms of thermal induced degradation or varying pH conditions.15,20–22 The dicarbonyl bridge in the curcumin molecule is its most vulnerable structural feature. Compounds such as vanillin, vanillic acid, protocatechuic acid, and further volatiles are the end products of this degradation process, some being of high relevance for the flavor of curcumin. With regard to the chemical structures of these compounds, the first carbon atom of the alkyl chain connecting the two catechol moieties seems therefore being the most sensitive one to heat application.16,22–24 The thermal decomposition kinetics and thermodynamics of curcumin have been investigated by Chen et al.:25 the kinetics, determined as apparent activation energy (Eα) values, run through two stages being both single-step processes.Prior to the formation of volatiles, oxidative cleavage of the curcumin molecule occurs, which leads to the intermediary ferulic acid and its further degradation product 4-vinyl guaiacol as an important reaction product (Fig. 2). This reaction is a typical decarboxylation reaction that is accelerated by free exposure to air and higher temperatures.16 Under the conditions investigated, a heat treatment of 50 minutes resulted in the highest formation 4-vinyl guaiacol, whereas longer roasting times were accompanied by a decrease in the concentration of 4-vinyl guaiacol which in turn is further degraded or reconstituted to the so called “deketene curcumin” (1,5-bis(4-hydroxy-3-methoxyphenyl)-1,4-pentadiene-3-one).15 Thus fine tuning processing parameters may be a critical step in optimizing the biological activity of plant bioactives including curcumin.26
Processing of food may modulate the biological activity of plant bioactives.26,27 For instance, it has been shown that the degree of roasting correlates with the efficiency of counteracting inflammation and inducing Nrf2 activity in coffee.28 Furthermore, it has been recently shown that cooking modulates the anticarcinogenic activity of curcumin due to pyrolytic formation of the already mentioned “deketene curcumin” (1,5-bis(4-hydroxy-3-methoxyphenyl)-1,4-pentadiene-3-one) as suggested by Dahmke et al.15 Herein is reported that roasted curcumin and its major degradation product 4-vinyl guaiacol exhibit, by in large, a similar Nrf2 inducing activity like curcumin per se. Thus thermal processing of curcumin, as far as roasting is concerned, does not seem to be associated with a loss of its biological activity in terms of Nrf2 signaling. In the present cell culture assays, curcumin was used as a positive control in order to induce Nrf2 and PON1. However, the concentration of curcumin was higher than those curcumin concentrations reported in human plasma following curcumin supplementation.2,29 Furthermore, little is known regarding plasma concentration of 4-vinyl guaiacol. Thus, future bioavailability studies are needed to take also 4-vinyl guaiacol and other curcumin degradation products into account.
Under the conditions investigated, 4-vinyl guaiacol and roasted curcumin did not only induce Nrf2 but also PON1 in a dose-dependent manner. Data of the present study indicate that 4-vinyl guaiacol is formed chemically due thermal processing of curcumin. However, it has been shown that decarboxylation of ferulic acid (the monomer of curcumin) due to microbial enzymes may yield 4-vinyl guaiacol levels, as well.30 Thus, 4-vinyl guaiacol could be possibly also produced by the human gut microbiota in response to a curcumin-rich diet.
Little is known about the anti-inflammatory activity of roasting products derived from curcumin. The present study shows that 4-vinyl guaiacol and roasted curcumin downregulated the mRNA levels of IL-6 in LPS-stimulated murine macrophages. Interestingly, roasted curcumin was slightly more potent than 4-vinyl guaiacol in inhibiting pro-inflammatory gene expression (Fig. 6).
Future studies are needed to pinpoint the underlying cellular and molecular mechanisms by which curcumin degradation products downregulates inflammatory gene expression.
Conclusion
Overall, the present study indicates that curcumin undergoes degradation due to roasting and its degradation products (i.e. 4-vinyl guaiacol) may exhibit significant biological activity such as antioxidant and anti-inflammatory properties in cultured cells. Data regarding the biological activity of curcumin degradation products in cultured cells in vitro should be verified in appropriate in vivo models in the future.Conflict of interest statement
Authors have no conflict of interest.Acknowledgements
The authors would like to thank Vivien Schmuck and Gaby Steinkamp for excellent technical assistance.References
- D. K. Agrawal and P. K. Mishra, Med. Res. Rev., 2010, 30, 818–860 CAS.
- T. Esatbeyoglu, P. Hübbe, I. M. A. Ernst, D. Chin, A. E. Wagner and G. Rimbach, Angew. Chem., Int. Ed., 2012, 51, 5308–5332 CrossRef CAS PubMed.
- B. B. Aggarwal, C. Sundaram, N. Malani and H. Ichikawa, Adv. Exp. Med. Biol., 2007, 595, 1–75 CrossRef.
- B. B. Aggarwal, S. C. Gupta and B. Sung, Br. J. Pharmacol., 2013, 169, 1672–1692 CrossRef CAS PubMed.
- D. Chin, P. Huebbe, K. Pallauf and G. Rimbach, Curr. Med. Chem., 2013, 20, 3955–3985 CrossRef CAS.
- G. P. Eckert, C. Schiborr, S. Hagl, R. Abdel-Kader, W. E. Muller, G. Rimbach and J. Frank, Neurochem. Int., 2013, 62, 595–602 CrossRef CAS PubMed.
- S. C. Gupta, G. Kismali and B. B. Aggarwal, BioFactors, 2013, 39, 2–13 CrossRef CAS PubMed.
- C. Schrader, C. Schiborr, J. Frank and G. Rimbach, Br. J. Nutr., 2011, 105, 167–170 CrossRef CAS PubMed.
- H. K. Na and Y. J. Surh, Free Radicals Biol. Med., 2014, 67, 353–365 CrossRef CAS PubMed.
- Y. J. Surh, J. K. Kundu and H. K. Na, Planta Med., 2008, 74, 1526–1539 CrossRef CAS PubMed.
- C. Schrader and G. Rimbach, Curr. Med. Chem., 2011, 18, 5624–5643 CrossRef CAS.
- C. Boesch-Saadatmandi, G. Rimbach, C. Schrader, B. M. Kofler, C. K. Armah and A. M. Minihane, Mol. Nutr. Food Res., 2010, 54, 1842–1850 CAS.
- M. R. Guimarães, F. R. Leite, L. C. Spolidorio, K. L. Kirkwood and C. Rossa Jr., Arch. Oral Biol., 2013, 58, 1309–1317 CrossRef PubMed.
- S. J. Kim, J. Periodontal Implant Sci., 2011, 41, 157–163 CrossRef CAS PubMed.
- I. N. Dahmke, S. P. Boettcher, M. Groh and U. Mahlknecht, Food Chem., 2014, 151, 514–519 CrossRef CAS PubMed.
- W. Fiddler, W. E. Parker, A. E. Wasserman and R. C. Doerr, J. Agric. Food Chem., 1967, 15, 757–761 CrossRef CAS.
- G. Valacchi, G. Rimbach, C. Saliou, S. U. Weber and L. Packer, Toxicology, 2001, 165, 225–234 CrossRef CAS.
- E. Borenfreund and J. A. Puerner, Toxicol. Lett., 1985, 24, 119–124 CrossRef CAS.
- A. Banning, S. Deubel, D. Kluth, Z. Zhou and R. Brigelius-Flohe, Mol. Cell. Biol., 2005, 25, 4914–4923 CrossRef CAS PubMed.
- A. Khurana and C.-T. Ho, J. Liq. Chromatogr., 1988, 11, 2295–2304 CrossRef CAS.
- Y. J. Wang, M. H. Pan, A. L. Cheng, L. I. Lin, Y. S. Ho, C. Y. Hsieh and J. K. Lin, J. Pharm. Biomed., 1997, 15, 1867–1876 CrossRef CAS.
- D. Suresh, K. N. Gurudutt and K. Srinivasan, Eur. Food Res. Technol., 2009, 228, 807–812 CrossRef CAS.
- H. Peleg, M. Naim, U. Zehavi, R. L. Rouseff and S. Nagy, J. Agric. Food Chem., 1992, 40, 764–767 CrossRef CAS.
- N. Vanbeneden, D. Saison, F. Delvaux and F. R. Delvaux, J. Agric. Food Chem., 2008, 56, 11983–11988 CrossRef CAS PubMed.
- Z. Chen, Y. Xia, S. Liao, Y. Huang, Y. Li, Y. He, Z. Tong and B. Li, Food Chem., 2014, 155, 81–86 CrossRef CAS PubMed.
- U. Boettler, K. Sommerfeld, N. Volz, G. Pahlke, N. Teller, V. Somoza, R. Lang, T. Hofmann and D. Marko, J. Nutr. Biochem., 2011, 22, 426–440 CrossRef CAS PubMed.
- U. Boettler, N. Volz, G. Pahlke, N. Teller, C. Kotyczka, V. Somoza, H. Stiebitz, G. Bytof, I. Lantz, R. Lang, T. Hofmann and D. Marko, Mol. Nutr. Food Res., 2011, 55, 798–802 CAS.
- I. Paur, T. R. Balstad and R. Blomhoff, Free Radicals Biol. Med., 2010, 48, 1218–1227 CrossRef CAS PubMed.
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Mol. Pharm., 2007, 4, 807–818 CrossRef CAS PubMed.
- W. Gu, X. Li, J. Huang, Y. Duan, Z. Meng, K. Q. Zhang and J. Yang, Appl. Microbiol. Biotechnol., 2011, 89, 1797–1805 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |