Open Access Article
Open Access ArticlePolyamine quinoline rhodium complexes: synthesis and pharmacological evaluation as antiparasitic agents against Plasmodium falciparum and Trichomonas vaginalis†
Tameryn
Stringer
a,
Dale
Taylor
b,
Hajira
Guzgay
c,
Ajit
Shokar
d,
Aaron
Au
d,
Peter J.
Smith
b,
Denver T.
Hendricks
c,
Kirkwood M.
Land
d,
Timothy J.
Egan
a and
Gregory S.
Smith
*a
aDepartment of Chemistry, University of Cape Town, Rondebosch 7701, South Africa. E-mail: gregory.smith@uct.ac.za
bDivision of Clinical Pharmacology, Department of Medicine, University of Cape Town Medical School, Observatory 7925, South Africa
cDivision of Medical Biochemistry, University of Cape Town, Rondebosch 7701, South Africa
dDepartment of Biological Sciences, University of the Pacific, Stockton, CA 95211, USA
First published on 30th July 2015
Abstract
A series of mono- and bis-salicylaldimine ligands and their corresponding Rh(I) complexes were prepared. The compounds were characterised using standard spectroscopic techniques including NMR, IR spectroscopy and mass spectrometry. The salicylaldimine ligands and complexes were screened for antiparasitic activity against two strains of Plasmodium falciparum i.e. the NF54 CQ-sensitive and K1 CQ-resistant strain as well as against the G3 isolate of Trichomonas vaginalis. The monomeric salicylaldimine quinolines exhibited good activity against the NF54 strain and the dimeric salicylaldimine quinolines exhibited no cross resistance across the two strains. The binuclear 5-chloro Rh(I) complex displayed the best activity against the Trichomonas vaginalis parasite, possibly a consequence of its enhanced lipophilicity. The compounds were also screened for cytotoxicity in vitro against WHCO1 oesophageal cancer cells. The monomeric salicylaldimine quinolines exhibited high selectivity towards malaria parasites compared to cancer cells, while the dimeric compounds were less selective.
Introduction
Malaria remains one of the most prevalent parasitic diseases worldwide.1 According to the World Health Organisation (WHO), 198 million cases of malaria were reported globally, of which 584![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths were documented in 2013.2 The most serious malarial infections are due to Plasmodium falciparum. This particular parasite rapidly develops resistance against various antimalarial treatments. Chloroquine, one of the most commonly used drugs, has been rendered less effective as a result.3 Currently, artemisinin combination therapy is the main treatment for this disease and involves the concurrent use of an artemisinin-based drug and a second drug. Combination therapy is employed to delay the onset of resistance, but there have been reports of artemisinin resistance in certain parts of the world.4–6 Since many of the current treatments are at risk of becoming obsolete, there is a need to discover alternative therapies. A metal-based candidate, ferroquine (FQ), has demonstrated the ability to overcome resistance experienced by its parent compound, chloroquine (CQ).7 Ferroquine exhibits potent in vitro and in vivo activity against various chloroquine-sensitive and chloroquine-resistant strains of Plasmodium.8,9 It has also very recently completed phase IIb clinical trials.10 The clinical success of ferroquine provides scope for the use of metal-based drugs in malaria therapy. Many analogues of ferroquine have since been reported.11–13
000 deaths were documented in 2013.2 The most serious malarial infections are due to Plasmodium falciparum. This particular parasite rapidly develops resistance against various antimalarial treatments. Chloroquine, one of the most commonly used drugs, has been rendered less effective as a result.3 Currently, artemisinin combination therapy is the main treatment for this disease and involves the concurrent use of an artemisinin-based drug and a second drug. Combination therapy is employed to delay the onset of resistance, but there have been reports of artemisinin resistance in certain parts of the world.4–6 Since many of the current treatments are at risk of becoming obsolete, there is a need to discover alternative therapies. A metal-based candidate, ferroquine (FQ), has demonstrated the ability to overcome resistance experienced by its parent compound, chloroquine (CQ).7 Ferroquine exhibits potent in vitro and in vivo activity against various chloroquine-sensitive and chloroquine-resistant strains of Plasmodium.8,9 It has also very recently completed phase IIb clinical trials.10 The clinical success of ferroquine provides scope for the use of metal-based drugs in malaria therapy. Many analogues of ferroquine have since been reported.11–13
Another metal-based chloroquine complex that exhibited promising antiplasmodial activity against Plasmodium parasites is [RhCl(COD)CQ], where COD = 1,5-cyclooctadiene (Fig. 1a). This particular complex was one of the first organometallic complexes screened against malarial parasites and was reported by Sánchez-Delgado et al.14 This complex reduced parasitemia more than CQ when examined in vivo.14 Since then, only a few examples of Rh(I)–CQ complexes have been reported in literature.11–13 For example, rhodium complexes of ferroquine display moderate activity when evaluated in vitro.15 Ruthenium quinoline complexes have also generated interest as potential antimalarials. A binuclear ruthenium chloroquine complex [RuCl2(CQ)]2 was evaluated and found to display potency against P. berghei, showing greater activity than CQ.14 Ruthenium p-cymene quinoline complexes have also been tested against various plasmodial strains.16 In these cases, the ruthenium moiety is not bonded to the quinoline nitrogen, but is coordinated to the ligand in a bidentate manner in the side chain. One of the complexes, a salicylaldimine quinoline, shown in Fig. 1b, exhibited promising activity against a CQ-sensitive strain of P. falciparum.16
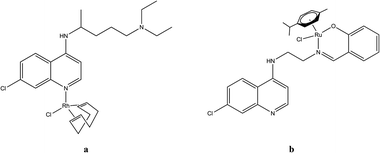 | ||
| Fig. 1 Structures of [RhCl(COD)CQ]14 and [RuCl(p-cymene)quinoline]16 that exhibit antiplasmodial activity. | ||
Trichomoniasis is a very common sexually transmitted parasitic disease caused by Trichomonas vaginalis. The disease is easily treatable but can spread, causing individuals to be susceptible to other diseases, including cancer and HIV.17–21 Drugs such as metronidazole and tinidazole are commonly used 5-nitroimidazoles often utilised to treat these infections.22,23 However, the current FDA-approved treatment, metronidazole, has been found to be less effective in some patients due to resistance.24,25 There has recently been an interest in obtaining suitable alternatives for treating this infection.25 Metal complexes have also recently generated interest as potential antitrichomonal agents. These include complexes of palladium, ruthenium and rhodium.26,27 Multinuclear metal complexes have also been observed to exhibit antiparasitic activity against the T. vaginalis parasite.28
Encouraged by the afore-mentioned results, we investigated the antiparasitic activity of a series of mononuclear and binuclear rhodium(I) chloroquine complexes. It has been observed for many systems that increasing the number of active moieties results in enhanced biological activity.29–34 The binuclear complexes prepared in this study are based on a polyamine scaffold. Polyamine-based compounds have been observed to exhibit enhanced uptake in plasmodium-infected erythrocytes.35 It was expected that the polyamine-containing systems impart beneficial effects on antiplasmodial activity, specifically in CQ-resistant strains of P. falciparum. The polyamine-based systems could possibly also positively influence antitrichomonal activity, as increased lipophilicity appears to coincide with favourable activity.27,28 This study aimed to address the influence of various characteristics on the potency and ability of the compounds to overcome resistance. These characteristics include the introduction of polyamine scaffolds, multinuclearity, varying substituents on the salicylaldimine moieties, size and lipophilicity. In addition, β-haematin inhibition studies were carried out to gain deeper insight into a possible mechanism of action. The cytotoxicity of these compounds has also been investigated in order to establish parasite selectivity.
Results and discussion
Synthesis and characterisation
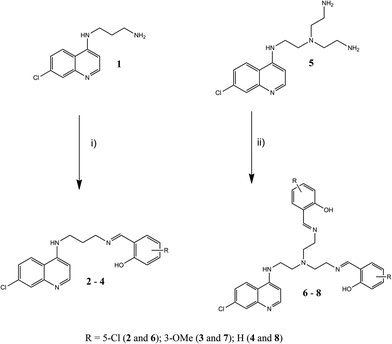
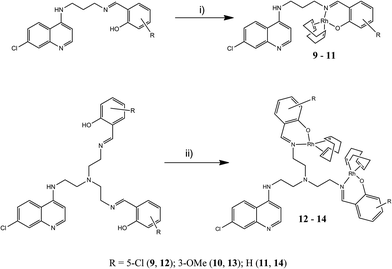
Rhodium(I)–quinoline complexes were prepared from mono- and bis-salicylaldimine ligands. Salicylaldehydes including 5-chlorosalicylaldehyde, 3-methoxysalicylaldehyde and salicylaldehyde were used to afford three monomeric and three dimeric salicylaldimine ligands (Scheme 1). The mono-salicylaldimines were prepared by Schiff-base condensation between N′-(7-chloroquinolin-4-yl)-propane-1,3-diamine36 (1) and the various aldehydes in diethyl ether to afford 2–4 as yellow amorphous solids. The bis-salicylaldimines were prepared from N-(7-chloroquinolin-4-yl)-tris(2-aminoethyl)amine37 (5) and the afore-mentioned aldehydes in ethanol to afford 6–8 as yellow powders. Since the quinoline nitrogen plays an important role in antiplasmodial activity,38 it was decided to incorporate the metal fragment in the lateral side chain of these ligands. Since the phenolic protons of the ligand are only mildly acidic (pKa ∼ 9, estimated using MarvinSketch V5.9.4), stirring the ligand in the presence of a strong base, such as NaH results in deprotonation of the phenolic proton. Upon addition of the metal precursor, the expected bidentate coordination is afforded. Complexes 9–14 were obtained in moderate yields (52–79%) by reaction of 2–4 and 6–8 with [RhCl(COD)]2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 (where COD = 1,5-cyclooctadiene) (Scheme 2). The mononuclear complexes (9–11) were obtained by reacting 1 equivalent of the rhodium dimer with 2 equivalents of the appropriate ligand, while the dinuclear complexes 12–14 were obtained by reaction of the ligand and dimer in a 1
39 (where COD = 1,5-cyclooctadiene) (Scheme 2). The mononuclear complexes (9–11) were obtained by reacting 1 equivalent of the rhodium dimer with 2 equivalents of the appropriate ligand, while the dinuclear complexes 12–14 were obtained by reaction of the ligand and dimer in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio.
1 stoichiometric ratio.
The formation of Schiff-base ligands 2–4 and 6–8 was confirmed by the presence of a singlet in the 1H NMR spectra of these ligands in the region of 7.9–8.3 ppm attributed to the imine proton. Complexation was confirmed by an upfield shift of the imine signal compared to the free ligand. The resonance for the phenolic proton was absent in the spectra of the complexes. This suggests coordination in a bidentate manner to the imine nitrogen and the phenolic oxygen. In addition to this, in the 1H NMR spectra of complexes 9–14, four distinct resonances for the protons of the COD moiety were observed. This is very different compared to the spectrum of the [RhCl(COD)]2 precursor, which only exhibits three signals for the COD protons. In these complexes, the methylene protons of the COD moiety exhibit non-equivalence. The exo and endo methylene resonances were assigned at approximately 1.8 and 2.3 ppm, respectively. Two multiplets for the olefinic protons of the COD moiety appeared at approximately 3.6 and 4.5 ppm. The appearance of two multiplets rather than one (in the precursor), further supports N,O coordination. The appearance of the two multiplets was attributed to the trans effect induced by the different N and O environments. The spectra of the complexes were consistent with similar N,O-Rh(I) complexes.40–44
In the 13C{1H} NMR spectra, signals for the aliphatic carbon atoms of the COD moiety appeared at approximately 28 and 32 ppm for the complexes 9–14. The olefinic carbon atoms appeared as two doublets at 71 and 86 ppm, respectively. Coupling constants of approximately 14 and 12 Hz were observed for these signals, which is consistent with data observed for similar Rh-N,O complexes.40–44
The most obvious difference between the infrared spectra of the ligands and the complexes was that two absorption bands were observed for the two different C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N environments in the ligand spectra. An absorption band for the Schiff-base C
N environments in the ligand spectra. An absorption band for the Schiff-base C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching frequency appeared between 1631 and 1648 cm−1, while the absorption band for the quinoline C
N stretching frequency appeared between 1631 and 1648 cm−1, while the absorption band for the quinoline C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching frequency appeared between 1609 and 1615 cm−1. In the case of the complexes, one absorption band was observed upon coordination. The absorption band that was initially at a higher frequency (∼1630 cm−1 (C
N stretching frequency appeared between 1609 and 1615 cm−1. In the case of the complexes, one absorption band was observed upon coordination. The absorption band that was initially at a higher frequency (∼1630 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine)) in the ligands, shifted towards the frequency of the band associated with the C
Nimine)) in the ligands, shifted towards the frequency of the band associated with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nquinoline, resulting in the appearance of one absorption band.
Nquinoline, resulting in the appearance of one absorption band.
Both ESI (recorded in the positive mode) and EI mass spectrometry was used to analyse these compounds. The molecular ion peak ([M]+) or a peak for the protonated form ([M + H]+) was observed in the spectra of these compounds confirming preparation of the desired compounds.
In vitro antiparasitic activity against Plasmodium falciparum
The antiplasmodial activity of the salicylaldimine ligands 2–4, 6–8 and their corresponding complexes 9–14 were evaluated in vitro against two strains of P. falciparum, i.e. the NF54 chloroquine-sensitive and K1 chloroquine-resistant strain in order to identify any effects of the polyamine scaffold, multinuclearity and salicylaldimine substituent on antiplasmodial activity. Chloroquine (CQ) and ferroquine (FQ) were used as reference drugs. The data obtained from this study are given in Table 1.| Compound | IC50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a ± SE NF54b (nM) a ± SE NF54b (nM) |
IC50 ± SE K1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c (nM) c (nM) |
Resistance index (RI)d | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) f f |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 D7.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) f f |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D4.7 D4.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) f f |
|---|---|---|---|---|---|---|
a IC50 represents the nanomolar equivalents required to inhibit parasite growth by 50%.
b NF54 chloroquine-sensitive strain of P. falciparum; n = number of data sets averaged, n = 3.
c K1 chloroquine-resistant strain of P. falciparum.
d Resistance index (RI) = IC50K1/IC50NF54.
e Literature value.45
f Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and log P and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D values predicted using MarvinSketch V5.9.4.
g nd = not determined. D values predicted using MarvinSketch V5.9.4.
g nd = not determined.
|
||||||
| 2 | 52 ± 1 | 720 ± 89 | 13.7 | 4.32 | 4.02 | 2.01 |
| 3 | 55 ± 1 | 640 ± 83 | 11.8 | 3.56 | 3.30 | 0.97 |
| 4 | 72 ± 6 | 680 ± 17 | 9.4 | 3.71 | 3.43 | 0.95 |
| 6 | 4940 ± 280 | 2260 ± 260 | 0.4 | 6.33 | 5.47 | 1.78 |
| 7 | 4930 ± 370 | 1760 ± 180 | 0.4 | 4.81 | 4.01 | 0.07 |
| 8 | 7000 ± 480 | 4520 ± 780 | 0.6 | 5.12 | 4.31 | 0.15 |
| 9 | 98 ± 9 | 866 ± 55 | 8.8 | ndg | nd | nd |
| 10 | 160 ± 20 | 1260 ± 190 | 7.7 | nd | nd | nd |
| 11 | 91 ± 9 | 1790 ± 61 | 19.6 | nd | nd | nd |
| 12 | 7960 ± 370 | 2940 ± 350 | 0.4 | nd | nd | nd |
| 13 | 4820 ± 75 | 2190 ± 120 | 0.4 | nd | nd | nd |
| 14 | 3960 ± 120 | 1530 ± 140 | 0.4 | nd | nd | nd |
| CQ | 25 ± 5 | 300 ± 38 | 11.8 | 4.63e | nd | nd |
| FQ | 33 ± 10 | 14e | 0.5 | 5.1e | nd | nd |
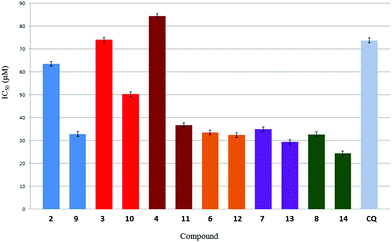
The monomeric salicylaldimine ligands 2–4 showed good activity against the NF54 strain of P. falciparum, exhibiting IC50 values well below 1 μM. The activity observed for these compounds are comparable to chloroquine (IC50 = 25 nM) and ferroquine (IC50 = 33 nM) in this strain. The dimeric salicylaldimine ligands (6–8) were less active than their monomeric counterparts (2–4). The corresponding rhodium(I) COD complexes were prepared in order to establish whether incorporation of the metal moiety is beneficial for antiplasmodial activity. This was done in the light of the Rh(I) complex shown in Fig. 1a, which reduced parasitemia to a greater extent to CQ in vivo and which further supports the use of Rh(I) in this investigation.14 Other Rh(I) complexes of CQ have also exhibited promising in vitro behaviour against both CQ-sensitive and CQ-resistant strains of P. Falciparum.15,46,47 In general, the mononuclear complexes were slightly less active than their corresponding ligands, yet the compounds still maintained good activity. Complex 14 was approximately 1.8 times more active than its corresponding ligand and is the most active binuclear complex.
Although the activity observed for the complexes prepared in this study cannot be compared directly to that of the CQ derivative evaluated by Sànchez-Delgado et al.14 (since different species of Plasmodium were used), it was observed that the mononuclear complexes 9–11 showed comparable activity to this complex (IC50 values between 91 and 160 nM). Thus, the quinoline moiety appears to be the pharmacophore responsible for the activity of these compounds, since the free ligands also exhibited good activity.
Similar trends in activity were observed for the K1 CQ-resistant strain. The monomeric ligands 2–4 were the most active, while the dimeric ligands, mononuclear complexes and binuclear complexes exhibited activity in the same range (1300–2900 nM). Overall, the dimeric ligands and binuclear complexes showed improved activity in the resistant strain compared to the sensitive strain. Table 1 gives the resistance indices (RI) for all of the tested compounds. Ligands 2–4 and complexes 9–11, which contain one salicylaldimine moiety, exhibited larger RI values compared to the dimeric salicylaldimine compounds 6–8 and 12–14. Ligands 2–4 and their corresponding complexes 9–11 displayed similar antiplasmodial behaviour to CQ. This suggests that incorporation of the polyamine scaffold is beneficial for overcoming CQ-resistance. The dimeric (polyamine) compounds showed similar or enhanced activity in the resistant strain compared to the sensitive strain which appear to follow a similar activity profile to ferroquine. The improved activity of ferroquine against resistant strains is a consequence of its more lipophilic character.9,48
The predicted log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and log
P and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D (pH 7.4 = erythrocyte; pH 4.7 = digestive vacuole) values of selected compounds are presented in Table 1. Generally, the monomeric ligands 2–4 exhibit lower log
D (pH 7.4 = erythrocyte; pH 4.7 = digestive vacuole) values of selected compounds are presented in Table 1. Generally, the monomeric ligands 2–4 exhibit lower log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and log
P and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 values compared to their dimeric counterparts 6–8. The addition of the metal fragment to the ligands results in an overall increase in the lipophilicity of the complexes compared to the free ligands. The log
D7.4 values compared to their dimeric counterparts 6–8. The addition of the metal fragment to the ligands results in an overall increase in the lipophilicity of the complexes compared to the free ligands. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of the 1,5-cyclooctadiene moiety was predicted to be 2.83 using MarvinSketch V5.9.4. Once the compounds reach the digestive vacuole (DV) of the parasite, protonation of the amino groups can occur, which in turn lowers the lipophilicity of these compounds. In comparison, the monomeric ligands have greater log
P value of the 1,5-cyclooctadiene moiety was predicted to be 2.83 using MarvinSketch V5.9.4. Once the compounds reach the digestive vacuole (DV) of the parasite, protonation of the amino groups can occur, which in turn lowers the lipophilicity of these compounds. In comparison, the monomeric ligands have greater log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D4.7 values than the dimeric systems. This can potentially allow for greater efflux of these monomers out of the DV compared to the dimeric compounds, and therefore they have lower activity in the resistant strain compared to the sensitive strain.
D4.7 values than the dimeric systems. This can potentially allow for greater efflux of these monomers out of the DV compared to the dimeric compounds, and therefore they have lower activity in the resistant strain compared to the sensitive strain.
β-Haematin inhibition activity
Haemozoin remains an important target for the development of potential antimalarials. Since many quinoline compounds, including chloroquine, are believed to inhibit haemozoin crystal growth, it was expected that the compounds prepared in this study would act by the same mechanism. When chloroquine reaches the digestive vacuole, it binds to a toxic product of haemoglobin degradation known as haematin (ferriprotoporphyrin IX) and prevents conversion into haemozoin, which is less toxic. A build-up of haematin results in damage to the parasite. Some studies suggest that haemozoin itself is the target of antimalarials.49,50 An NP-40 detergent-mediated assay was used to establish if these compounds inhibit β-haematin (synthetic haemozoin) formation.51 The amount of β-haematin formed was quantified using a colorimetric pyridine ferrochrome method developed by Ncokazi and Egan.52 Research suggests that haemozoin formation is not a spontaneous process and that haem crystallization occurs in the presence of neutral lipids.53,54 The neutral detergent, NP-40, was used to mimic lipids and mediates β-haematin formation in the assay.The ligands (2–4 and 6–8) and complexes (9–14) were screened in order to elucidate a possible mechanism for their antiplasmodial activity and the results are depicted in Fig. 2. The monomeric ligands 2–4 exhibited the lowest β-haematin inhibition activity of all of the tested compounds and showed similar β-haematin inhibition activity to chloroquine. The dimeric ligands (6–8) were approximately twice as active (IC50 = 33–35 μM) compared to their monomeric counterparts. The R group appeared to have little to no significant effect on β-haematin inhibition in the case of the dimeric compounds. The higher activity confirms that these compounds inhibit β-haematin crystallization to a greater extent compared to the monomeric ligands in this assay.
The mononuclear complexes 9–11 exhibited enhanced activity compared to their corresponding ligands. The binuclear complexes 12–14, showed similar activity to their respective ligands. Most of the mononuclear complexes showed similar activity to the binuclear complexes. It appears that incorporation of the metal fragment as part of these systems results in enhanced β-haematin inhibition activity. This may be a consequence of the geometry of the metal fragment and increased lipophilicity. The complexes possess a square planar geometry about the metal centre, which introduces a planar system about the salicylaldimine moiety. This may favour interactions with haem, which may be a reason for the increased β-haematin inhibition activity.
A plot comparing log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50 and molecular weight is shown in Fig. 3. Compounds with similar molecular weights behave similarly. The monomeric ligands (2–4) have molecular weights in the range of 339–373 g mol−1. These compounds were the least active and gave log
IC50 and molecular weight is shown in Fig. 3. Compounds with similar molecular weights behave similarly. The monomeric ligands (2–4) have molecular weights in the range of 339–373 g mol−1. These compounds were the least active and gave log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50 values of approximately 1.9. Compounds 6–11, which are the dimeric salicylaldimine ligands and mononuclear complexes, have molecular weights in the range of 515 and 583 g mol−1. These compounds gave log
IC50 values of approximately 1.9. Compounds 6–11, which are the dimeric salicylaldimine ligands and mononuclear complexes, have molecular weights in the range of 515 and 583 g mol−1. These compounds gave log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50 values of approximately 1.5. Compounds 13 and 14 were the most active and had large molecular weights. Based on this, a relationship between the size of the compound and its ability to inhibit β-haematin formation is postulated. This has been observed for multimeric quinoline thioureas as well.28 The mononuclear complexes showed improved β-haematin inhibition in comparison with their ligands while the dimeric ligands, mononuclear complexes and binuclear complexes exhibited comparable β-haematin inhibition activity. No simple correlation between β-haematin inhibition and antiplasmodial activity was observed, however. The monomeric ligands (2–4) were the most active compounds in both the NF54 CQ-sensitive and K1 CQ-resistant strains, but these compounds were the least effective β-haematin inhibitors. In addition to this, the least active compounds (dimeric salicylaldimines) in vitro displayed the best β-haematin inhibition. Although it might be expected that the antiplasmodial activity would be related to a compound's ability to inhibit haemozoin formation, it must be remembered that the ability of the drug to be transported into the digestive vacuole and accumulate therein is an important factor that influences antiplasmodial activity.55 The strongest inhibitors of β-haematin formation in this assay are not necessarily the best inhibitors inside of the DV. The concentration of the compounds in the DV is greatly affected by its log
IC50 values of approximately 1.5. Compounds 13 and 14 were the most active and had large molecular weights. Based on this, a relationship between the size of the compound and its ability to inhibit β-haematin formation is postulated. This has been observed for multimeric quinoline thioureas as well.28 The mononuclear complexes showed improved β-haematin inhibition in comparison with their ligands while the dimeric ligands, mononuclear complexes and binuclear complexes exhibited comparable β-haematin inhibition activity. No simple correlation between β-haematin inhibition and antiplasmodial activity was observed, however. The monomeric ligands (2–4) were the most active compounds in both the NF54 CQ-sensitive and K1 CQ-resistant strains, but these compounds were the least effective β-haematin inhibitors. In addition to this, the least active compounds (dimeric salicylaldimines) in vitro displayed the best β-haematin inhibition. Although it might be expected that the antiplasmodial activity would be related to a compound's ability to inhibit haemozoin formation, it must be remembered that the ability of the drug to be transported into the digestive vacuole and accumulate therein is an important factor that influences antiplasmodial activity.55 The strongest inhibitors of β-haematin formation in this assay are not necessarily the best inhibitors inside of the DV. The concentration of the compounds in the DV is greatly affected by its log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P as well as the pKa of protonatable sites. In this case, the lipophilicity at pH 7.4 would affect the transport of the compounds across membranes. Since it is higher compared to their monomers, less of the compound would expected to be taken up due to increased membrane–drug interactions. This may be why low activity is observed in the parasite, but a greater β-haematin inhibition activity is observed in the detergent-mediated assay. Lack of correlation has been observed for similar quinoline systems but the mechanism responsible for the activity is believed to be via β-haematin inhibition.47,48
P as well as the pKa of protonatable sites. In this case, the lipophilicity at pH 7.4 would affect the transport of the compounds across membranes. Since it is higher compared to their monomers, less of the compound would expected to be taken up due to increased membrane–drug interactions. This may be why low activity is observed in the parasite, but a greater β-haematin inhibition activity is observed in the detergent-mediated assay. Lack of correlation has been observed for similar quinoline systems but the mechanism responsible for the activity is believed to be via β-haematin inhibition.47,48
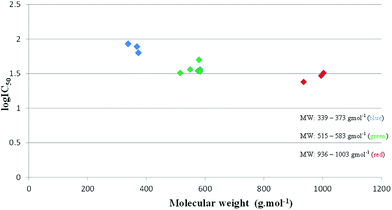 | ||
Fig. 3 Relationship between the molecular weight of the tested compounds and β-haematin inhibition log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50. IC50. | ||
In vitro cytotoxicity
All of the synthesized compounds and CQ were also screened against WHCO1 oesophageal cancer cells, to determine the cytotoxicity of these compounds. To be considered as antiplasmodial, the compounds should exhibit selectivity towards malaria parasites, with minimal cytotoxicity directed at host cells. The results pertinent to this study are depicted in Table 2. The tested compounds showed moderate cytotoxicity against this cell-line in the same range as cisplatin, a commonly used chemotherapeutic agent (IC50 = 13.0 μM).56 CQ also exhibited similar cytotoxicity to the synthesised compounds. The dimeric salicylaldimine compounds 6, 8 and 13 exhibited the highest cytotoxicity. With the exception of the monomeric 3-OMe salicylaldimine ligand (3), the monomeric salicylaldimines exhibited slightly lower cytotoxicity compared to the dimeric derivatives. Studies carried out on polyamines reveal that these systems are capable of transporting cytotoxic drugs into tumor cells.57 Tumor cells are able to import polyamines by means of a polyamine transporter (PAT) in order to sustain their growth. The transporter is also able to tolerate modified polyamines, therefore drug-incorporated polyamines may be able to penetrate tumor cells via the PAT.58 The dimeric salicylaldimines are considered to be polyamines because they contain more than one amino group. This could be a reason for the enhanced activity of some of these compounds in this cell-line. Many traditional quinoline-containing antimalarials, including chloroquine, have also been investigated as antitumor agents against human breast cancer cells and exhibited moderate activity.59| Compound | IC50 (μM) | 95% confidence interval | Selectivity index (SI)a | Selectivity index (SI)b |
|---|---|---|---|---|
| a (IC50WHCO1/IC50NF54). b (IC50WHCO1/IC50K1). c IC50 value obtained for cisplatin against WHCO1 cancer cells.56 | ||||
| 2 | 8.5 | 7.5–9.2 | 163 | 11.8 |
| 3 | 8.8 | 7.8–9.9 | 160 | 13.5 |
| 4 | 8.4 | 7.2–9.6 | 116 | 12.3 |
| 6 | 5.9 | 5.2–6.8 | 1.20 | 2.63 |
| 7 | 11.1 | 9.7–12.8 | 2.25 | 6.31 |
| 8 | 4.7 | 4.3–5.2 | 0.674 | 1.04 |
| 9 | 8.7 | 7.9–9.7 | 89.2 | 10.1 |
| 10 | 11.3 | 9.5–13.6 | 68.7 | 8.97 |
| 11 | 9.8 | 8.8–10.9 | 107 | 5.45 |
| 12 | 7.8 | 6.3–9.5 | 0.975 | 2.63 |
| 13 | 3.9 | 3.4–4.4 | 0.802 | 1.76 |
| 14 | 7.6 | 7.2–8.0 | 1.91 | 4.96 |
| CQ | 6.3 | 5.7–6.9 | 252 | 21 |
| Cisplatin | 13.0c | — | — | — |
There are examples of systems where anticancer activity appears to be a size-dependent phenomenon. Dendritic ruthenium arene compounds have been shown to display a similar effect. The multimeric systems show enhanced activity to monomeric systems.30,31 Multimeric quinoline thioureas also exhibited a size dependent increase in activity.28 Complex 13 is the most active compound of the series. Generally, the metal complexes exhibited slightly lower cytotoxicity compared to the free ligands. Selectivity indices (SI) were calculated for these compounds for both the chloroquine-sensitive strain (NF54) and the chloroquine-resistant strain (K1) of P. falciparum (Table 2). The monomeric ligands (2–4), complexes (9–11) and CQ showed high selectivity towards the NF54 strain, giving rise to large SI values. These values are lowered when compared to the K1 strain as these compounds exhibit decreased activity in this strain. The dimeric ligands and complexes displayed low SI values less than 10 which suggests that the antiplasmodial activity is comparable to the activity against the WHCO1 cell-line, suggesting little to no selectivity. This insinuates that these compounds may act by a cytotoxic mechanism in the parasite as well.
In vitro antiparasitic activity against Trichomonas vaginalis
The salicylaldimine ligands and their complexes were screened against the G3 strain of Trichomonas vaginalis (T. vaginalis). The data obtained for this study are presented in Table 3. All of the compounds, including metronidazole, were screened at a concentration of 50 μM. Five of the compounds exhibited more than 50% parasite inhibition at the tested concentration. The 5-Cl salicylaldimine compounds 6, 9 and 12 exhibited the best activity in each respective series, with the binuclear complex (12) exhibiting the best activity overall, comparable to metronidazole at the tested concentration. Comparing the activity of the ligands, it appears that incorporation of the tris(2-amino)ethyl amine scaffold is only beneficial in the case of the 5-Cl derivative. There appears to be a synergistic effect between this scaffold and the 5-Cl moiety.| Compound | % inhibition | IC50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (μM) ± SE a (μM) ± SE |
|---|---|---|
| a IC50 represents the micromolar equivalents of test compounds required to inhibit parasite growth by 50%. b nd = not determined. c IC50 value taken from ref. 27. | ||
| 2 | 40.81 ± 1.06 | ndb |
| 3 | 49.08 ± 7.42 | nd |
| 4 | 49.08 ± 3.71 | nd |
| 6 | 77.14 ± 1.10 | nd |
| 7 | 10.88 ± 4.62 | nd |
| 8 | 46.07 ± 8.75 | nd |
| 9 | 85.21 ± 5.30 | 12.00 ± 0.06 |
| 10 | 46.35 ± 6.05 | nd |
| 11 | 44.58 ± 2.95 | nd |
| 12 | 100 | 4.80 ± 0.54 |
| 13 | 97.28 ± 1.83 | 6.10 ± 0.88 |
| 14 | 67.62 ± 4.40 | nd |
| Metronidazole | 100 | 0.72c |
The compounds containing the 5-Cl motif (with the exception of 2) showed enhanced activity compared to the compounds where R = 3-OMe and R = H. Metronidazole is a 5-nitroimidazole compound and its activity appears to be dependent on the presence of the electron-withdrawing NO2 group.60 The electron-withdrawing 5-Cl moiety may impart a similar biological effect as the 5-NO2 group for metronidazole. It has been suggested that free radical production is a possible mode of action for this drug.60 The chlorido substituent has been shown to enhance the antiparasitic activity of some metal-containing compounds towards trichomoniasis. For example, palladium(II) thiosemicarbazones containing this group showed considerable parasite inhibition against the T1 strain of T. vaginalis.26 In a separate study, ruthenium–arene thiosemicarbazone complexes possessing a chlorido moiety on the aryl ring showed promising activity against the G3 strain.27 Incorporation of the cyclooctadiene moiety as part of the complexes also further increases lipophilic nature which may in part be attributed to the enhanced activity of selected complexes. A study of the biological activity of compounds against T. vaginalis reveals lipophilicity to play an important role. This has been observed in cases where ferrocene-containing compounds showed enhanced parasite inhibition against T. vaginalis compared to analogous organic derivatives.28 2-Pyridyl pyrimidines were tested for their antiplasmodial and antileishmanial activity and showed a correlation between increased lipophilicity and higher activity.61 In a separate study, lipophilic tetracyclines exhibited enhanced parasite growth inhibition over non-lipophilic derivatives against various strains of T. vaginalis.62 In these cases, the increased lipophilicity aids in the transport of the compounds across membranes, with a concomitant accumulation of the drug.
Compounds 9, 12 and 13 showed parasite inhibition greater than 80%. These compounds were further evaluated in order to obtain IC50 values (Table 3) to compare their activity to metronidazole. Complex 12 exhibited the highest activity giving an IC50 value of 4.8 μM, while complex 13 displayed similar activity to 12. The 5-Cl mononuclear complex (9) exhibited activity 2-fold lower than 13.
Conclusions
A series of mono- (2–4) and bis-salicylaldimine (6–8) quinoline ligands were prepared using template procedures. Their corresponding neutral Rh(I) 1,5-cyclooctadiene complexes (9–14) were also synthesized. The compounds were characterized using standard spectroscopic and analytical techniques. The data confirms coordination of the metal to the ligand in a bidentate manner to the imine nitrogen and the phenolic oxygen and not at the quinoline nitrogen. The compounds were screened for their antiplasmodial activity in vitro against two strains of P. falciparum. In the chloroquine-sensitive strain (NF54), the monomeric ligands (2–4) and the mononuclear complexes (9–11) exhibited enhanced activity compared to their dimeric counterparts. In the K1 strain, the monomeric ligands and complexes experienced a loss of activity as was evident by the higher RI values, suggesting that these complexes possess a similar activity profile to that of chloroquine. The dimeric derivatives exhibit comparable activity in the resistant and the sensitive strain, suggesting that the polyamine is beneficial for maintaining activity in the resistant strain. The increased number of protonation sites in the dimeric systems could allow for less efflux of these molecules out of the DV and therefore similar activity is observed in both strains. The monomeric systems were less effective at inhibiting β-haematin formation while the dimeric salicylaldimine ligands and complexes showed enhanced activity, possibly a consequence of their increased lipophilicity. Most of the dimeric derivatives showed increased cytotoxicity compared to their monomeric counterparts. The monomeric compounds exhibited substantial selectivity towards malaria parasites than WHCO1 cancer cells while the dimeric compounds showed lower selectivity, suggesting that cytotoxicity may be a reason for their antiplasmodial activity. The 5-Cl bis-salicylaldimine Rh(I) complex (12) showed the highest activity against the G3 isolate of T. vaginalis. Increased lipophilicity due to the polyamine scaffold and 5-Cl substituent appeared to promote enhanced activity. Despite the fact that the tested compounds did not possess activity comparable to commonly used antiparasitics, some valuable insight has been obtained from this study. Polyamine scaffolds could be used in order to overcome cross-resistance experienced by many quinoline-based compounds in different strains P. falciparum. The multinuclear nature of the complexes may also result in maintaining similar activity in CQ-sensitive and CQ-resistant strains of P. falciparum. Furthermore, the chlorido group plays an important role in antiparasitic activity, including activity against T. vaginalis, as well as β-haematin inhibition activity. Furthermore, lipophilicity is an important factor to consider when designing potential antiparasitic agents as this may largely influence the behaviour of these compounds in vitro. Perhaps in silico studies can be used to design similar derivatives with characteristics that are more effective in vitro.Experimental
General details
Synthetic reactions were performed under an argon atmosphere at ambient temperatures unless otherwise stated. All reagents were purchased from Sigma Aldrich and used as received. Solvents were dried over Fluka Molecular Sieves with indicator. Nuclear magnetic resonance (NMR) spectra were recorded on a Varian Unity XR400 spectrometer (1H: 399.95 MHz, 13C{1H}: 100.58 MHz), Varian Mercury XR300 spectrometer (1H: 300.08 MHz, 13C{1H}: 75.46 MHz) or Bruker Ultrashield 400 Plus spectrometer (1H: 400.20 MHz, 13C{1H}: 100.60 MHz) at 30.0 °C using tetramethylsilane (TMS) as the internal standard. Infrared (IR) absorptions were measured on a PerkinElmer Spectrum One FT-IR spectrometer and samples were analysed in the solid state as KBR pellets. Elemental analyses were carried out using a Fisions EA 110 elemental analyser. Melting points were recorded using a Reichert-Jung Thermovar melting point apparatus and are uncorrected. Electron impact (EI), high resolution (HR) and low resolution ESI-mass spectrometry was used to further characterise all new compounds and determinations were carried out using a JEOL GCmateII apparatus or a Waters API Quattro instrument in the positive mode. N-(7-Chloroquinolin-4-yl)-propane-1,3-diamine (1),36N-(7-chloroquinolin-4-yl) tris(2-aminoethyl)amine (5)37 and [RhCl(COD)]2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 were synthesised following literature methods.
39 were synthesised following literature methods.
General procedure to prepare 4-amino-7-chloroquinoline mono-imines 2–4
(N′-(7-Chloroquinolin-4-yl)-propane-1,3-diamine) (1) was suspended in diethyl ether (10 ml). To this, a slight excess of the desired salicylaldehyde in diethyl ether (10 ml) was added dropwise and the resulting mixture stirred at room temperature for 16 hours. The resulting precipitate was filtered using a Büchner funnel and washed with diethyl ether to remove excess aldehyde. The product was then dried in vacuo.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 8.52 (1H, d, 3JH,H = 5.36, Ar–H), 13.18 (1H, br s, OH). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 29.9 (CH2); 41.3 (CH2); 57.4 (CH2); 99.1 (Cquin); 117.1; 118.6 (CAr); 119.3; 120.7 (Cquin); 123.5; 125.4 (Cquin); 128.9 (Cquin); 130.4 (CAr); 132.4 (CAr); 134.9; 149.1; 149.5; 151.9 (Cquin); 159.5; 164.6 (HC
N); 8.52 (1H, d, 3JH,H = 5.36, Ar–H), 13.18 (1H, br s, OH). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 29.9 (CH2); 41.3 (CH2); 57.4 (CH2); 99.1 (Cquin); 117.1; 118.6 (CAr); 119.3; 120.7 (Cquin); 123.5; 125.4 (Cquin); 128.9 (Cquin); 130.4 (CAr); 132.4 (CAr); 134.9; 149.1; 149.5; 151.9 (Cquin); 159.5; 164.6 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3238 (N–H); 1637 (C
N). IR (KBr) v, cm−1: 3238 (N–H); 1637 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1615 (C
N); 1615 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nquin). ESI-MS+: m/z 374.0824 ([M + H]+). Anal. Calcd for C19H17Cl2N3O: C 60.97; H 4.58%; N 11.23%, found: C 61.11; H 4.88; N 11.33%.
Nquin). ESI-MS+: m/z 374.0824 ([M + H]+). Anal. Calcd for C19H17Cl2N3O: C 60.97; H 4.58%; N 11.23%, found: C 61.11; H 4.88; N 11.33%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 8.50 (1H, d, 3JH,H = 5.32, Ar–H); 13.69 (1H, br s, OH). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 29.9 (CH2); 41.2 (CH2); 56.2 (OMe); 56.9 (CH2); 99.1 (Cquin); 114.3 (CAr); 117.2; 118.3 (CAr); 118.5; 120.9 (Cquin); 122.9 (CAr); 125.3 (Cquin); 128.8 (Cquin); 134.9; 148.5; 149.2; 149.5; 151.5; 152.0 (Cquin); 165.9 (HC
N); 8.50 (1H, d, 3JH,H = 5.32, Ar–H); 13.69 (1H, br s, OH). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 29.9 (CH2); 41.2 (CH2); 56.2 (OMe); 56.9 (CH2); 99.1 (Cquin); 114.3 (CAr); 117.2; 118.3 (CAr); 118.5; 120.9 (Cquin); 122.9 (CAr); 125.3 (Cquin); 128.8 (Cquin); 134.9; 148.5; 149.2; 149.5; 151.5; 152.0 (Cquin); 165.9 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3424 (N–H); 1631 (C
N). IR (KBr) v, cm−1: 3424 (N–H); 1631 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1611 (C
N); 1611 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nquin). ESI-MS+: m/z 370.1319 ([M + H]+). Anal. Calcd for C20H20ClN3O2·2H2O: C 59.19; H 5.96; N 10.35%, found: C 59.13; H 5.59; N 10.60%.
Nquin). ESI-MS+: m/z 370.1319 ([M + H]+). Anal. Calcd for C20H20ClN3O2·2H2O: C 59.19; H 5.96; N 10.35%, found: C 59.13; H 5.59; N 10.60%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 8.52 (1H, d, 3JH,H = 5.34, Ar–H); 13.21 (1H, br s, OH). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 30.1 (CH2); 41.5 (CH2); 57.4 (CH2); 99.2 (Cquin); 117.1 (CAr); 117.2; 118.7; 118.8 (CAr); 120.7 (Cquin); 125.4 (Cquin); 129.0 (Cquin); 131.3 (CAr); 132.5 (CAr); 134.9; 149.4; 149.5; 152.0 (Cquin); 161.0; 165.8 (HC
N); 8.52 (1H, d, 3JH,H = 5.34, Ar–H); 13.21 (1H, br s, OH). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 30.1 (CH2); 41.5 (CH2); 57.4 (CH2); 99.2 (Cquin); 117.1 (CAr); 117.2; 118.7; 118.8 (CAr); 120.7 (Cquin); 125.4 (Cquin); 129.0 (Cquin); 131.3 (CAr); 132.5 (CAr); 134.9; 149.4; 149.5; 152.0 (Cquin); 161.0; 165.8 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3260 (N–H); 1634 (C
N). IR (KBr) v, cm−1: 3260 (N–H); 1634 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1613 (C
N); 1613 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nquin). EI-MS+: m/z 339 ([M]+). Anal. Calcd for C19H18ClN3O: C 67.15; H 5.33; N 12.37%, found: C 66.90; H 5.07; N 12.70%.
Nquin). EI-MS+: m/z 339 ([M]+). Anal. Calcd for C19H18ClN3O: C 67.15; H 5.33; N 12.37%, found: C 66.90; H 5.07; N 12.70%.
General procedure for the synthesis of 4-amino-7-chloroquinoline bis-imines 6–8
N-(7-Chloroquinolin-4-yl)-tris(2-aminoethyl)amine (5) was dissolved in EtOH (5 ml). To this, the appropriate aldehyde in EtOH (10 ml) was added and the resulting solution stirred at room temperature for 16 hours. The solvent was reduced under pressure and the product precipitated with diethyl ether. The resulting solid was then dried in vacuo.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 8.49 (1H, d, 3JH,H = 5.22, Ar–H); 13.59 (2H, s, OH). 13C{1H} NMR (100.64 MHz, DMSO-d6) δ, ppm: 41.3 (CH2); 52.6 (CH2); 54.9 (CH2); 56.4 (CH2); 99.2 (Cquin); 117.5; 119.4 (CAr); 119.5; 121.7; 124.0 (Cquin); 124.8 (Cquin); 127.1 (Cquin); 130.8 (CAr); 132.5 (CAr); 134.3; 148.3; 150.8; 151.3 (Cquin); 161.3; 165.6 (HC
N); 8.49 (1H, d, 3JH,H = 5.22, Ar–H); 13.59 (2H, s, OH). 13C{1H} NMR (100.64 MHz, DMSO-d6) δ, ppm: 41.3 (CH2); 52.6 (CH2); 54.9 (CH2); 56.4 (CH2); 99.2 (Cquin); 117.5; 119.4 (CAr); 119.5; 121.7; 124.0 (Cquin); 124.8 (Cquin); 127.1 (Cquin); 130.8 (CAr); 132.5 (CAr); 134.3; 148.3; 150.8; 151.3 (Cquin); 161.3; 165.6 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3312 (N–H); 1648 (C
N). IR (KBr) v, cm−1: 3312 (N–H); 1648 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1609 (C
N); 1609 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nquin). ESI-MS+: m/z 584.1404 ([M + H]+); 292.5737 ([M + 2H]2+). Anal. Calcd for C29H28Cl3N5O2·2H2O: C 56.09; H 5.19; N 11.28%, found: C 55.66; H 4.96; N 11.21%.
Nquin). ESI-MS+: m/z 584.1404 ([M + H]+); 292.5737 ([M + 2H]2+). Anal. Calcd for C29H28Cl3N5O2·2H2O: C 56.09; H 5.19; N 11.28%, found: C 55.66; H 4.96; N 11.21%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 8.30 (1H, d, 3JH,H = 6.13, Ar–H). 13C{1H} NMR (100.64 MHz, DMSO-d6) δ, ppm: 41.9 (CH2); 52.9 (CH2); 55.2 (CH2); 55.9 (CH2); 56.4 (OMe); 99.2 (Cquin); 115.6 (CAr); 116.8; 117.2 (CAr); 118.5; 123.6 (Cquin); 123.8 (CAr); 125.2 (Cquin); 125.8 (Cquin); 136.1; 147.4; 147.8; 149.0; 153.2; 154.3 (Cquin); 166.7 (HC
N); 8.30 (1H, d, 3JH,H = 6.13, Ar–H). 13C{1H} NMR (100.64 MHz, DMSO-d6) δ, ppm: 41.9 (CH2); 52.9 (CH2); 55.2 (CH2); 55.9 (CH2); 56.4 (OMe); 99.2 (Cquin); 115.6 (CAr); 116.8; 117.2 (CAr); 118.5; 123.6 (Cquin); 123.8 (CAr); 125.2 (Cquin); 125.8 (Cquin); 136.1; 147.4; 147.8; 149.0; 153.2; 154.3 (Cquin); 166.7 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3414 (N–H); 1635 (C
N). IR (KBr) v, cm−1: 3414 (N–H); 1635 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1613 (C
N); 1613 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nquin). EI-MS+: m/z 575 ([M]+). Anal. Calcd for C31H34ClN5O4·2.5H2O: C 59.95; H 6.33; N 11.11%, found: C 59.52; H 6.05, N 11.53%.
Nquin). EI-MS+: m/z 575 ([M]+). Anal. Calcd for C31H34ClN5O4·2.5H2O: C 59.95; H 6.33; N 11.11%, found: C 59.52; H 6.05, N 11.53%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 13C{1H} NMR (100.64 MHz, DMSO-d6) δ, ppm: 41.9 (CH2); 52.4 (CH2); 54.9 (CH2); 56.6 (CH2); 99.0 (Cquin); 117.0 (CAr); 118.5 (CAr); 118.8; 121.0; 125.7 (Cquin); 126.5 (Cquin); 129.6 (Cquin); 131.9 (CAr); 132.7 (CAr); 136.8; 137.4; 146.6; 154.7 (Cquin); 161.6; 166.8 (HC
N). 13C{1H} NMR (100.64 MHz, DMSO-d6) δ, ppm: 41.9 (CH2); 52.4 (CH2); 54.9 (CH2); 56.6 (CH2); 99.0 (Cquin); 117.0 (CAr); 118.5 (CAr); 118.8; 121.0; 125.7 (Cquin); 126.5 (Cquin); 129.6 (Cquin); 131.9 (CAr); 132.7 (CAr); 136.8; 137.4; 146.6; 154.7 (Cquin); 161.6; 166.8 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3414 (N–H); 1633 (C
N). IR (KBr) v, cm−1: 3414 (N–H); 1633 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1615 (C
N); 1615 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nquin). EI-MS+: m/z 515 ([M]+). Anal. Calcd for C29H30ClN5O2·2.5H2O: C 62.08; H 6.29; N 12.48%, found: C 62.00; H 5.89; N 12.68%.
Nquin). EI-MS+: m/z 515 ([M]+). Anal. Calcd for C29H30ClN5O2·2.5H2O: C 62.08; H 6.29; N 12.48%, found: C 62.00; H 5.89; N 12.68%.
General procedure to prepare 4-amino-7-chloroquinoline mono-imine Rh(I) complexes 9–11
The appropriate ligand was dissolved in dry DCM (30 ml) and to this NaH was added in excess. The mixture was stirred at room temperature for 1.5 hours under argon. A solution of [RhCl(COD)]2 in DCM (5 ml) was then added dropwise to the mixture and stirred for 3 hours at room temperature. After this time, water (20 ml) was added to the reaction mixture and the organic extract washed with water (3 × 20 ml). The organic layer was dried over anhydrous MgSO4. The drying agent was removed by filtration and the solvent of the resulting filtrate reduced under vacuum. The product was precipitated with n-hexane and filtered using a Büchner funnel.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 4.54 (2H, m, CH
CH (COD)); 4.54 (2H, m, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 5.08 (1H, br s, NH); 6.41 (1H, d, 3JH,H = 5.25, Ar–H); 6.78 (1H, d, 3JH,H = 9.04, Ar–H); 7.05 (1H, d, 4JH,H = 2.72, Ar–H); 7.21 (1H, dd, 4JH,H = 2.83, 3JH,H = 9.36, Ar–H); 7.34 (1H, dd, 4JH,H = 2.13, 3JH–H = 8.98, Ar–H); 7.64 (1H, d, 3JH,H = 8.90, Ar–H); 7.87 (1H, s, HC
CH (COD)); 5.08 (1H, br s, NH); 6.41 (1H, d, 3JH,H = 5.25, Ar–H); 6.78 (1H, d, 3JH,H = 9.04, Ar–H); 7.05 (1H, d, 4JH,H = 2.72, Ar–H); 7.21 (1H, dd, 4JH,H = 2.83, 3JH,H = 9.36, Ar–H); 7.34 (1H, dd, 4JH,H = 2.13, 3JH–H = 8.98, Ar–H); 7.64 (1H, d, 3JH,H = 8.90, Ar–H); 7.87 (1H, s, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 7.97 (1H, d, 4JH,H = 2.03, Ar–H); 8.56 (1H, d, 3JH,H = 5.18, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.8 (COD); 31.6 (COD); 32.7 (CH2); 40.9 (CH2); 57.2 (CH2); 71.7 (d, 1JRh–C = 14.09, COD); 86.0 (d, 1JRh–C = 11.86, COD); 99.2 (Cquin); 117.2; 118.4; 119.5; 121.0 (Cquin); 123.2 (CAr); 125.5 (Cquin); 128.9 (Cquin); 132.9 (CAr); 134.8 (CAr); 135.0; 149.2; 149.3; 152.0 (Cquin); 158.2; 164.9 (HC
N); 7.97 (1H, d, 4JH,H = 2.03, Ar–H); 8.56 (1H, d, 3JH,H = 5.18, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.8 (COD); 31.6 (COD); 32.7 (CH2); 40.9 (CH2); 57.2 (CH2); 71.7 (d, 1JRh–C = 14.09, COD); 86.0 (d, 1JRh–C = 11.86, COD); 99.2 (Cquin); 117.2; 118.4; 119.5; 121.0 (Cquin); 123.2 (CAr); 125.5 (Cquin); 128.9 (Cquin); 132.9 (CAr); 134.8 (CAr); 135.0; 149.2; 149.3; 152.0 (Cquin); 158.2; 164.9 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3435 (N–H); 1609 (C
N). IR (KBr) v, cm−1: 3435 (N–H); 1609 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). EI-MS+: m/z 583 ([M]+). Anal. Calcd for C27H28Cl2N3ORh: C 55.50; H 4.83; N 7.19%, found: C 55.15; H 5.34; N 7.54%.
N). EI-MS+: m/z 583 ([M]+). Anal. Calcd for C27H28Cl2N3ORh: C 55.50; H 4.83; N 7.19%, found: C 55.15; H 5.34; N 7.54%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 3.81 (3H, s, OMe) 4.66 (2H, m, CH
CH (COD)); 3.81 (3H, s, OMe) 4.66 (2H, m, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 5.14 (1H, br s, NH); 6.38 (1H, d, 3JH,H = 5.33, Ar–H); 6.46 (1H, t, 3JH,H = 7.80, Ar–H); 6.73 (1H, dd, 4JH,H = 1.55, 3JH,H = 8.14, Ar–H); 6.85 (1H, dd, 2JH,H = 1.58, 3JH,H = 7.52, Ar–H); 7.34 (1H, dd, 4JH,H = 2.05, 3JH,H = 8.87, Ar–H); 7.66 (1H, d, 3JH,H = 8.95, Ar–H); 7.95 (2H, m, HC
CH (COD)); 5.14 (1H, br s, NH); 6.38 (1H, d, 3JH,H = 5.33, Ar–H); 6.46 (1H, t, 3JH,H = 7.80, Ar–H); 6.73 (1H, dd, 4JH,H = 1.55, 3JH,H = 8.14, Ar–H); 6.85 (1H, dd, 2JH,H = 1.58, 3JH,H = 7.52, Ar–H); 7.34 (1H, dd, 4JH,H = 2.05, 3JH,H = 8.87, Ar–H); 7.66 (1H, d, 3JH,H = 8.95, Ar–H); 7.95 (2H, m, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, Ar–H); 8.53 (1H, d, 3JH,H = 5.30, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.8 (COD); 31.7 (COD); 32.7 (CH2); 40.9 (CH2); 56.7 (CH2); 57.0 (OMe); 71.2 (d, 1JRh,C = 14.12, COD); 85.5 (d, 1JRh,C = 11.78, COD); 99.1 (Cquin); 113.6 (CAr); 115.8 (CAr); 117.3; 119.1; 121.2 (Cquin); 125.5 (Cquin); 126.8 (C-8′); 128.8 (Cquin); 132.2; 134.9; 149.2; 149.3; 151.2; 152.0 (Cquin); 165.8 (HC
N, Ar–H); 8.53 (1H, d, 3JH,H = 5.30, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.8 (COD); 31.7 (COD); 32.7 (CH2); 40.9 (CH2); 56.7 (CH2); 57.0 (OMe); 71.2 (d, 1JRh,C = 14.12, COD); 85.5 (d, 1JRh,C = 11.78, COD); 99.1 (Cquin); 113.6 (CAr); 115.8 (CAr); 117.3; 119.1; 121.2 (Cquin); 125.5 (Cquin); 126.8 (C-8′); 128.8 (Cquin); 132.2; 134.9; 149.2; 149.3; 151.2; 152.0 (Cquin); 165.8 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3439 (N–H); 1613 (C
N). IR (KBr) v, cm−1: 3439 (N–H); 1613 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). EI-MS+: m/z 579 ([M]+). Anal. Calcd for C28H31ClN3O2Rh·0.5H2O: C 57.10; H 5.48; N 7.25%, found: C 57.19; H 6.11; N 7.36%.
N). EI-MS+: m/z 579 ([M]+). Anal. Calcd for C28H31ClN3O2Rh·0.5H2O: C 57.10; H 5.48; N 7.25%, found: C 57.19; H 6.11; N 7.36%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 4.54 (2H, m, CH
CH (COD)); 4.54 (2H, m, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 5.12 (1H, br s, NH); 6.39 (1H, d, 3JH,H = 5.36, Ar–H); 6.53 (1H, t, 3JH,H = 7.88, Ar–H); 6.85 (1H, d, 3JH,H = 8.51, Ar–H); 7.09 (1H, dd, 4JH,H = 1.76, 3JH,H = 7.93, Ar–H); 7.27–7.34 (2H, m, Ar–H); 7.65 (1H, d, 3JH,H = 8.98, Ar–H); 7.94 (2H, m, HC
CH (COD)); 5.12 (1H, br s, NH); 6.39 (1H, d, 3JH,H = 5.36, Ar–H); 6.53 (1H, t, 3JH,H = 7.88, Ar–H); 6.85 (1H, d, 3JH,H = 8.51, Ar–H); 7.09 (1H, dd, 4JH,H = 1.76, 3JH,H = 7.93, Ar–H); 7.27–7.34 (2H, m, Ar–H); 7.65 (1H, d, 3JH,H = 8.98, Ar–H); 7.94 (2H, m, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, Ar–H); 8.54 (1H, d, 3JH,H = 5.29, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.8 (COD); 31.7 (COD); 32.7 (CH2); 40.9 (CH2); 57.0 (CH2); 71.3 (d, 1JRh,C = 14.08, COD); 85.7 (d, 1JRh,C = 11.96, COD); 99.1 (Cquin); 114.6 (CAr); 117.3; 119.0; 121.1 (Cquin); 121.6 (CAr); 125.5 (Cquin); 128.9 (Cquin); 134.9 (CAr); 135.0 (CAr); 149.2; 149.3; 152.1 (Cquin); 166.0 (HC
N, Ar–H); 8.54 (1H, d, 3JH,H = 5.29, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.8 (COD); 31.7 (COD); 32.7 (CH2); 40.9 (CH2); 57.0 (CH2); 71.3 (d, 1JRh,C = 14.08, COD); 85.7 (d, 1JRh,C = 11.96, COD); 99.1 (Cquin); 114.6 (CAr); 117.3; 119.0; 121.1 (Cquin); 121.6 (CAr); 125.5 (Cquin); 128.9 (Cquin); 134.9 (CAr); 135.0 (CAr); 149.2; 149.3; 152.1 (Cquin); 166.0 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 166.4. IR (KBr) v, cm−1: 3431 (N–H); 1607 (C
N); 166.4. IR (KBr) v, cm−1: 3431 (N–H); 1607 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). EI-MS+: m/z 549 ([M]+). Anal. Calcd for C27H29ClN3ORh: C 58.97; H 5.32, N 7.64%, found: C 58.61; H 5.64; N 7.30%.
N). EI-MS+: m/z 549 ([M]+). Anal. Calcd for C27H29ClN3ORh: C 58.97; H 5.32, N 7.64%, found: C 58.61; H 5.64; N 7.30%.
General procedure to prepare 4-amino-7-chloroquinoline bis-imine Rh(I) complexes 12–14
The appropriate ligand was dissolved in dry DCM (30 ml) and to this NaH was added in excess. The mixture was stirred at room temperature for 1.5 hours under argon. A solution of [RhCl(COD)]2 in DCM (5 ml) was then added dropwise to the mixture and stirred for 5–6 hours at room temperature. After this time, water (20 ml) was added to the reaction mixture and the organic extract washed with water (3 × 20 ml). The organic layer was dried over anhydrous MgSO4. The drying agent was removed by filtration and the solvent of the resulting filtrate reduced under vacuum. The product was precipitated with n-hexane and filtered using a Büchner funnel.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 4.52 (4H, m, CH
CH (COD)); 4.52 (4H, m, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 5.23 (1H, br s, NH); 6.37 (1H, d, 3JH,H = 5.52, Ar–H); 6.72 (2H, d, 3JH,H = 9.02, Ar–H); 7.03 (2H, d, 4JH,H = 2.61, Ar–H); 7.18 (2H, dd, 4JH,H = 2.64, 3JH,H = 9.05, Ar–H) 7.26–7.31 (2H, m, Ar–H); 7.71 (2H, s, HC
CH (COD)); 5.23 (1H, br s, NH); 6.37 (1H, d, 3JH,H = 5.52, Ar–H); 6.72 (2H, d, 3JH,H = 9.02, Ar–H); 7.03 (2H, d, 4JH,H = 2.61, Ar–H); 7.18 (2H, dd, 4JH,H = 2.64, 3JH,H = 9.05, Ar–H) 7.26–7.31 (2H, m, Ar–H); 7.71 (2H, s, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 7.93 (1H, m, Ar–H); 8.53 (1H, d, 3JH,H = 5.01, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.9 (COD); 31.7 (COD); 40.9 (CH2); 53.9 (CH2); 57.9 (CH2); 58.2 (CH2); 71.5 (d, 1JRh,C = 14.03, COD); 86.1 (d, 1JRh,C = 11.53, COD); 99.3 (Cquin); 118.6; 119.5; 120.7 (Cquin); 120.9; 123.30 (CAr); 125.7 (Cquin); 128.9 (Cquin); 132.8 (CAr); 134.9 (CAr); 135.0; 149.2; 149.3; 151.9 (Cquin); 159.2; 164.8 (HC
N); 7.93 (1H, m, Ar–H); 8.53 (1H, d, 3JH,H = 5.01, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.9 (COD); 31.7 (COD); 40.9 (CH2); 53.9 (CH2); 57.9 (CH2); 58.2 (CH2); 71.5 (d, 1JRh,C = 14.03, COD); 86.1 (d, 1JRh,C = 11.53, COD); 99.3 (Cquin); 118.6; 119.5; 120.7 (Cquin); 120.9; 123.30 (CAr); 125.7 (Cquin); 128.9 (Cquin); 132.8 (CAr); 134.9 (CAr); 135.0; 149.2; 149.3; 151.9 (Cquin); 159.2; 164.8 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3427 (N–H); 1607 (C
N). IR (KBr) v, cm−1: 3427 (N–H); 1607 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). ESI-MS+ (HR): m/z 1004.1231 ([M + H]+ requires 1004.1218). Anal. Calcd for C45H50Cl3N5O2Rh2: C 53.88; H 5.02; N 6.98%, found: C 54.08; H 5.50; N 7.09%.
N). ESI-MS+ (HR): m/z 1004.1231 ([M + H]+ requires 1004.1218). Anal. Calcd for C45H50Cl3N5O2Rh2: C 53.88; H 5.02; N 6.98%, found: C 54.08; H 5.50; N 7.09%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 3.77 (6H, s, OMe); 4.64 (4H, m, CH
CH (COD)); 3.77 (6H, s, OMe); 4.64 (4H, m, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 5.19 (1H, br s, NH); 6.32 (1H, d, 3JH,H = 5.46, Ar–H); 6.46 (2H, t, 3JH,H = 7.75, Ar–H); 6.64 (2H, dd, 4JH,H = 1.42, 3JH,H = 8.07, Ar–H); 6.84 (2H, dd, 4JH,H = 1.54, 3JH,H = 7.58, Ar–H); 7.06–7.20 (2H, m, Ar–H); 7.75 (2H, s, HC
CH (COD)); 5.19 (1H, br s, NH); 6.32 (1H, d, 3JH,H = 5.46, Ar–H); 6.46 (2H, t, 3JH,H = 7.75, Ar–H); 6.64 (2H, dd, 4JH,H = 1.42, 3JH,H = 8.07, Ar–H); 6.84 (2H, dd, 4JH,H = 1.54, 3JH,H = 7.58, Ar–H); 7.06–7.20 (2H, m, Ar–H); 7.75 (2H, s, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 7.90 (1H, m, Ar–H); 8.48 (1H, d, 3JH,H = 5.15, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.9 (COD); 31.7 (COD); 41.1 (CH2); 53.6 (CH2); 56.6 (CH2); 58.1 (CH2, OMe); 71.0 (d, 1JRh,C = 14.18, COD); 85.5 (d, 1JRh,C = 11.68, COD); 99.1 (Cquin); 113.7 (CAr); 115.6 (CAr); 117.3; 119.1; 121.2 (Cquin); 125.3 (Cquin); 126.8 (CAr); 128.7 (Cquin); 134.8; 149.2; 149.4; 151.3; 151.9 (Cquin); 158.0; 165.6 (HC
N); 7.90 (1H, m, Ar–H); 8.48 (1H, d, 3JH,H = 5.15, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.9 (COD); 31.7 (COD); 41.1 (CH2); 53.6 (CH2); 56.6 (CH2); 58.1 (CH2, OMe); 71.0 (d, 1JRh,C = 14.18, COD); 85.5 (d, 1JRh,C = 11.68, COD); 99.1 (Cquin); 113.7 (CAr); 115.6 (CAr); 117.3; 119.1; 121.2 (Cquin); 125.3 (Cquin); 126.8 (CAr); 128.7 (Cquin); 134.8; 149.2; 149.4; 151.3; 151.9 (Cquin); 158.0; 165.6 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). IR (KBr) v, cm−1: 3362 (N–H); 1604 (C
N). IR (KBr) v, cm−1: 3362 (N–H); 1604 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). ESI-MS+ (HR): m/z 996.2199 ([M + H]+, requires 996.2209). Anal. Calcd for C47H56ClN5O4Rh2·H2O: C 55.66; H 5.76; N 6.90%, found: C 55.20; H 5.93; N 7.32%.
N). ESI-MS+ (HR): m/z 996.2199 ([M + H]+, requires 996.2209). Anal. Calcd for C47H56ClN5O4Rh2·H2O: C 55.66; H 5.76; N 6.90%, found: C 55.20; H 5.93; N 7.32%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 4.53 (4H, m, CH
CH (COD)); 4.53 (4H, m, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH (COD)); 5.28 (1H, br s, NH); 6.34 (1H, d, 3JH,H = 5.05, Ar–H); 6.53 (2H, t, 3JH,H = 7.47, Ar–H); 6.80 (2H, d, 3JH,H = 8.53, Ar–H); 7.01 (2H, d, 3JH,H = 7.64, Ar–H); 7.19–7.34 (4H, m, Ar–H); 7.76 (2H, s, HC
CH (COD)); 5.28 (1H, br s, NH); 6.34 (1H, d, 3JH,H = 5.05, Ar–H); 6.53 (2H, t, 3JH,H = 7.47, Ar–H); 6.80 (2H, d, 3JH,H = 8.53, Ar–H); 7.01 (2H, d, 3JH,H = 7.64, Ar–H); 7.19–7.34 (4H, m, Ar–H); 7.76 (2H, s, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 7.92 (1H, m, Ar–H); 8.51 (1H, d, 3JH,H = 5.64, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.9 (COD); 31.7 (COD); 40.9 (CH2); 53.6 (CH2); 57.9 (CH2); 58.1 (CH2); 71.1 (d, 1JRh,C = 13.69, COD); 85.7 (d, 1JRh,C = 11.67, COD); 99.2 (Cquin); 114.7 (CAr); 117.3; 119.0; 121.1 (Cquin); 121.7 (CAr); 125.4 (Cquin); 128.8 (Cquin); 134.9 (CAr); 149.2; 149.4; 152.0 (Cquin); 165.8 (HC
N); 7.92 (1H, m, Ar–H); 8.51 (1H, d, 3JH,H = 5.64, Ar–H). 13C{1H} NMR (100.64 MHz, CDCl3) δ, ppm: 28.9 (COD); 31.7 (COD); 40.9 (CH2); 53.6 (CH2); 57.9 (CH2); 58.1 (CH2); 71.1 (d, 1JRh,C = 13.69, COD); 85.7 (d, 1JRh,C = 11.67, COD); 99.2 (Cquin); 114.7 (CAr); 117.3; 119.0; 121.1 (Cquin); 121.7 (CAr); 125.4 (Cquin); 128.8 (Cquin); 134.9 (CAr); 149.2; 149.4; 152.0 (Cquin); 165.8 (HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 166.4. IR (KBr) v, cm−1: 3418 (N–H); 1606 (C
N); 166.4. IR (KBr) v, cm−1: 3418 (N–H); 1606 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). ESI-MS+ (HR): m/z 936.2004 ([M + H]+, requires 936.1998). Anal. Calcd for C45H52ClN5O2Rh2·2H2O: C 55.59; H 5.81; N 7.20%, found: C 55.81; H 5.80; N 6.89%.
N). ESI-MS+ (HR): m/z 936.2004 ([M + H]+, requires 936.1998). Anal. Calcd for C45H52ClN5O2Rh2·2H2O: C 55.59; H 5.81; N 7.20%, found: C 55.81; H 5.80; N 6.89%.
Antiplasmodial assay
Samples were screened in triplicate on one occasion against the chloroquine-sensitive NF54 strain and chloroquine-resistant K1 strains of Plasmodium falciparum. Continuous in vitro cultures of asexual erythrocyte stages of P. falciparum were maintained using a modified version of the method of Trager and Jensen.63 Quantitative assessment of antiplasmodial activity in vitro was determined via the parasite lactate dehydrogenase assay using a modified method of that described by Makler et al.64 The samples were prepared as a 20 mg ml−1 stock solution using DMSO and sonicated to enhance solubility. Samples were tested as a suspension if not completely dissolved. Stock solutions were stored at −20 °C. Further dilutions were prepared on the day of the experiment. Chloroquine was used as the reference drug in all experiments. A full dose–response measurement was performed for all compounds to determine the concentration inhibiting 50% of parasite growth (IC50 value). The samples were tested at a starting concentration of 1000 ng ml−1, which was then serially diluted 2-fold in complete medium to give 10 concentrations; with the lowest concentration being approximately 2 ng ml−1. The same dilution technique was used for all samples. The highest concentration of solvent to which the parasites were exposed to had no measurable effect on the parasite viability. The IC50 values were obtained using a non-linear dose–response curve fitting analysis via Graph Pad Prism v.4.0 software.β-Haematin inhibition assay
The β-haematin assay was adapted from the method described by Wright and co-workers.51 Compounds were prepared as a 10 mM stock solution in DMSO. The samples were tested at various concentrations between 5 and 500 μM. The stock solution was serially diluted to give 12 concentrations in a 96 well flat-bottom assay plate. NP-40 detergent was added to mediate the formation of β-haematin (305.5 μM). A 25 mM stock solution of haematin was prepared by dissolving haemin (16.3 mg) in dimethyl sulfoxide (1 ml). A 177.76 μl aliquot of haematin stock was suspended in 20 ml of a 2 M acetate buffer at pH 4.7. The suspension was then added to the plate to give a final haematin concentration of 100 μM. The plate was then incubated for 16 hours at 37 °C. The assay was analysed using the pyridine-ferrochrome method developed by Ncokazi and Egan.52 32 μl of a solution of 50% pyridine, 20% acetone, 20% water and 10% 2 M HEPES buffer (pH 7.4) was added to each well. To this, 60 μl acetone was added to each well and mixed. The absorbance of the resulting complex was measured at 405 nm on a SpectraMax 340PC plate reader. The IC50 values were obtained using a non-linear dose–response curve fitting analysis via Graph Pad Prism v.5.0 software.Cytotoxicity (MTT) assay
The oesophageal cancer cell line WHCO1, derived from a primary oesophageal squamous cell carcinoma, was provided by Professor Rob Veale (University of the Witwatersrand, Johannesburg, South Africa). IC50 determinations were carried out using an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay.65 3000 cells were seeded per well in 96-well plates. Plates were incubated at 37 °C under 5% CO2 (24 hours), after which aqueous DMSO solutions of each compound (10 μL, with a constant final concentration of DMSO of 0.2%) were plated at various concentrations. After 48 hours incubation, observations were made, and MTT (10 μL) solution added to each well. After 4 hours of incubation, solubilisation solution (100 μL) was added to each well, and incubated overnight. Plates were read at 595 nm on a BioTek microplate reader, and IC50 values calculated using Graph Pad Prism v.4.0. Package of GraphPad Software, San Diego, USA.Antitrichomonal assay
Cultures of T. vaginalis G3 isolate were grown in 5 ml complete TYM Diamond's media in a 37 °C incubator for 24 hours. 50 mM stock solutions of the compounds were made in DMSO and were screened. Cells untreated and inoculated with 5 μl DMSO were used as controls. 5 μl of 50 mM stocks of compound library were inoculated for a final concentration of 50 μM. Results were calculated based on counts utilising a haemocytometer after 24 hours.Acknowledgements
Financial support from the National Research Foundation (NRF) of South Africa, the University of Cape Town and the South African Medical Research Council (MRC) is gratefully acknowledged. Even though the work was supported by the MRC, the views and opinions expressed are not those of the MRC but of the authors of the material produced or published. Kirkwood M. Land acknowledges support from the University of the Pacific. Professor Ebbe Nordlander is gratefully acknowledged for helpful discussions.Notes and references
- The World Health Organisation, Malaria <http://www.int.features/factfiles/malaria>, (accessed 28 January 2011).
- The World Health Organisation, World Malaria Report 2014, http://www.who.int/malaria/media/world_malaria_report_2014/en/ (accessed 22 January 2015).
- P. L. Trigg and A. V. Kondrachine, in Malaria. Parasite biology, pathogenesis and protection, ed. I. W. Sherman, ASM Press, Washington, DC, 1998, p. 11 Search PubMed.
- M. Enserink, Science, 2010, 328, 844 CrossRef CAS PubMed.
- World Health Organisation, Global Plan for Artemisinin Resistance Containment (GPARC), WHO Press, Geneva, 2011 Search PubMed.
- A. M. Dondorp, F. Nosten, P. Yi, D. Das, A. P. Phyo, J. Tarning, K. M. Lwin, F. Ariey, W. Hanpithakpong, S. J. Lee, P. Ringwald, K. Silamut, M. Imwong, K. Chotivanich, P. Lim, T. Herdman, S. An, S. Yeung, P. Singhasivanon, N. P. J. Day, N. Lindegardh, D. Socheat and N. J. N. White, New Engl. J. Med., 2009, 361, 455 CrossRef CAS PubMed.
- C. Biot, G. Glorian, L. A. Maciejewski, J. S. Brocard, O. Domarle, G. Blampain, P. Millet, A. J. Georges, H. Abessolo and D. Dive, J. Med. Chem., 1997, 40, 3715 CrossRef CAS PubMed.
- F. Dubar, J. Khalife, J. Brocard, D. Dive and C. Biot, Molecules, 2008, 13, 2900 CrossRef CAS PubMed.
- F. Dubar, T. J. Egan, B. Pradines, D. Kuter, K. K. Ncokazi, D. Forge, J. F. Paul, C. Pierrot, H. Kalamou, J. Khalife, E. Buisine, C. Rogier, H. Vezin, I. Forfar, C. Slomianny, X. Trivelli, S. Kapishnikov, L. Leiserowitz, D. Dive and C. Biot, Chem. Biol., 2011, 6, 275 CAS.
- G. Mombo-Ngoma, C. Supan, M. P. Dal-Bianco, M. A. Missinou, P.-B. Matsiegui, C. L. O. Salazar, S. Issifou, D. Ter-Minassian, M. Ramharter, M. Kombila, P. G. Kremsner and B. Lell, Malar. J., 2011, 10, 53 CrossRef CAS PubMed.
- C. Biot, W. Castro, C. Y. Botte and M. Navarro, Dalton Trans., 2012, 41, 6335 RSC.
- M. Navarro, W. Castro and C. Biot, Organometallics, 2012, 31, 5715 CrossRef CAS.
- P. F. Salas, C. Herrmann and C. Orvig, Chem. Rev., 2013, 113, 3450 CrossRef CAS PubMed.
- R. A. Sánchez-Delgado, M. Navarro, H. Perez and J. A. Urbina, J. Med. Chem., 1996, 39, 1095 CrossRef PubMed.
- M. A. L. Blackie, P. Beagley, K. Chibale, C. Clarkson, J. R. Moss and P. J. Smith, J. Organomet. Chem., 2003, 688, 144 CrossRef CAS PubMed.
- L. Glans, A. Ehnbom, C. de Kock, A. Martínez, J. Estrada, P. J. Smith, M. Haukka, R. A. Sánchez-Delgado and E. Nordlander, Dalton Trans., 2012, 41, 2764 RSC.
- D. Petrin, K. Delgaty, R. Bhatt and G. Garber, Clin. Microbiol. Rev., 1998, 11, 300 CAS.
- Z. F. Zhang, S. Graham, S. Z. Yu, J. Marshall, M. Zielezny, Y. X. Chen, M. Sun, S. L. Tang, C. S. Liao, J. L. Xu and X. Z. Yang, Ann. Epidemiol., 1995, 5, 325 CrossRef CAS.
- C. M. Ryan, N. De Miguel and P. J. Johnson, Essays Biochem., 2011, 51, 161 CAS.
- S. Sutcliffe, E. Giovannucci, J. F. Alderete, T.-H. Chang, C. A. Gaydos, J. M. Zenilman, A. M. De Marzo, W. C. Willett and E. A. Platz, Cancer Epidemiol. Biomarkers Prev., 2006, 15, 939 CAS.
- P. Kissinger and A. Adamski, Sex. Transm. Infect., 2013, 89, 426 CrossRef PubMed.
- R. Anjaeyulu, S. A. Gupte and D. B. Desai, J. Int. Med. Res., 1977, 5, 438 CAS.
- K. Madhuri, M. S. Kumar and L. Kalyani, Int. J. Pharm. Chem. Biol. Sci., 2011, 1, 38 CAS.
- S. L. Cudmore, K. L. Delgaty, S. F. Hayword-McClelland, D. P. Petrin and G. E. Garber, Clin. Microbiol. Rev., 2004, 17, 783 CrossRef CAS PubMed.
- J. R. Schwebke and D. Burgess, Clin. Microbiol. Rev., 2004, 17, 794 CrossRef PubMed.
- P. Chellan, T. Stringer, A. Shokar, P. J. Dornbush, G. Vazquez-Anaya, K. M. Land, K. Chibale and G. S. Smith, J. Inorg. Biochem., 2011, 105, 1562 CrossRef CAS PubMed.
- M. Adams, Y. Li, H. Khot, C. de Kock, P. J. Smith, K. M. Land, K. Chibale and G. S. Smith, Dalton Trans., 2013, 42, 4677 RSC.
- T. Stringer, D. Taylor, C. de Kock, H. Guzgay, A. Au, S. H. An, B. Sanchez, R. O'Connor, N. Patel, K. M. Land, P. J. Smith, D. T. Hendricks, T. J. Egan and G. S. Smith, Eur. J. Med. Chem., 2013, 69, 90 CrossRef CAS PubMed.
- P. Govender, N. C. Antonels, J. Mattsson, A. K. Renfrew, P. J. Dyson, J. R. Moss, B. Therrien and G. S. Smith, J. Organomet. Chem., 2009, 694, 3470 CrossRef CAS PubMed.
- P. Govender, A. K. Renfrew, C. M. Clavel, P. J. Dyson, B. Therrien and G. S. Smith, Dalton Trans., 2011, 40, 1158 RSC.
- P. Govender, L. C. Sudding, C. M. Clavel, P. J. Dyson, B. Therrien and G. S. Smith, Dalton Trans., 2013, 42, 1267 RSC.
- S. D. Khanye, J. Gut, P. J. Rosenthal, K. Chibale and G. S. Smith, J. Organomet. Chem., 2011, 696, 3296 CrossRef CAS PubMed.
- P. Govender, H. Lemmerhirt, A. T. Hutton, B. Therrien, P. J. Bednarski and G. S. Smith, Organometallics, 2014, 33, 5535 CrossRef CAS.
- M. Auzias, B. Therrien, G. Süss-Fink, P. Stepnicka, W. H. Ang and P. J. Dyson, Inorg. Chem., 2008, 47, 578 CrossRef CAS PubMed.
- I. B. Muller, R. Das Gupta, K. Lüersen, C. Wrenger and R. D. Walter, Mol. Biochem. Parasitol., 2008, 160, 1 CrossRef PubMed.
- C. C. Musonda, S. Little, V. Yardley and K. Chibale, Bioorg. Med. Chem. Lett., 2007, 17, 4733 CrossRef CAS PubMed.
- D. P. Iwaniuk, E. D. Whetmore, N. Rosa, K. Ekoue-Kovi, J. Alumasa, A. C. de Dios, P. D. Roepe and C. Wolf, Bioorg. Med. Chem., 2009, 17, 6560 CrossRef CAS PubMed.
- T. J. Egan, R. Hunter, C. H. Kaschula, H. M. Marques, A. Misplon and J. Walden, J. Med. Chem., 2000, 43, 283 CrossRef CAS PubMed.
- G. Giordano and R. H. Crabtree, Inorg. Synth., 1990, 28, 88 CrossRef CAS PubMed.
- M. Enamullah, A. K. M. Royhan Uddin, G. Hogarth and C. Janiak, Inorg. Chim. Acta, 2012, 387, 173 CrossRef CAS PubMed.
- C. Janiak, A.-C. Chamayou, A. K. M. Royhan Uddin, M. Uddin, K. S. Hagen and M. Enamullah, Dalton Trans., 2009, 3698 RSC.
- M. Enamullah, A. K. M. Royhan Uddin, A.-C. Chamayou and C. Janial, Z. Naturforsch, B: Chem. Sci., 2007, 62, 807 CrossRef CAS.
- M. Enamullah, A. Sharmin, M. Hasegawa, T. Hoshi, A.-C. Chamayou and C. Janiak, Eur. J. Inorg. Chem., 2006, 2146 CrossRef CAS PubMed.
- M. Enamullah, M. Uddin and W. Linert, J. Coord. Chem., 2007, 60, 2309 CrossRef CAS PubMed.
- P. F. Salas, C. Herrmann, J. F. Cawthray, C. Nimphius, A. Kenkel, J. Chen, C. de Kock, P. J. Smith, B. O. Patrick, M. J. Adam and C. Orvig, J. Med. Chem., 2013, 56, 1596 CrossRef CAS PubMed.
- Y. Li, C. de Kock, P. J. Smith, K. Chibale and G. S. Smith, Organometallics, 2014, 33, 4345 CrossRef CAS.
- Y. Li, C. de Kock, P. J. Smith, H. Guzgay, D. T. Hendricks, K. Naran, V. Mizrahi, D. F. Warner, K. Chibale and G. S. Smith, Organometallics, 2013, 32, 141 CrossRef CAS.
- C. Biot, N. Chavain, F. Dubar, B. Pradines, X. Trivelli, J. Brocard, I. Forfar and D. Dive, J. Organomet. Chem., 2009, 694, 845 CrossRef CAS PubMed.
- N. Sunduru, K. Srivastava, S. Rajakumar, S. K. Puri, J. K. Saxena and P. M. S. Chauhan, Bioorg. Med. Chem. Lett., 2009, 19, 2570 CrossRef CAS PubMed.
- R. Buller, M. L. Peterson, O. Almarrson and L. Leiserowitz, Cryst. Growth Des., 2002, 2, 553 CAS.
- R. D. Sandlin, M. D. Carter, P. J. Lee, J. M. Auschwitz, S. E. Leed, J. D. Johnson and D. W. Wright, Antimicrob. Agents Chemother., 2011, 55, 3363 CrossRef CAS PubMed.
- K. K. Ncokazi and T. J. Egan, Anal. Biochem., 2005, 338, 306 CrossRef CAS PubMed.
- A. N. Hoang, K. K. Ncokazi, K. A. de Villiers, D. W. Wright and T. J. Egan, Dalton Trans., 2010, 39, 1235 RSC.
- A. N. Hoang, R. D. Sandlin, A. Omar, T. J. Egan and D. W. Wright, Biochemistry, 2010, 49, 10107 CrossRef CAS PubMed.
- C. H. Kaschula, T. J. Egan, R. Hunter, N. Basilico, S. Parapini, D. Taramelli, E. Pasini and D. Monti, J. Med. Chem., 2002, 45, 3531 CrossRef CAS PubMed.
- J. Rajput, J. R. Moss, A. T. Hutton, D. T. Hendricks, C. E. Arendse and C. Imrie, J. Organomet. Chem., 2004, 689, 1553 CrossRef CAS PubMed.
- C. Wang, J. G. Delcros, L. Cannon, F. Konate, H. Carias, J. Biggerstaff, R. A. Gardner and O. Phanstiel, J. Med. Chem., 2003, 46, 5129 CrossRef CAS PubMed.
- Z. Tian, S. Xie, Z. Mei, J. Zhao, W. Gao and C. Wang, Org. Biomol. Chem., 2009, 7, 4651 CAS.
- A. R. Martirosyan, R. Rahim-Bata, A. B. Freeman, C. D. Clarke, R. L. Howard and J. S. Strobl, Biochem. Pharmacol., 2004, 68, 1729 CrossRef CAS PubMed.
- I. S. Adagu, D. Nolder, D. C. Warhurst and J.-F. Rossignol, J. Antimicrob. Chemother., 2002, 49, 103 CrossRef CAS PubMed.
- C. C. Musonda, G. A. Whitlock, M. J. Witty, R. Brun and M. Kaiser, Bioorg. Med. Chem. Lett., 2009, 19, 401 CrossRef CAS PubMed.
- S. K. Katiyar and T. D. Edlind, Antimicrob. Agents Chemother., 1991, 35, 2198 CrossRef CAS.
- W. Trager and J. B. Jensen, Science, 1976, 193, 673 CAS.
- M. T. Makler, J. M. Ries, J. A. Williams, J. E. Bancroft, R. C. Piper, B. L. Gibbins and D. J. Hinrichs, Am. J. Trop. Med. Hyg., 1993, 48, 739 CAS.
- J. van Meerloo, G. J. L. Kaspers and J. Cloos, in Cancer Cell Culture: Methods and Protocols (Methods in Molecular Biology), ed. I. A. Cree, Humana Press, 2011, p. 237 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5dt02378e |
| This journal is © The Royal Society of Chemistry 2015 |