Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
O,N,N-Pincer ligand effects on oxidatively induced carbon–chlorine coupling reactions at palladium†
Luka A.
Wright
a,
Eric G.
Hope
a,
Gregory A.
Solan
*a,
Warren B.
Cross
ab and
Kuldip
Singh
a
aDepartment of Chemistry, University of Leicester, University Road, Leicester LE1 7RH, UK. E-mail: gas8@leicester.ac.uk
bSchool of Science and Technology, Nottingham Trent University, Clifton Lane, Nottingham NG11 8NS, UK
First published on 18th February 2015
Abstract
The syntheses of two families of sterically tuneable O,N,N pro-ligands are reported, namely the 2-(phenyl-2′-ol)-6-imine-pyridines, 2-(C6H4-2′-OH),6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NAr)C5H3N [Ar = 4-i-PrC6H4 (HL1a), 2,6-i-Pr2C6H3 (HL1b)] and the 2-(phenyl-2′-ol)-6-(amino-prop-2-yl)pyridines, 2-(C6H4-2′-OH),6-(CMe2NHAr)C5H3N [Ar = 4-i-PrC6H4 (HL2a), 2,6-i-Pr2C6H3 (HL2b)], using straightforward synthetic approaches and in reasonable overall yields. Interaction of HL1a/c and HL2a/b with palladium(II) acetate affords the O,N,N-pincer complexes, [{2-(C6H4-2′-O)-6-(CMe
NAr)C5H3N [Ar = 4-i-PrC6H4 (HL1a), 2,6-i-Pr2C6H3 (HL1b)] and the 2-(phenyl-2′-ol)-6-(amino-prop-2-yl)pyridines, 2-(C6H4-2′-OH),6-(CMe2NHAr)C5H3N [Ar = 4-i-PrC6H4 (HL2a), 2,6-i-Pr2C6H3 (HL2b)], using straightforward synthetic approaches and in reasonable overall yields. Interaction of HL1a/c and HL2a/b with palladium(II) acetate affords the O,N,N-pincer complexes, [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (1a), 2,6-i-Pr2C6H3 (1b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (2a), 2,6-i-Pr2C6H3 (2b)), which can be readily converted to their chloride derivatives, [{2-(C6H4-2′-O)-6-(CMe
NAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (1a), 2,6-i-Pr2C6H3 (1b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (2a), 2,6-i-Pr2C6H3 (2b)), which can be readily converted to their chloride derivatives, [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (3a), 2,6-i-Pr2C6H3 (3b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (4a), 2,6-i-Pr2C6H3 (4b)), respectively, on reaction with an aqueous sodium chloride solution. Treating each of 3a, 3b, 4a and 4b with two equivalents of di-p-tolyliodonium triflate at 100 °C in a toluene/acetonitrile mixture affords varying amounts of 4-chlorotoluene along with the 4-iodotoluene by-product with the conversions highly dependent on the steric and backbone properties of the pincer complex employed (viz.4a > 3a > 4b > 3b); notably, the least sterically bulky and most flexible amine-containing 4a reaches 90% conversion to 4-chlorotoluene in 15 h as opposed to 17% for imine-containing 3b. In the case of 3a, the inorganic palladium species recovered from the reaction has been identified as the Pd(II) salt [{2-(C6H4-2′-O)-6-(CMe
NAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (3a), 2,6-i-Pr2C6H3 (3b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (4a), 2,6-i-Pr2C6H3 (4b)), respectively, on reaction with an aqueous sodium chloride solution. Treating each of 3a, 3b, 4a and 4b with two equivalents of di-p-tolyliodonium triflate at 100 °C in a toluene/acetonitrile mixture affords varying amounts of 4-chlorotoluene along with the 4-iodotoluene by-product with the conversions highly dependent on the steric and backbone properties of the pincer complex employed (viz.4a > 3a > 4b > 3b); notably, the least sterically bulky and most flexible amine-containing 4a reaches 90% conversion to 4-chlorotoluene in 15 h as opposed to 17% for imine-containing 3b. In the case of 3a, the inorganic palladium species recovered from the reaction has been identified as the Pd(II) salt [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N(4-i-PrC6H4)C5H3N}Pd(NCMe)][O3SCF3] (5a), which was independently prepared by the reaction of 3a with silver triflate in acetonitrile. Single crystal X-ray structures are reported for HL1a, HL2a, 1a, 1b, 2a, 2b, 3a and 5a.
N(4-i-PrC6H4)C5H3N}Pd(NCMe)][O3SCF3] (5a), which was independently prepared by the reaction of 3a with silver triflate in acetonitrile. Single crystal X-ray structures are reported for HL1a, HL2a, 1a, 1b, 2a, 2b, 3a and 5a.
Introduction
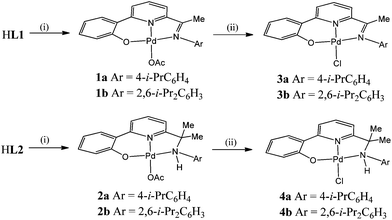
While hypervalent iodine salts of the type [Ar2I][X] (X = OTf, BF4) have been widely used in Pd(0)/(II) cross coupling reactions,1 their application in Pd(II)/(IV) and/or Pd(II)/(III) chemistry has only started to emerge over the last decade.2,3 With regard to the Pd(II)/(IV) couple, stable palladium(IV) species have been characterised,4 computationally modelled5 and highlight the ability of the I(III) reagent to transfer an “Ar+” group to the palladium(II) centre; decomposition can ensue via reductive elimination of an aryl-containing product. The chlorination of Pd(II)–C and Pd(II)–Cl containing complexes with PhICl2 represents another transformation that has been more extensively studied and these reactions are considered to proceed via a facile C–Cl bond forming reductive elimination from a Pd(IV) intermediate.6,7 For example, van Koten has spectroscopically characterised a transient Pd(IV) species from the reaction of an Pd(II) chloride N,Cph,N-pincer complex with PhICl2, which is presumed to then undergo C–Cl bond forming reductive elimination with the phenyl moiety of the pincer ligand.8 Indeed, a variety of pincer ligand frameworks including symmetrical (e.g., N,C,N5,8) and unsymmetrical (e.g., C,N,N,9O,N,C,10O,N,N11) variations have proved conducive to promoting the formation of related electron deficient Pd(IV) intermediates, a feature that is likely to be attributable to the electron supplying nature of the tridentate manifold.In this article we report the stoichiometric reactivity of a range of palladium(II) chloride O,N,N-pincer complexes towards di-p-tolyliodonium triflate with a view to monitoring the effect that the O,N,N-spectator ligand has on the anticipated formation of 4-chlorotoluene. In particular, we target two families of pyridine-based O,Npy,N pincers in order to investigate how structural features within their respective O,Npy,N ligand manifold influence the C–Cl bond forming process; the effects of imine (L1) vs. amine (L2) nitrogen donor and steric factors within the N-aryl group (Ar = 4-i-PrC6H4, 2,6-i-Pr2C6H3) will be examined (Fig. 1). Full details of the synthetic and characterisation data for the pro-ligands, 2-(phenyl-2′-ol)-6-ketimine-pyridines (HL1) and 2-(phenyl-2′-ol)-6-(amino-prop-2-yl)pyridines (HL2), will be reported as will the corresponding data for their palladium(II) acetate (1 and 2) and chloride (3, 4) complexes.
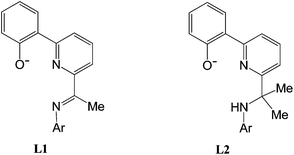 | ||
| Fig. 1 Monoanionic 2-(phenyl-2′-olate)-6-ketimine-pyridine (L1) and 2-(phenyl-2′-olate)-6-(amino-prop-2-yl)pyridine (L2) pincer ligands. | ||
Results and discussion
(a) Preparation of pro-ligands HL1 and HL2
The 2-(phenyl-2′-ol)-6-imine-pyridines, 2-(C6H4-2′-OH),6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NAr)C5H3N [Ar = 4-i-PrC6H4 (HL1a), 2,6-i-Pr2C6H3 (HL1b)], have been prepared in modest to good yield via sequential Suzuki coupling and condensation reactions from 2-hydroxyphenylboronic acid and 2-bromo-6-acetyl pyridine (Scheme 1). As a slight modification to the reported synthesis of ketone precursor, 2-(C6H4-2′-OH),6-(CMe
NAr)C5H3N [Ar = 4-i-PrC6H4 (HL1a), 2,6-i-Pr2C6H3 (HL1b)], have been prepared in modest to good yield via sequential Suzuki coupling and condensation reactions from 2-hydroxyphenylboronic acid and 2-bromo-6-acetyl pyridine (Scheme 1). As a slight modification to the reported synthesis of ketone precursor, 2-(C6H4-2′-OH),6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C5H3N, it was found that the cross coupling proceeds more efficiently and over a shorter reaction time using a catalyst composed of Pd(OAc)2 and PPh3 in a reaction vessel open to the air.12 Treatment of HL1a and HL1b with trimethylaluminium in toluene at elevated temperature followed by hydrolysis gave the 2-(phenyl-2′-ol)-6-(amino-prop-2-yl)pyridines, 2-(C6H4-2′-OH),6-(CMe2NHAr)C5H3N [Ar = 4-i-PrC6H4 (HL2a), 2,6-i-Pr2C6H3 (HL2b)], in good yield. The new compounds, HL1a, HL2a and HL2b, have been characterised by a combination of 1H, 13C{1H} NMR, IR spectroscopy and ESI mass spectrometry (see Experimental).
O)C5H3N, it was found that the cross coupling proceeds more efficiently and over a shorter reaction time using a catalyst composed of Pd(OAc)2 and PPh3 in a reaction vessel open to the air.12 Treatment of HL1a and HL1b with trimethylaluminium in toluene at elevated temperature followed by hydrolysis gave the 2-(phenyl-2′-ol)-6-(amino-prop-2-yl)pyridines, 2-(C6H4-2′-OH),6-(CMe2NHAr)C5H3N [Ar = 4-i-PrC6H4 (HL2a), 2,6-i-Pr2C6H3 (HL2b)], in good yield. The new compounds, HL1a, HL2a and HL2b, have been characterised by a combination of 1H, 13C{1H} NMR, IR spectroscopy and ESI mass spectrometry (see Experimental).
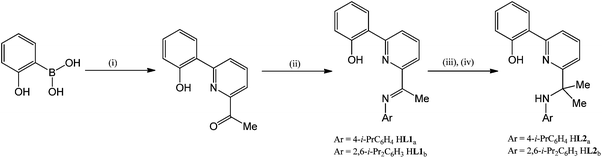 | ||
| Scheme 1 Reagents and conditions: (i) 2-Br-6-{MeC(O)}C5H3N, cat. Pd(OAc)2/PPh3, toluene, 90 °C, 12 h; (ii) ArNH2, MeOH, cat. CH3COOH, reflux; (iii) AlMe3, toluene, 110 °C, 12 h; (iv) H2O. | ||
Compounds, HL1a, HL2a and HL2b, all display protonated molecular ions peaks in their electrospray mass spectra and downfield shifted signals for the phenolic protons (range: δ 14.18–14.60) in their 1H NMR spectra. For HL1a, the imine methyl substituent is seen as a singlet at δ 2.32 in the 1H NMR spectrum while the IR spectrum reveals a characteristic ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imine stretch at 1635 cm−1. For amine-containing HL2a and HL2b, broad singlets are visible for the NH protons between δ 3.3–4.0 in their 1H NMR spectra along with sharp singlets for the equivalent gem-dimethyl protons. Further confirmation of the composition of HL1a and HL2a was achieved using single crystal X-ray diffraction.
N)imine stretch at 1635 cm−1. For amine-containing HL2a and HL2b, broad singlets are visible for the NH protons between δ 3.3–4.0 in their 1H NMR spectra along with sharp singlets for the equivalent gem-dimethyl protons. Further confirmation of the composition of HL1a and HL2a was achieved using single crystal X-ray diffraction.
Perspective views of HL1a and HL2a are depicted in Fig. 2a and b; selected bond distances and angles for both structures are listed in Table 1. Each structure consists of a central pyridine ring that is substituted at its 2-position by a phenyl-2′-ol group but differs at the 6-position with a trans-configured N-arylimine unit for HL1a [C(12)–N(2) 1.2692(19) Å] or a saturated CMe2NH(4-i-PrC6H4) unit for HL2a [C(11)–C(12)–N(2) 108.97(16)°]. In general, the pyridine nitrogen atoms adopt a cis conformation with respect to the neighbouring phenol oxygen as a result of a hydrogen-bonding interaction between the phenol hydrogen atom and the pyridine nitrogen [O(1)⋯N(1) 2.563 (HL1a), 2.537 Å (HL2a)], a conformation that has been observed in related structures.12–14
| HL1a | HL2b | |
|---|---|---|
| Bond lengths | ||
| C(1)–O(1) | 1.3455(19) | 1.353(2) |
| C(12)–N(2) | 1.2692(19) | 1.460(2) |
| C(6)–C(7) | 1.466(2) | 1.480(2) |
| C(11)–C(12) | 1.482(2) | 1.530(3) |
| Bond angles | ||
| C(11)–C(12)–N(2) | 115.71(15) | 108.97(16) |
| C(12)–N(2)–C(14) | 123.06(15) | 125.80(16) |
(b) Palladium(II) complexes of L1 and L2
Interaction of HL1a/b and HL2a/b with palladium(II) acetate affords the O,N,N-pincer complexes, [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (1a), 2,6-i-Pr2C6H3 (1b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (2a), 2,6-i-Pr2C6H3 (2b)), in good yield (Scheme 2). Compounds 1 and 2 can be readily converted to their chloride analogues [{2-(C6H4-2′-O)-6-(CMe
NAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (1a), 2,6-i-Pr2C6H3 (1b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}Pd(OAc)] (Ar = 4-i-PrC6H4 (2a), 2,6-i-Pr2C6H3 (2b)), in good yield (Scheme 2). Compounds 1 and 2 can be readily converted to their chloride analogues [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (3a), 2,6-i-Pr2C6H3 (3b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (4a), 2,6-i-Pr2C6H3 (4b)) by treating their chloroform or dichloromethane solutions with aqueous sodium chloride. Alternatively, 1a can be prepared more conveniently by the template reaction of 2-(C6H4-2′-OH),6-(CMe
NAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (3a), 2,6-i-Pr2C6H3 (3b)) and [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}PdCl] (Ar = 4-i-PrC6H4 (4a), 2,6-i-Pr2C6H3 (4b)) by treating their chloroform or dichloromethane solutions with aqueous sodium chloride. Alternatively, 1a can be prepared more conveniently by the template reaction of 2-(C6H4-2′-OH),6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C5H3N, Pd(OAc)2 and 4-isopropylaniline in toluene. Complexes 1–4 are air stable and have been characterised using a combination of mass spectrometry (FAB, ESI and ToF), IR and NMR (1H and 13C) spectroscopy and elemental analyses (see Experimental section). In addition, crystals of 1a, 1b, 2a, 2b and 3a have been the subject of single crystal X-ray diffraction studies.
O)C5H3N, Pd(OAc)2 and 4-isopropylaniline in toluene. Complexes 1–4 are air stable and have been characterised using a combination of mass spectrometry (FAB, ESI and ToF), IR and NMR (1H and 13C) spectroscopy and elemental analyses (see Experimental section). In addition, crystals of 1a, 1b, 2a, 2b and 3a have been the subject of single crystal X-ray diffraction studies.
 | ||
| Scheme 2 Reagents and conditions: (i) Pd(OAc)2, toluene, 75–80 °C; (ii) NaCl(aq.), CHCl3 or CH2Cl2, RT. | ||
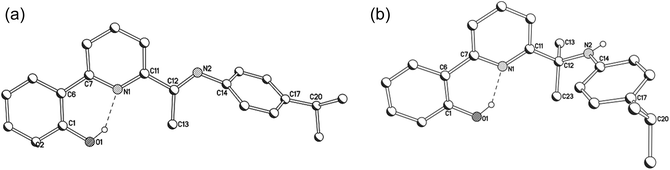
The molecular structures of imine-based 1a, 1b and 3a are closely related and will be discussed together; amine-containing 2a and 2b will be discussed later. Views of 1b and 3a are given in Fig. 3 and 4; selected bond distances and angles are collected for all three structures in Table 2. There are four independent molecules for 1a in the unit cell (molecules A–D) which differ most noticeably in the relative inclinations of the adjacent phenolate and pyridine rings (vide infra). The structures (1a, 1b and 3a) each consist of a single palladium(II) centre bound by a tridentate monoanionic 2-(phenyl-2′-olate)-6-ketimine-pyridine ligand along with a monodentate O-bound acetate (1) or chloride (3) to complete a distorted square planar geometry. Both 5- and 6-membered chelate rings are present within the complexes with the bite angle for the 6-membered ring being slightly more compatible with the geometrical requirements of the palladium(II) centre [O(1)–Pd(1)–N(2)6-membered: 96.4(4)av. (1a), 94.4(1) (1b), 93.8 (2)° (3a) vs. N(2)–Pd(1)–N(1)5-membered 82.1(4)av. (1a), 81.7(1) (1b), 81.7(2)° (3a)]. In all cases some twisting of the phenolate unit with respect to the pyridyl plane is apparent [tors. N(2)–C(13)–C(14)–C(15) 0.0(3)A, 2.5(3)B, 5.7(3)C, 9.5(3)D (1a), 14.1(3) (1b), 22.1(3)° (3a)]. In general, the Pd–Nimine bond distance is the longest of the three metal–ligand interactions involving the O,N,N-ligand followed by the Pd–Npyridine distance and then by the Pd–Ophenolate distance which is best exemplified for complex 3a [Pd(1)–N(1)imine 2.011(4) > Pd(1)–N(2)pyridine 1.972(4) > Pd(1)–O(1)phenolate 1.961(3) Å]. Replacing an O-bound acetate for a chloride has little effect on the trans Pd–Npyridine distance [1.972(4) Å (3a) vs. 1.980(10)av. (1a)]. The N-aryl group in 1b is inclined towards orthogonality with regard to the neighbouring C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine vector [tors. C(7)–N(2)–C(1)–C(2) 86.1(3)°], while in the less sterically bulky 1a and 3a the aryl group is tilted [tors. C(7)–N(2)–C(1)–C(2) 66.4(4)av (1a), 57.8(6) (3a)°]. There are no intermolecular contacts of note. The structural features resemble related aldimine-based palladium complexes [{2-(3-C12H8-2-O)-6-(CH
Nimine vector [tors. C(7)–N(2)–C(1)–C(2) 86.1(3)°], while in the less sterically bulky 1a and 3a the aryl group is tilted [tors. C(7)–N(2)–C(1)–C(2) 66.4(4)av (1a), 57.8(6) (3a)°]. There are no intermolecular contacts of note. The structural features resemble related aldimine-based palladium complexes [{2-(3-C12H8-2-O)-6-(CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NAr)C5H3N}PdX] (X = OAc, Cl) reported elsewhere.14,15
NAr)C5H3N}PdX] (X = OAc, Cl) reported elsewhere.14,15
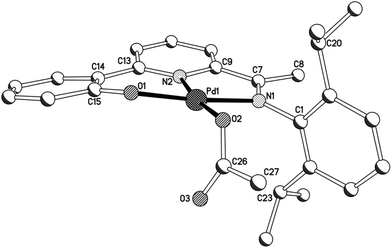 | ||
| Fig. 3 Molecular structure of 1b including a partial atom numbering scheme. All hydrogen atoms have been omitted for clarity. | ||
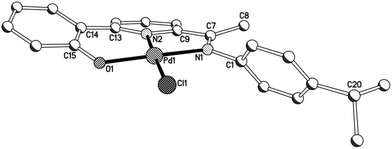 | ||
| Fig. 4 Molecular structure of 3a including a partial atom numbering scheme. All hydrogen atoms have been omitted for clarity. | ||
| 1a | 1b | 3a | ||||
|---|---|---|---|---|---|---|
| Molecule A | Molecule B | Molecule C | Molecule D | |||
| Bond lengths | ||||||
| Pd(1)–O(1) | 1.947(7) | 1.928(8) | 1.951(8) | 1.934(8) | 1.953(3) | 1.961(3) |
| Pd(1)–N(1) | 1.972(9) | 1.980(9) | 1.978(10) | 1.961(10) | 2.006(3) | 2.011(4) |
| Pd(1)–N(2) | 1.961(9) | 1.972(9) | 1.980(9) | 2.005(10) | 1.969(3) | 1.972(4) |
| Pd(1)–Cl(1) | — | — | — | — | — | 2.3039(14) |
| Pd(1)–O(2) | 2.038(8) | 2.033(8) | 2.016(8) | 2.025(8) | 2.036(3) | — |
| C(7)–N(1) | 1.319(12) | 1.295(13) | 1.303(13) | 1.302(14) | 1.292(5) | 1.301(6) |
| C(7)–C(8) | 1.484(13) | 1.496(14) | 1.515(15) | 1.514(15) | 1.509(5) | 1.497(7) |
| C(15)–O(1) | 1.306(12) | 1.310(13) | 1.347(13) | 1.321(14) | 1.317(5) | 1.317(6) |
| Bond angles | ||||||
| N(1)–Pd(1)–N(2) | 82.9(4) | 82.2(4) | 81.8(4) | 81.9(4) | 81.68(13) | 81.65(17) |
| N(1)–Pd(1)–O(1) | 177.5(4) | 177.5(4) | 177.2(4) | 178.2(4) | 174.49(12) | 174.56(16) |
| N(2)–Pd(1)–O(1) | 95.2(4) | 96.1(4) | 96.2(4) | 96.4(4) | 94.35(12) | 93.84(16) |
| N(2)–Pd(1)–Cl(1) | — | — | — | — | — | 177.97(13) |
| N(2)–Pd(1)–O(2) | 176.9(3) | 175.0(4) | 175.8(4) | 176.1(4) | 172.47(12) | — |
A view of amine-based 2a is given in Fig. 5; selected bond distances and angles are given for both 2a and 2b in Tables 3. The structures are similar to imine-containing 1a and 1b with a distorted square planar palladium(II) centre bound by a monoanionic O,N,N ligand and a monodentate O-bound acetate. In this case the more flexible 2-(phenyl-2′-olate)-6-(amino-prop-2-yl)pyridine acts as the O,N,N ligand again forming both 5-membered and 6-membered chelate rings. The presence of both a gem-dimethyl sp3-hybridised carbon (N(1)–C(7)–C(10) 108.9(8) (2a) and 109.7(2)° (2b)) and secondary amine nitrogen donor results in some puckering of the 5-membered chelate ring while the 6-membered chelate ring shows similar properties to those observed in 1a, 1b and 3a with some twisting of the phenolate unit with respect to the pyridyl plane evident [tors. N(2)–C(14)–C(15)–C(16) 18.3(3) (2a), 21.6° (2b)]. The Pd–Ophenolate and Pd–Npyridine distances are comparable to those in 1a, 1b and 3a while the Pd–Namine length is ca. 0.05 Å longer than the average Pd–Nimine distance in 1a, 1b and 3a consistent with the poorer donor characteristics of an amine. The pendant oxygen atom on the acetate ligand undergoes an intramolecular hydrogen bond interaction with the amine hydrogen atom [O(3)⋯N(1) 2.750 (2a), 2.895 (2b) Å]. It is worthy of note that the isopropyl group on C(2) in 2b occupies a position above the axial site of the N(1)–N(2)–O(1)–Pd(1) square plane (vide infra). There are no intermolecular contacts of note.
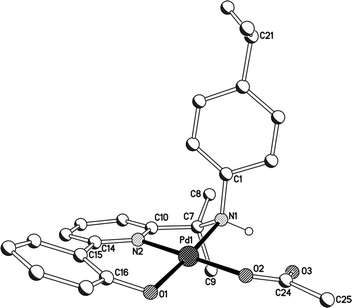 | ||
| Fig. 5 Molecular structure of 2a including a partial atom numbering scheme. All hydrogen atoms, apart from H1, have been omitted for clarity. | ||
| 2a | 2b | |
|---|---|---|
| Bond lengths | ||
| Pd(1)–O(1) | 1.951(6) | 1.9541(19) |
| Pd(1)–N(1) | 2.061(6) | 2.045(2) |
| Pd(1)–N(2) | 1.983(7) | 1.972(2) |
| Pd(1)–O(2) | 2.008(6) | 2.034(2) |
| C(7)–C(8) | 1.519(11) | 1.533(4) |
| C(7)–C(9) | 1.557(11) | 1.529(4) |
| C(7)–N(1) | 1.467(10) | 1.525(4) |
| Bond angles | ||
| N(1)–Pd(1)–N(2) | 81.8(3) | 84.46(9) |
| N(1)–Pd(1)–O(1) | 176.1(3) | 179.16(9) |
| N(2)–Pd(1)–O(1) | 94.5(3) | 94.99(9) |
| N(1)–Pd(1)–O(2) | 96.2(3) | 94.71(9) |
| N(2)–Pd(1)–O(2) | 176.8(3) | 174.90(8) |
| O(1)–Pd(1)–O(2) | 87.6(2) | 85.90(8) |
| N(1)–C(7)–C(10) | 108.9(8) | 109.7(2) |
Complexes 1–4, display either molecular ion peaks and/or fragmentation peaks corresponding to the loss of an acetate or a chloride in their mass spectra. For imine-based 1 and 3, the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imine stretch shifts by ca. 35 cm−1 to lower wavenumber when compared to those for the corresponding free HL1, supportive of imine coordination.16 In 1b and 3b two distinct doublets are seen for the isopropyl methyl groups in their 1H NMR spectra consistent with restricted rotation about the N-aryl or Ar–i-Pr bonds in solution. In contrast, there are four distinct doublets in 2b and 4b implying all four methyl groups are now inequivalent in the amine-based pincer complexes. The N–H protons in 2a and 2b are downfield shifted (between δ 8.7–9.9) consistent with the NH⋯Oacetate hydrogen bonding as seen in the solid state, whilst in their chloride derivatives, 4a and 4b, the corresponding protons are found more upfield (between δ 6.1–6.7). The acetate methyl groups in 1 and 2 can be seen at δ ca. 1.6 in their 1H NMR spectra with the MeC(O)O carbon atoms observable at δ ca. 178.8 in their 13C NMR spectra. In addition strong bands assignable to the symmetric and asymmetric ν(COO) vibrations in 1 and 2, are in agreement with those expected for monodentate acetate ligands.17
N)imine stretch shifts by ca. 35 cm−1 to lower wavenumber when compared to those for the corresponding free HL1, supportive of imine coordination.16 In 1b and 3b two distinct doublets are seen for the isopropyl methyl groups in their 1H NMR spectra consistent with restricted rotation about the N-aryl or Ar–i-Pr bonds in solution. In contrast, there are four distinct doublets in 2b and 4b implying all four methyl groups are now inequivalent in the amine-based pincer complexes. The N–H protons in 2a and 2b are downfield shifted (between δ 8.7–9.9) consistent with the NH⋯Oacetate hydrogen bonding as seen in the solid state, whilst in their chloride derivatives, 4a and 4b, the corresponding protons are found more upfield (between δ 6.1–6.7). The acetate methyl groups in 1 and 2 can be seen at δ ca. 1.6 in their 1H NMR spectra with the MeC(O)O carbon atoms observable at δ ca. 178.8 in their 13C NMR spectra. In addition strong bands assignable to the symmetric and asymmetric ν(COO) vibrations in 1 and 2, are in agreement with those expected for monodentate acetate ligands.17
(c) Reactivity of 3 and 4 towards [p-tolyl2I][O3SCF3]
All four palladium(II) chloride pincer complexes, 3a, 3b, 4a and 4b, were assessed on their ability to undergo oxidation with a hypervalent iodonium reagent and mediate the formation of a carbon–chlorine coupled product. Typically, 3 and 4 were treated with two equivalents of di-p-tolyliodonium triflate at 100 °C in a mixture of toluene–acetonitrile and their reaction mixtures monitored by gas chromatography using an internal standard to quantify the conversions (Scheme 3).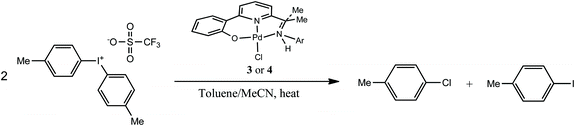 | ||
| Scheme 3 Oxidation of 3 and 4 with di-p-tolyliodonium triflate to give 4-chlorotoluene and 4-iodotoluene. | ||
The results of the screening are collected in Table 4. Several points emerge from inspection of the data. Firstly, all the palladium pincer complexes screened afford 4-chlorotoluene in varying amounts along with the expected 4-iodotoluene by-product. Secondly, two structure/reactivity relationships are apparent namely: (i) within each N,N,O family the least sterically bulky N-aryl group promotes the highest conversions to 4-chorotoluene, e.g., 4a (93%, entry 6) vs.4b (26%, entry 7) and 3a (80%, entry 4) vs.3b (17%, entry 5); (ii) amine-containing 4a and 4b yield higher conversions than their direct imine counterparts 3a and 3b, respectively. Thirdly, periodic monitoring of the conversion for 3a reveals a rapid initial reaction (33% in 1 h, entry 1) which reaches a plateau over time.
| Entry | Pd(II) chloride pincer | Time/h | Conversion/% to 4-chlorotolueneb | Conversion/% to 4-iodotolueneb |
|---|---|---|---|---|
| a Conditions: 3 or 4 (0.05 mmol), [(p-tol)2I][OTf] (0.1 mmol), ([Pd]/[(p-tol)2IOTf] = 2), toluene/MeCN, 100 °C. b Determined using gas chromatography using naphthalene as an internal standard. | ||||
| 1 | 3a | 1 | 33 | 27 |
| 2 | 3a | 2.5 | 57 | 42 |
| 3 | 3a | 6 | 74 | 67 |
| 4 | 3a | 15 | 80 | 71 |
| 5 | 3b | 15 | 17 | 7 |
| 6 | 4a | 15 | 93 | 89 |
| 7 | 4b | 15 | 26 | 10 |
It is uncertain as to the origin of these ligand effects but it would seem likely that the sterically bulky 2,6-i-Pr2Ph substitution pattern in 3b and 4b is inhibiting the oxidative transfer of the aryl group to the palladium centre. Indeed, work-up of the reaction between imine-containing 3b and di-p-tolyliodonium triflate at 100 °C over 15 hours (entry 5) gave unreacted starting materials as the major identifiable inorganic components. The increased flexibility of the ligand manifold in amine-containing 4 may, in part, contribute to the improved performance over the corresponding imine.
Unfortunately we were unable to prove or disprove the involvement of a transient Pd(IV) species (e.g., [(ONN)PdCl(p-tolyl)(NCMe)][O3SCF3]) by NMR spectroscopy due to the poor solubility of the reaction mixtures at lower temperatures. Nevertheless, we were able, in one case, to identify the palladium-containing decomposition product of the presumed reductive elimination event. Solid residues isolated from the reaction of 3a with di-p-tolyliodonium triflate (entry 4) could be extracted into acetonitrile and found to contain unreacted di-p-tolyliodonium triflate and the Pd(II) salt [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N(4-i-PrC6H4)C5H3N}Pd(NCMe)][O3SCF3] (5a). Confirmation of the presence of 5a was obtained through spiking an 1H NMR solution of the mixture with a genuine sample of 5a (prepared from the reaction of 3a with AgO3SCF3 in acetonitrile). Indeed 5a has been fully characterised by mass spectrometry, IR and NMR (1H, 19F and 13C) spectroscopy and has been the subject of a single crystal X-ray diffraction study.
N(4-i-PrC6H4)C5H3N}Pd(NCMe)][O3SCF3] (5a). Confirmation of the presence of 5a was obtained through spiking an 1H NMR solution of the mixture with a genuine sample of 5a (prepared from the reaction of 3a with AgO3SCF3 in acetonitrile). Indeed 5a has been fully characterised by mass spectrometry, IR and NMR (1H, 19F and 13C) spectroscopy and has been the subject of a single crystal X-ray diffraction study.
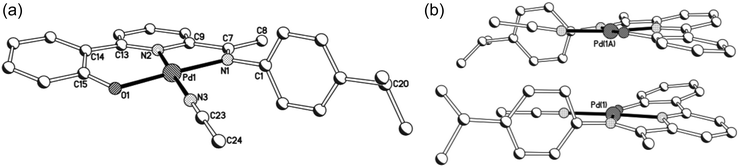
A view of 5a is given in Fig. 6a; selected bond distances and angles are collected in Table 5. There are two independent cations and associated anions in the unit cell with the main differences between the cations being the inclinations of N-aryl groups. The structure of 5a comprises a cationic palladium(II) unit charged balanced by a non-coordinating triflate anion. The cationic unit adopts a distorted square planar geometry [max. distortion: N(1)–Pd(1)–N(2) 82.0(3)A, 82.5(3)B°] with the 2-(phenyl-2′-olate)-6-ketimine-pyridine ligand occupying three coordination sites and the η1-N acetonitrile molecule the fourth. The structural parameters displayed by the pincer ligand closely mirror the features observed in neutral precursor 3a with the Pd–Nimine distance again the longest [Pd(1)–N(1) 2.017(8) Å, 1.997(8) Å] of the three donor atoms. Interestingly, the independent cations assemble in such a way as to maintain the Pd(II) centres in close proximity (Pd(1)⋯Pd(1A) 3.313 Å) and only slightly further apart than the sum of the van der Waals radii (3.26 Å) (Fig. 6b). Further confirmation of the salt-like nature of 5a comes from the positive ESI mass spectrum (recorded in MeCN) which reveals peaks corresponding to the cationic unit while the negative spectrum the triflate anion. The 19F NMR spectrum (in CD3CN) displays a single peak at δ −79.3 comparable with that observed in related triflate salts of Pd-acetonitrile species.18
| Molecule A | Molecule B | |
|---|---|---|
| Bond lengths | ||
| Pd(1)–N(1) | 2.017(8) | 1.997(8) |
| Pd(1)–N(2) | 1.953(8) | 1.951(8) |
| Pd(1)–N(3) | 2.007(9) | 1.994(10) |
| Pd(1)–O(1) | 1.959(7) | 1.979(7) |
| C(7)–N(1) | 1.277(13) | 1.297(13) |
| C(9)–C(7) | 1.515(14) | 1.473(15) |
| C(23)–N(3) | 1.138(13) | 1.176(14) |
| Range S(1)–Otriflate | 1.416(9)–1.434(11) | |
| Bond angles | ||
| N(1)–Pd(1)–N(2) | 82.0(3) | 82.5(3) |
| N(1)–Pd(1)–O(1) | 175.0(3) | 176.1(3) |
| N(1)–Pd(1)–N(3) | 95.9(3) | 94.5(3) |
| N(2)–Pd(1)–O(1) | 94.5(3) | 95.0(3) |
| N(2)–Pd(1)–N(3) | 177.3(3) | 174.9(3) |
Conclusions
Two families of palladium(II) chloride O,N,N pincer complexes (3 and 4), differing in the type of exterior nitrogen donor and, within each family, the steric properties of the N-aryl ring, have been prepared via their respective acetate analogues (1 and 2) and fully characterised. Oxidation of 3 and 4 with di-p-tolyliodonium triflate leads in all cases to carbon–chloride coupling to give 4-chlorotoluene with the conversion highly dependent on the O,N,N pincer framework employed; the recovery of 5a with an intact pincer framework highlights the robustness of the ligand manifold to oxidation. Notably, the least sterically hindered member of each family (3a and 4a) leads to the highest conversion with amine-containing 4a the highest. These observations set the stage for an investigation of these and related pincer systems in various Pd(II)/(IV)-mediated C–X coupling reactions. These results will be reported in due course.Experimental
General
All operations, unless otherwise stated, were carried out under an inert atmosphere of dry, oxygen-free nitrogen using standard Schlenk and cannular techniques or in a nitrogen purged glove box. Solvents were distilled under nitrogen from appropriate drying agents19 or were employed directly from a Solvent Purification System (Innovative Technology, Inc). The electrospray (ESI) mass spectra were recorded using a micromass Quattra LC mass spectrometer with acetonitrile or methanol as the matrix. FAB mass spectra (including high resolution) were recorded on a Kratos Concept spectrometer with NBA as matrix or on a Water Xevo QToF mass spectrometer equipped with an atmospheric solids analysis probe (ASAP). The infrared spectra were recorded in the solid state with Universal ATR sampling accessories on a Perkin Elmer Spectrum One FTIR instrument. NMR spectra were recorded on a Bruker DPX 300 spectrometer operating at 300.03 (1H) and 75.4 MHz (13C) or a Bruker DRX400 spectrometer at 400.13 (1H), 376.46 (19F) and 100.61 MHz (13C) or a Bruker Avance III 500 spectrometer at 125 MHz (13C), at ambient temperature unless otherwise stated; chemical shifts (ppm) are referred to the residual protic solvent peaks and coupling constants are expressed in hertz (Hz). Melting points (mp) were measured on a Gallenkamp melting point apparatus (model MFB-595) in open capillary tubes and were uncorrected. Elemental analyses were performed at the Science Technical Support Unit, London Metropolitan University. The reagents 2,6-diisopropylaniline, 4-isopropylaniline, silver triflate and trimethylaluminium (2 M solution in toluene) were purchased from Aldrich Chemical Co. and used without further purification. The compounds 2-hydroxyphenylboronic acid,12 2-bromo-6-acetyl pyridine20 and di-p-tolyliodonium triflate21 and HL1b12 were prepared using literature procedures. All other chemicals were obtained commercially and used without further purification.Synthesis of 2-(phenyl-2′-ol)-6-acetyl-pyridine
A round-bottomed flask equipped with stirrer bar and reflux condenser, open to the air, was loaded with 2-bromo-6-acetylpyridine (2.10 g, 10.00 mmol), Pd(OAc)2 (0.047 g, 0.21 mmol), triphenylphosphine (0.110 mg, 0.42 mmol) and 2-hydroxyphenyl boronic acid (1.88 g, 13.7 mmol). Toluene (40 mL), ethanol (22 mL) and aqueous 2 M K2CO3 (13 mL, 26.00 mmol) were added and the mixture heated to 90 °C for 12 h. The resultant black reaction mixture was cooled to room temperature followed by the addition of 1 mL H2O2 (30% in water) and stirred for a further 30 min. The organic phase was separated and the aqueous phase washed with toluene (3 × 10 mL). The combined organic extracts were washed with water (3 × 30 mL) and brine (10 mL) and concentrated to afford a brown solid. This solid was slurried in methanol (10 mL) for 1 h and the resultant solid filtered and washed with methanol (3 mL) and dried under reduced pressure. 2-(Phenyl-2′-ol)-6-acetyl-pyridine was collected as a yellow solid (1.885 g, 84%). 1H NMR (CDCl3, 400 MHz): δ 2.71 (s, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 6.90 (ddd, 3JHH 8.4, 3JHH 7.4, 4JHH 1.4, 1H, Ar–H), 7.00 (dd, 3JHH 8.3, 4JHH 1.3, 1H, Ar–H), 7.30 (ddd, 3JHH 8.5, 3JHH 7.5, 4JHH 1.7, 1H, Ar–H), 7.78 (dd, 3JHH 8.1, 4JHH 1.7, 1H, Ar–H), 7.94 (m, 2H, Py–H), 8.06 (dd, 3JHH 7.1, 4JHH 2.1, 1H, Py–H), 13.64 (s, 1H, O–H). ESIMS m/z: 214 [M + H]+. The data was consistent with that reported in ref. 13.
O), 6.90 (ddd, 3JHH 8.4, 3JHH 7.4, 4JHH 1.4, 1H, Ar–H), 7.00 (dd, 3JHH 8.3, 4JHH 1.3, 1H, Ar–H), 7.30 (ddd, 3JHH 8.5, 3JHH 7.5, 4JHH 1.7, 1H, Ar–H), 7.78 (dd, 3JHH 8.1, 4JHH 1.7, 1H, Ar–H), 7.94 (m, 2H, Py–H), 8.06 (dd, 3JHH 7.1, 4JHH 2.1, 1H, Py–H), 13.64 (s, 1H, O–H). ESIMS m/z: 214 [M + H]+. The data was consistent with that reported in ref. 13.
Synthesis of 2-(C6H4-2′-OH),6-{CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) N(4-i-PrC6H4)}C5H3N (HL1a)
N(4-i-PrC6H4)}C5H3N (HL1a)
2-(Phenyl-2′-ol)-6-acetyl-pyridine (0.405 g, 1.90 mmol), 4-isopropyl aniline (0.473 g, 3.50 mmol) and MgSO4 (2.76 g, 23.0 mmol) were suspended in bench methanol (10 mL) and one drop of acetic acid added. The mixture was stirred and heated at reflux for 9 days whereupon a further drop of acetic acid was added and the mixture stirred at reflux for an additional 12 h. On cooling to room temperature the reaction mixture was filtered and the MgSO4 washed with chloroform (30 mL) and the filtrate concentrated under reduced pressure. The resultant solid was heated in MeOH (10 mL), cooled to room temperature and the suspension collected by filtration and dried under reduced pressure affording HL1a as yellow solid (0.381 g, 61%). Single crystals suitable for an X-ray determination were grown by slow cooling of a saturated solution of HL1a in EtOH. Mp: 123–125 °C. 1H NMR (CDCl3, 300 MHz): δ 1.20 (d, 3JHH 7.1, 6H, CHMe2), 2.32 (s, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 2.84 (sept, 3JHH 7.1, 1H, CHMe2), 6.69 (d, 3JHH 8.4, 2H, Armipp–H), 6.85 (app. td, 3JHH 8.1, 4JHH 1.2, 1H, Arphenol–H), 6.96 (dd, 3JHH 8.2, 4JHH 1.2, 1H, Arphenol–H), 7.16 (d, 3JHH 8.3, 2H, Armipp–H), 7.25 (app. td, 3JHH 8.2, 4JHH 1.5, 1H, Arphenol–H), 7.75 (dd, 3JHH 8.1, 4JHH 1.4, 1H, Arphenol–H), 7.79–7.91 (m, 2H, Py–H), 8.12 (dd, 3JHH 7.6, 4JHH 1.1, 1H, Py–H), 14.18 (s, 1H, O–H). 13C{1H} NMR (CDCl3, 75 MHz): δ 15.5 (CH3C
N), 2.84 (sept, 3JHH 7.1, 1H, CHMe2), 6.69 (d, 3JHH 8.4, 2H, Armipp–H), 6.85 (app. td, 3JHH 8.1, 4JHH 1.2, 1H, Arphenol–H), 6.96 (dd, 3JHH 8.2, 4JHH 1.2, 1H, Arphenol–H), 7.16 (d, 3JHH 8.3, 2H, Armipp–H), 7.25 (app. td, 3JHH 8.2, 4JHH 1.5, 1H, Arphenol–H), 7.75 (dd, 3JHH 8.1, 4JHH 1.4, 1H, Arphenol–H), 7.79–7.91 (m, 2H, Py–H), 8.12 (dd, 3JHH 7.6, 4JHH 1.1, 1H, Py–H), 14.18 (s, 1H, O–H). 13C{1H} NMR (CDCl3, 75 MHz): δ 15.5 (CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 23.1 (CHMe2), 32.6 (CHMe2), 117.4 (CH), 117.7 (C), 118.0 (CH), 118.3 (CH), 118.8 (CH), 119.1 (CH), 125.4 (CH), 125.9 (CH), 130.6 (CH), 137.2 (CH), 143.6 (C), 147.2 (C), 152.7 (C), 155.6 (C), 158.6 (C), 163.6 (C
N), 23.1 (CHMe2), 32.6 (CHMe2), 117.4 (CH), 117.7 (C), 118.0 (CH), 118.3 (CH), 118.8 (CH), 119.1 (CH), 125.4 (CH), 125.9 (CH), 130.6 (CH), 137.2 (CH), 143.6 (C), 147.2 (C), 152.7 (C), 155.6 (C), 158.6 (C), 163.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine). IR (cm−1): ν(C
Nimine). IR (cm−1): ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imine 1635, ν(C
N)imine 1635, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)pyridine 1587. ESIMS m/z: 331 [M + H]+, 329 [M − H]. HRMS (ASAP): Calc. for C22H23N2O [M + H]+ 331.1810, found 331.1803. Anal calc. for (C22H22N2O) C 79.97, N 8.48, H 6.71. Found: C 79.97, N 8.41, H 6.64%.
N)pyridine 1587. ESIMS m/z: 331 [M + H]+, 329 [M − H]. HRMS (ASAP): Calc. for C22H23N2O [M + H]+ 331.1810, found 331.1803. Anal calc. for (C22H22N2O) C 79.97, N 8.48, H 6.71. Found: C 79.97, N 8.41, H 6.64%.
Synthesis of 2-(C6H4-2′-OH),6-(CMe2NHAr)C5H3N (HL2)
(a) Ar = 4-i-PrC6H4 (HL2a): A Schlenk flask equipped with stir bar was evacuated and backfilled with nitrogen. The vessel was loaded with HL1a (0.510 g, 1.50 mmol) and toluene (20 ml) and trimethylaluminium (2.0 ml, 4.00 mmol, 2 M solution in toluene) introduced dropwise. The solution was then stirred and heated to reflux for 12 h before being cooled to room temperature and concentrated under reduced pressure. Petroleum ether (20 ml, 40/60) was added and the solution cooled to 5 °C prior to the slow addition of water (20 ml). The mixture was then stirred for 1 h at room temperature before the organic phase was isolated. The aqueous phase was extracted with chloroform (4 × 50 ml) and the combined organic extracts washed with water (3 × 10 mL) and brine (1 × 10 mL) and then dried over MgSO4. The solvent was removed under reduced pressure to provide HL2a as an orange oil which solidified slowly over time (0.500 g, 96%). Single crystals suitable for an X-ray determination were grown by slow cooling of a saturated solution of HL2a in ethanol. Mp: 109–112 °C. 1H NMR (CDCl3, 400 MHz): δ 1.07 (d, 3JHH 7.0, 6H, CHMe2), 1.66 (s, 6H, N–C(CH3)2), 2.66 (sept, 3JHH 7.0, 1H, CHMe2), 3.97 (br s, 1H, N–H), 6.19 (d 3JHH 8.6, 2H, Armipp–H), 6.81 (d, 3JHH 8.6, 2H, Armipp–H), 6.86 (app. td, 3JHH 8.1, 4JHH 1.2, 1H, Ar–H), 6.96 (dd, 3JHH 8.3, 4JHH 1.2, 1H, Ar–H), 7.25 (ddd, 3JHH 8.5, 3JHH 7.2, 4JHH 1.6, 1H, Ar–H), 7.49–7.53 (1H, m, Ar–H), 7.69–7.73 (2H, m, Ar–H), 7.77 (dd, 3JHH 8.0, 4JHH 1.6, 1H, Py–H), 14.55 (s, 1H, O–H). 13C{1H} NMR (CDCl3, 100 MHz): δ 23.06 (CHMe2), 28.2 (N–C(CH3)2), 32.0 (CHMe2), 56.5 (C–N), 114.3 (CH), 115.8 (CH), 117.4 (CH), 117.7 (C), 177.8 (CH), 118.1 (CH), 125.2 (CH), 125.7 (CH), 130.4 (CH), 137.1 (C), 137.6 (CH), 142.2 (C), 155.9 (C), 158.9 (C), 162.8 (C). IR (cm−1): 1592 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)pyridine. ESIMS m/z: 347 [M + H]+. HRMS (EI): Calc. for: C23H27N2O [M + H]+ 347.2123, found: 347.2140.
N)pyridine. ESIMS m/z: 347 [M + H]+. HRMS (EI): Calc. for: C23H27N2O [M + H]+ 347.2123, found: 347.2140.
(b) Ar = 2,6-i-Pr2C6H3 (HL2b): A similar procedure to that described for HL2a was followed using HL1b (0.601 g, 2.70 mmol), toluene (20 ml) and trimethylaluminium (3.40 ml, 6.70 mmol 2 M solution in toluene). On work-up, HL2b was afforded as an orange oil which solidified slowly over time (0.549 g, 88%). Mp: 70–72 °C. 1H NMR (CDCl3, 400 MHz): δ 0.98 (d, 3JHH 7.0, 12H, CHMe2), 1.49 (s, 6H, N–C(CH3)2), 2.95 (sept, 3JHH 7.0, 2H, CHMe2), 3.34 (br s, 1H, N–H), 6.85 (ddd, 3JHH 8.2, 3JHH 7.4, 4JHH 1.3, 1H, Ar–H), 6.94 (dd, 3JHH 8.2, 4JHH 1.2, 1H, Ar–H), 6.98 (m (app. s), 3H, Ar–H), 7.23 (ddd, 3JHH 8.4, 3JHH 7.2, 4JHH 1.6, 1H, Ar–H), 7.59 (dd, 3JHH 7.4, 4JHH 1.2, 1H, Py–H), 7.72–7.79 (3H, m, Ar–H), 14.60 (s, 1H, O–H). 13C{1H} NMR (CDCl3, 100 MHz): δ 22.8 (CHMe2), 27.4 (CHMe2), 28.2 (N–C(CH3)2), 58.1 (C–N), 115.7 (CH), 117.2 (CH), 117.4 (CH), 117.7 (CH), 118.1 (C), 122.1 (CH), 123.5 (CH), 125.3 (CH), 130.3 (CH), 137.0 (CH), 138.7 (C), 144.3 (C), 155.5 (C), 159.0 (C), 165.1 (C). IR (cm−1): 1591 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)pyridine. ESIMS m/z: 389 [M + H]+. HRMS (EI): Calc. for C26H33N2O [M + H]+ 389.2593, found 389.2606.
N)pyridine. ESIMS m/z: 389 [M + H]+. HRMS (EI): Calc. for C26H33N2O [M + H]+ 389.2593, found 389.2606.
Synthesis of [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) NAr)C5H3N}Pd(OAc)] (1)
NAr)C5H3N}Pd(OAc)] (1)
(a) Ar = 4-i-PrC6H4 (1a): A Schlenk flask equipped with stir bar was evacuated and backfilled with nitrogen. The vessel was loaded with HL1a (0.100 g, 0.300 mmol), Pd(OAc)2 (0.068 g, 0.300 mmol) and toluene (10 ml) and then stirred and heated at 80 °C for 12 h. On cooling to room temperature the volatiles were removed under reduced pressure. The resultant solid was dissolved in dichloromethane (5 mL) and hexane (100 mL) introduced affording 1a as a red solid (0.136 g, 90%). Single crystals suitable for an X-ray determination were grown by slow diffusion of hexane into a solution of 1a in chloroform. Mp: >240 °C (decomp.). 1H NMR (CDCl3, 400 MHz): δ 1.22 (d, 3JHH 6.9, 6H, CHMe2), 1.48 (s, 3H, CH3C(O)O–), 2.07 (s, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 2.90 (sept, 3JHH 6.9, 1H, CHMe2), 6.61 (ddd, 3JHH 8.2, 3JHH 6.6, 4JHH 1.5, 1H, Arphenolate–H), 7.04 (dd, 3JHH 8.5, 4JHH 1.3, 1H, Ar–H), 7.09–7.14 (m, 4H, Ar–H), 7.23 (d, 3JHH 8.2, 2H, Armipp–H), 8.02 (d, 3JHH 8.5, 1H, Py–H), 8.06 (dd, 3JHH 8.5, 3JHH 8.5, 1H, Py–H), 8.97 (d, 3JHH 8.7, 1H, PyH). 13C{1H} NMR (CDCl3, 100 MHz): δ 16.5 (CH3C
N), 2.90 (sept, 3JHH 6.9, 1H, CHMe2), 6.61 (ddd, 3JHH 8.2, 3JHH 6.6, 4JHH 1.5, 1H, Arphenolate–H), 7.04 (dd, 3JHH 8.5, 4JHH 1.3, 1H, Ar–H), 7.09–7.14 (m, 4H, Ar–H), 7.23 (d, 3JHH 8.2, 2H, Armipp–H), 8.02 (d, 3JHH 8.5, 1H, Py–H), 8.06 (dd, 3JHH 8.5, 3JHH 8.5, 1H, Py–H), 8.97 (d, 3JHH 8.7, 1H, PyH). 13C{1H} NMR (CDCl3, 100 MHz): δ 16.5 (CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 21.7 (CH3C(O)O–), 22.9 (CHMe2), 32.9 (CHMe2), 114.5 (CH), 118.2 (C), 122.3 (CH), 122.3 (CH), 122.4 (CH), 125.6 (CH), 126.2 (CH), 128.5 (CH), 130.6 (CH), 137.5 (CH), 141.2 (C), 147.5 (C), 150.0 (C), 162.0 (C), 172.4 (C
N), 21.7 (CH3C(O)O–), 22.9 (CHMe2), 32.9 (CHMe2), 114.5 (CH), 118.2 (C), 122.3 (CH), 122.3 (CH), 122.4 (CH), 125.6 (CH), 126.2 (CH), 128.5 (CH), 130.6 (CH), 137.5 (CH), 141.2 (C), 147.5 (C), 150.0 (C), 162.0 (C), 172.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine), 177.0 (C
Nimine), 177.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). IR (cm−1): 1613 (C
O). IR (cm−1): 1613 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imine, 1590 (COOasymm/C
N)imine, 1590 (COOasymm/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Npyridine), 1456 (COOsymm). FABMS m/z: 435 [M − OAc]+. Anal calc. for (C24H24N2O3Pd): C 58.25; H 4.89; N 5.66 Found: C 58.12; H 4.83; N 5.67%.
Npyridine), 1456 (COOsymm). FABMS m/z: 435 [M − OAc]+. Anal calc. for (C24H24N2O3Pd): C 58.25; H 4.89; N 5.66 Found: C 58.12; H 4.83; N 5.67%.
(b) Ar = 2,6-i-Pr2C6H3 (1b): A similar procedure to that described for 1a was followed using HL1b (0.100 g, 0.27 mmol), Pd(OAc)2 (0.061 g, 0.27 mmol) afforded 1b as a red solid (0.135 g, 93%). Crystals suitable for an X-ray determination were grown by slow diffusion of hexane into a solution of 1b in chloroform. Mp: >240 °C (decomp.). 1H NMR (CDCl3, 400 MHz): δ 1.04 (d, 3JHH 6.9, 6H, CHMe2), 1.41 (d, 3JHH 6.7, 6H, CHMe2), 1.43 (s, 3H, CH3C(O)C–), 2.24 (s, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 3.22 (sept, 3JHH 6.8, 2H, CHMe2), 6.64 (ddd, 3JHH 8.3, 3JHH 6.3, 4JHH 1.9, 1H, Arphenolate–H), 7.14–7.22 (4H, m, under CHCl3), 7.28 (dd, 3JHH 8.2, 3JHH 7.3, 1H, Ar–H), 7.60 (dd, 3JHH 7.5, 4JHH 1.0, 1H, Py–H), 7.79 (d, 3JHH 8.6, 1H, Arphenolate–H), 8.08 (dd, 3JHH 8.8, 3JHH 7.5, 1H, Py–H), 8.43 (d, 3JHH 8.7, 1H, Py–H). 13C{1H} NMR (CDCl3, 125 MHz): δ 18.5 (CHMe2), 22.5 (CHMe2), 23.7 (CH3C(O)O–), 24.4 (CH3C
N), 3.22 (sept, 3JHH 6.8, 2H, CHMe2), 6.64 (ddd, 3JHH 8.3, 3JHH 6.3, 4JHH 1.9, 1H, Arphenolate–H), 7.14–7.22 (4H, m, under CHCl3), 7.28 (dd, 3JHH 8.2, 3JHH 7.3, 1H, Ar–H), 7.60 (dd, 3JHH 7.5, 4JHH 1.0, 1H, Py–H), 7.79 (d, 3JHH 8.6, 1H, Arphenolate–H), 8.08 (dd, 3JHH 8.8, 3JHH 7.5, 1H, Py–H), 8.43 (d, 3JHH 8.7, 1H, Py–H). 13C{1H} NMR (CDCl3, 125 MHz): δ 18.5 (CHMe2), 22.5 (CHMe2), 23.7 (CH3C(O)O–), 24.4 (CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 28.8 (CHMe2), 115.8 (CH), 119.2 (C), 122.6 (CH), 123.8 (CH), 123.9 (CH), 126.6 (CH), 128.4 (CH), 128.5 (CH), 132.3 (CH), 137.1 (CH), 139.5 (C), 140.8 (C), 152.7 (C), 154.2 (C), 164.1 (C), 174.2 (C
N), 28.8 (CHMe2), 115.8 (CH), 119.2 (C), 122.6 (CH), 123.8 (CH), 123.9 (CH), 126.6 (CH), 128.4 (CH), 128.5 (CH), 132.3 (CH), 137.1 (CH), 139.5 (C), 140.8 (C), 152.7 (C), 154.2 (C), 164.1 (C), 174.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine), 177.3 (C
Nimine), 177.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). IR (cm−1): 1600 (C
O). IR (cm−1): 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine/COOasymm/C
Nimine/COOasymm/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Npyridine), 1456 (COOsymm). ESIMS m/z: 477 [M − OAc]+, 518 [(M − OAc + MeCN]+. HRMS (ASAP): Calc. for: C27H30N2O3Pd [M]+ 536.1291 Found 536.1333.
Npyridine), 1456 (COOsymm). ESIMS m/z: 477 [M − OAc]+, 518 [(M − OAc + MeCN]+. HRMS (ASAP): Calc. for: C27H30N2O3Pd [M]+ 536.1291 Found 536.1333.
Synthesis of [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}Pd(OAc)] (2)
(a) Ar = 4-i-PrC6H4 (2a): A Schlenk flask equipped with a stir bar was evacuated, back-filled with nitrogen and then loaded with HL2a (0.040 g, 0.12 mmol), Pd(OAc)2 (0.026 g, 0.12 mmol) and toluene (4 mL). After stirring and heating at 75 °C for 12 h, the reaction mixture was allowed to cool to room temperature and the volatiles removed under reduced pressure. The residue was dissolved in dichloromethane (1 mL) before hexane (20 mL) was added to precipitate the product. The product was collected on a Celite plug, washed with hexane (10 mL) before being dissolved in dichloromethane (10 mL) and the solution collected. On evaporation of the volatile components, 2a was obtained as a red powder (0.057 g, 93%). Single crystals suitable for an X-ray determination were grown by slow diffusion of hexane into a solution of 2a in chloroform. Mp: >240 °C (decomp.). 1H NMR (CDCl3, 400 MHz): δ 1.09 (d, 3JHH 6.9, 6H, CHMe2), 1.36 (s, 3H, N–C(CH3)2), 2.00 (s, 3H, CH3C(O)C–), 2.40 (s, 3H, N–C(CH3)2), 2.72 (sept, 3JHH 6.9, 1H, CHMe2), 6.60 (ddd, 3JHH 8.1, 3JHH 6.5, 4JHH 1.8, 1H, Arphenolate–H), 6.67 (d, 3JHH 8.4, 2H, Armipp–H), 6.87 (dd, 3JHH 6.1, 4JHH 2.3, 1H, Py–H), 6.94 (d, 3JHH 8.3, 2H, Armipp–H), 7.06–7.14 (m, 2H, Arphenolate–H), 7.57 (d, 3JHH 8.5, 1H, Arphenolate–H), 7.80 (d, 3JHH 8.5, 1H, Py–H), 7.82 (dd, 3JHH 8.5, 3JHH 6.2, 1H, Py–H), 9.92 (br s, 1H, NH). 13C{1H} NMR (CDCl3, 100 MHz): δ 23.8 (CHMe2), 24.1 (CH3C(O)O–), 24.4 (N–C(CH3)2), 33.6 (CHMe2), 33.6 (N–C(CH3)2), 70.2 (C–N), 116.0 (CH), 116.3 (CH), 121.4 (CH), 121.8 (C), 122.9 (CH), 123.0 (CH), 127.5 (CH), 128.9 (CH), 132.3 (CH), 138.9 (C), 139.7 (C), 147.2 (C), 153.5 (C), 164.4 (C), 168.0 (C), 181.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). IR (cm−1): 3400 (NH), 1574 (COOasymm/C
O). IR (cm−1): 3400 (NH), 1574 (COOasymm/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Npyridine), 1448 (COOsymm). ESIMS: m/z 510 [M]+, 592 [M − OAc + MeCN]+. HRMS (FAB): m/z Calc. for C25H28N2O3Pd [M]+ 510.6296. Found 510.1125.
Npyridine), 1448 (COOsymm). ESIMS: m/z 510 [M]+, 592 [M − OAc + MeCN]+. HRMS (FAB): m/z Calc. for C25H28N2O3Pd [M]+ 510.6296. Found 510.1125.
(b) Ar = 2,6-i-Pr2C6H3 (2b): A similar procedure to that outlined for 2a was employed using HL2b (0.024 g, 0.61 mmol) and Pd(OAc)2 (0.014 g, 0.061 mmol) gave 2b as a yellow solid (0.033 g, 98%). Single crystals suitable for an X-ray determination were grown by slow diffusion of hexane into a solution of 2b in dichloromethane. Mp: >240 °C (decomp.). 1H NMR (CDCl3, 400 MHz): δ 0.72 (d, 3JHH 6.9, 3H, CHMe2), 1.18 (s, 3H, NC(CH3)2), 1.19 (d, 3JHH 6.9, 3H, CHMe2), 1.22 (d, 3JHH 6.9, 3H, CHMe2), 1.54 (d, 3JHH 6.7, 3H, CHMe2), 1.91 (s, 3H, CH3C(O)O–), 2.31 (s, 3H, N–C(CH3)2), 3.16 (sept, 3JHH 6.7, 1H, CH(Me)2), 3.72 (sept, 3JHH 6.8, 1H, CH(Me)2), 6.61 (ddd, 3JHH 8.5, 3JHH 6.4, 4JHH 2.0, 1H, Arphenolate–H), 6.84 (dd, 3JHH 6.9, 4JHH 2.1, 1H, Py H), 7.02–7.18 (m, 5H, Ar–H), 7.54 (d, 3JHH 8.3, 1H, Arphenolate–H), 7.74–7.80 (m, 2H, Py–H), 8.66 (br s, 1H, NH). 13C{1H} NMR (CDCl3 100 MHz): δ 21.9 (CHMe2), 22.3 (CH3C(O)O–), 23.7 (CH3), 24.1 (CH3), 24.5 (CHMe2), 24.6 (CH3), 27.4 (CHMe2), 27.7 (CHMe2), 32.3 (N–C(CH3)2), 70.8 (C–N), 115.0 (CH), 115.6 (CH), 110.0 (CH), 120.8 (C), 121.5 (CH), 124.2 (CH), 124.7 (CH), 126.9 (CH), 127.9 (CH), 131.4 (CH), 134.5 (C), 137.8 (CH), 143.0 (C), 143.5 (C), 152.3 (C), 163.1 (C), 169.3 (C), 179.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). IR (cm−1): 3064 (NH), 1590 (COOasymm/C
O). IR (cm−1): 3064 (NH), 1590 (COOasymm/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Npyridine), 1450 (COOsymm). TOFMS (ASAP): m/z 553 [M + H]+, 493 [M − OAc]+. Anal. calc. for (C28H34N2O3Pd·3CH2Cl2): C 46.09, H 4.99 N 3.47% Found: C 46.00, H 4.64, N 3.61%.
Npyridine), 1450 (COOsymm). TOFMS (ASAP): m/z 553 [M + H]+, 493 [M − OAc]+. Anal. calc. for (C28H34N2O3Pd·3CH2Cl2): C 46.09, H 4.99 N 3.47% Found: C 46.00, H 4.64, N 3.61%.
Synthesis of [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) NAr)C5H3N}PdCl] (3)
NAr)C5H3N}PdCl] (3)
(a) Ar = 4-i-PrC6H4 (3a): A round bottomed flask equipped with stirrer bar and open to the air was loaded with 1a (0.568 g, 1.15 mmol), chloroform (30 mL) and brine (30 mL). After stirring vigorously at room temperature for 1 h the organic phase was separated, washed with water (3 × 30 ml) and filtered through a Celite plug. The plug was washed with chloroform (10 mL) and the solution concentrated to a smaller volume (ca. 5 mL) before hexane (100 mL) was added to precipitate the title compound as dark red solid (0.537 g, 99%). Single crystals suitable for an X-ray determination were grown by slow diffusion of hexane into a solution of 3a in chloroform. Mp: >240 °C (decomp). 1H NMR (CDCl3, 400 MHz): δ 1.22 (d, 3JHH 6.9, 6H, CHMe2), 2.28 (s, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 2.88 (sept, 3JHH 6.9, 1H, CHMe2), 6.67 (ddd, 3JHH 8.3, 3JHH 6.1, 4JHH 2.0, 1H, Arphenolate–H), 7.04 (d, 3JHH 8.4, 2H, Armipp–H), 7.16–7.24 (m, 4H, Ar–H), 7.61 (dd, 3JHH 7.6, 4JHH 1.0, 1H, Py–H), 7.68 (d, 3JHH 8.4, 1H, Arphenolate–H), 7.89 (dd, 3JHH 8.6, 3JHH 7.5, 1H, Py–H), 8.18 (d, 3JHH 8.7, 1H, Py–H). 13C{1H} NMR (CDCl3, 125 MHz): δ 18.5 (CH3C
N), 2.88 (sept, 3JHH 6.9, 1H, CHMe2), 6.67 (ddd, 3JHH 8.3, 3JHH 6.1, 4JHH 2.0, 1H, Arphenolate–H), 7.04 (d, 3JHH 8.4, 2H, Armipp–H), 7.16–7.24 (m, 4H, Ar–H), 7.61 (dd, 3JHH 7.6, 4JHH 1.0, 1H, Py–H), 7.68 (d, 3JHH 8.4, 1H, Arphenolate–H), 7.89 (dd, 3JHH 8.6, 3JHH 7.5, 1H, Py–H), 8.18 (d, 3JHH 8.7, 1H, Py–H). 13C{1H} NMR (CDCl3, 125 MHz): δ 18.5 (CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 23.9 (CHMe2), 33.7 (CHMe2), 116.0 (CH), 119.1 (C), 123.2 (CH), 123.6 (CH), 124.1 (CH), 125.7 (CH), 126.5 (CH), 128.9 (CH), 132.1 (CH), 138.0 (CH), 143.7 (C), 148.2 (C), 150.5 (C), 154.7 (C), 162.4 (C), 175.7 (C
N), 23.9 (CHMe2), 33.7 (CHMe2), 116.0 (CH), 119.1 (C), 123.2 (CH), 123.6 (CH), 124.1 (CH), 125.7 (CH), 126.5 (CH), 128.9 (CH), 132.1 (CH), 138.0 (CH), 143.7 (C), 148.2 (C), 150.5 (C), 154.7 (C), 162.4 (C), 175.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine). IR (cm−1): ν(C
Nimine). IR (cm−1): ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imine 1598. FABMS m/z: 470 [M]+, 435 [M − Cl]+. Anal calc. for (C22H21N2OPdCl): C 56.07; H 4.49; N 5.94. Found: C 55.99; H 4.38; N 6.01%.
N)imine 1598. FABMS m/z: 470 [M]+, 435 [M − Cl]+. Anal calc. for (C22H21N2OPdCl): C 56.07; H 4.49; N 5.94. Found: C 55.99; H 4.38; N 6.01%.
(b) Ar = 2,6-i-Pr2C6H3 (3b): A similar procedure to that described for 3a was employed using 1b (0.289 g, 0.54 mmol) affording 3b as a red solid (0.221 g, 80%). Mp: >240 °C (decomp.). 1H NMR (CDCl3, 400 MHz): δ 1.06 (d, 3JHH 6.9, 6H, CHMe2), 1.39 (d, 3JHH 6.8, 6H, CHMe2), 2.22 (s, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 3.06 (sept, 3JHH 6.8, 2H, CHMe2), 6.69 (ddd, 3JHH 8.4, 3JHH 6.8, 4JHH 1.5, 1H, Arphenolate–H), 7.16 (d, 3JHH 7.9, 2H, Ardipp–H), 7.20–7.32 (m, 3H, Ar–H), 7.71 (dd, 3JHH 7.5, 4JHH 1.0, 1H, Py–H), 7.82 (dd, 3JHH 8.6, 4JHH 1.4, 1H, Arphenolate–H), 8.13 (dd, 3JHH 8.8, 3JHH 7.6, 1H, Py–H), 8.47 (d, 3JHH 8.8, 1H, Py–H). 13C{1H} NMR (CDCl3, 125 MHz): δ 18.2 (CHMe2), 23.7 (CHMe2), 23.9 (CH3C
N), 3.06 (sept, 3JHH 6.8, 2H, CHMe2), 6.69 (ddd, 3JHH 8.4, 3JHH 6.8, 4JHH 1.5, 1H, Arphenolate–H), 7.16 (d, 3JHH 7.9, 2H, Ardipp–H), 7.20–7.32 (m, 3H, Ar–H), 7.71 (dd, 3JHH 7.5, 4JHH 1.0, 1H, Py–H), 7.82 (dd, 3JHH 8.6, 4JHH 1.4, 1H, Arphenolate–H), 8.13 (dd, 3JHH 8.8, 3JHH 7.6, 1H, Py–H), 8.47 (d, 3JHH 8.8, 1H, Py–H). 13C{1H} NMR (CDCl3, 125 MHz): δ 18.2 (CHMe2), 23.7 (CHMe2), 23.9 (CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 28.9 (CHMe2), 116.2 (CH), 118.7 (C), 122.8 (CH), 123.8 (CH), 124.2 (CH), 127.0 (CH), 128.4 (CH), 128.6 (CH), 132.6 (CH), 137.2 (CH), 139.8 (C), 141.3 (C), 152.3 (C), 154.1 (C), 163.5 (C), 175.2 (C
N), 28.9 (CHMe2), 116.2 (CH), 118.7 (C), 122.8 (CH), 123.8 (CH), 124.2 (CH), 127.0 (CH), 128.4 (CH), 128.6 (CH), 132.6 (CH), 137.2 (CH), 139.8 (C), 141.3 (C), 152.3 (C), 154.1 (C), 163.5 (C), 175.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine). IR (cm−1): ν(C
Nimine). IR (cm−1): ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imine 1607. FABMS: m/z 512 [M]+, 477 [M − Cl]+. TOFMS (ASAP): m/z 513 [M + H]+, 477 [M − Cl]+. Anal. calc. for (C25H27N2OPdCl): C 58.49, H 5.30, N 5.46 Found: C 58.38, H 5.27, N 5.52%.
N)imine 1607. FABMS: m/z 512 [M]+, 477 [M − Cl]+. TOFMS (ASAP): m/z 513 [M + H]+, 477 [M − Cl]+. Anal. calc. for (C25H27N2OPdCl): C 58.49, H 5.30, N 5.46 Found: C 58.38, H 5.27, N 5.52%.
Synthesis of [{2-(C6H4-2′-O)-6-(CMe2NHAr)C5H3N}PdCl] (4)
(a) Ar = 4-i-PrC6H4 (4a): A round bottomed flask equipped with stirrer bar and open to the air was loaded with 2a (0.281 g, 0.55 mmol), dichloromethane (20 mL) and brine (20 mL). After stirring vigorously at room temperature for 12 h the organic phase was separated, washed with water (3 × 30 ml) and filtered through a Celite plug. Hexane (100 mL) was added to precipitate the product which was trapped on a Celite plug and washed with hexane (20 mL). The product was dissolved in dichloromethane and the solution collected. All volatiles were removed under reduced pressure affording 4a as a yellow solid (0.219 g, 82%). Mp: >240 °C (decomp.). 1H NMR (CDCl3, 400 MHz): δ 1.19 (d, 3JHH 7.0, 6H, CHMe2), 1.53 (s, 3H, N–C(CH3)2), 2.51 (s, 3H, N–C(CH3)2), 2.83 (sept, 3JHH 7.0, 1H, CHMe2), 6.67 (br, s, 1H, NH), 6.69–6.73 (m, 1H, Ar–H), 6.94–6.99 (m, 3H, Ar–H), 7.07 (d, 3JHH 8.7, 2H, Ar–H), 7.21 (d, 3JHH 4.3, 2H, Ar–H), 7.69 (d, 3JHH 8.4, 1H, Ar–H), 7.92–8.00 (m, 2H, Ar–H). 13C{1H} NMR (CDCl3, 125 MHz): δ 23.8 (CHMe2), 24.2 (N–C(CH3)2), 33.6 (CHMe2), 33.8 (N–C(CH3)2), 72.0 (C–N), 116.1 (CH), 116.3 (CH), 121.6 (CH), 121.9 (C), 123.0 (CH), 123.3 (CH), 127.5 (CH), 129.0 (CH), 132.4 (CH), 139.1 (CH), 139.4 (C), 147.5 (C), 152.6 (C), 164.2 (C), 167.0 (C). IR (cm−1): ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Npyridine) 1573, ν(NH) 3171. FABMS: m/z 486 [M]+, 451 [M − Cl]+. HRMS (ASAP): m/z Calc. for C23H26N2OPdCl [M + H]+ 487.0768. Found 487.0792. Calc. for C23H25N2OPd [M − Cl]+ 451.002. Found 451.1026. Calc. for (C23H25N2OPdCl·CHCl3): C 47.51; H 4.32; N 4.62 Found: C 47.54; H 4.19; N 4.71%.
Npyridine) 1573, ν(NH) 3171. FABMS: m/z 486 [M]+, 451 [M − Cl]+. HRMS (ASAP): m/z Calc. for C23H26N2OPdCl [M + H]+ 487.0768. Found 487.0792. Calc. for C23H25N2OPd [M − Cl]+ 451.002. Found 451.1026. Calc. for (C23H25N2OPdCl·CHCl3): C 47.51; H 4.32; N 4.62 Found: C 47.54; H 4.19; N 4.71%.
(b) Ar = 2,6-i-Pr2C6H3 (4b): A similar procedure to that described for 4a was employed using 2b (0.221 g, 0.40 mmol) affording 4b as a yellow solid (0.154 g, 73%). Mp: >240 °C (decomp). 1H NMR (CDCl3, 400 MHz): δ 0.83 (d, 3JHH 6.9, 3H, CHMe2), 1.23 (s, 3H, N–C(CH3)2), 1.27 (d, 3JHH 6.8, 3H, CHMe2), 1.41 (d, 3JHH 6.6, 3H, CHMe2), 1.51 (d, 3JHH 6.7, 3H, CHMe2), 2.16 (s, 3H, N–C(CH3)2), 3.02 (sept, 3JHH 6.7, 1H, CHMe2), 3.35 (sept, 3JHH 6.8, 1H, CHMe2), 6.10 (br, s, 1H, NH), 6.61 (ddd, 3JHH 8.2, 3JHH 6.3, 4JHH 2.1, 1H, Arphenolate–H), 6.86 (dd, 3JHH 7.5, 4JHH 1.2, 1H, Py–H), 7.05–7.08 (m, 2H, Ar–H), 7.03–7.17 (m, 3H, Ar–H), 7.55 (d, 3JHH 8.3, 1H, Arphenolate–H), 7.81 (dd, 3JHH 8.3, 3JHH 7.4, 1H, Py–H), 7.88 (d, 3JHH 8.4, 1H, Py–H). 13C{1H} NMR (CDCl3, 100 MHz): δ 22.6 (CHMe2), 24.3 (N–C(CH3)2), 24.4 (CHMe2), 24.9 (CHMe2), 25.5 (CHMe2), 28.9 (CHMe2), 29.3 (CHMe2), 34.3 (N–C(CH3)2), 72.0 (C–N), 116.1 (CH), 116.1 (CH), 121.4 (C), 123.1 (CH), 124.5 (CH), 125.7 (CH), 128.0 (CH), 129.0 (CH), 132.3 (CH), 135.7 (C), 138.9 (CH), 142.2 (C), 143.0 (C), 153.4 (C), 164.0 (C), 169.0 (C). IR (cm−1): ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Npyridine): 1573. FABMS: m/z 528 [M]+, 493 [M − Cl]+. HRMS (ASAP) m/z: Calc. for C26H32N2OPdCl [M + H]+ 529.1238. Found 529.1235. Calc. for C26H31N2OPd [M − Cl]+ 493.1471 Found 493.1413. Anal calc. for (C26H31N2OPdCl·0.5CHCl3): C 54.03; H 5.39; N 4.76 Found: C 54.44, H 5.75, N 4.78%.
Npyridine): 1573. FABMS: m/z 528 [M]+, 493 [M − Cl]+. HRMS (ASAP) m/z: Calc. for C26H32N2OPdCl [M + H]+ 529.1238. Found 529.1235. Calc. for C26H31N2OPd [M − Cl]+ 493.1471 Found 493.1413. Anal calc. for (C26H31N2OPdCl·0.5CHCl3): C 54.03; H 5.39; N 4.76 Found: C 54.44, H 5.75, N 4.78%.
Synthesis of [{2-(C6H4-2′-O)-6-(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) N{(4-i-PrC6H4)}C5H3N}Pd(NCMe)][O3SCF3] (5a)
N{(4-i-PrC6H4)}C5H3N}Pd(NCMe)][O3SCF3] (5a)
A Schlenk flask was loaded in the glovebox and 3a (0.124 g, 0.264 mmol) along with AgOSO2CF3 (68 mg, 0.264 mmol) introduced. On removal from the glovebox, MeCN (10 mL) was added and the reaction mixture stirred at room temperature for 12 h in the absence of light. The resultant slurry was allowed to settle before the insoluble components were removed by cannular filtration and the filtrate collected in a second dry Schlenk flask. The solvent was removed under reduced pressure affording 5a as a hygroscopic orange solid (0.160 g, 97%). Single crystals suitable for an X-ray determination were obtained by layering of a solution of 5a in MeCN–toluene (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 v/v) with hexane. 1H NMR (CD3CN, 400 MHz): δ 1.31 (d, 3JHH 7.0, 6H, CHMe2), 2.45 (s, 3H, CH3C
95 v/v) with hexane. 1H NMR (CD3CN, 400 MHz): δ 1.31 (d, 3JHH 7.0, 6H, CHMe2), 2.45 (s, 3H, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 3.05 (sept, 3JHH 7.0, 1H, CHMe2), 6.93 (ddd, 3JHH 8.4, 3JHH 7.0, 4JHH 1.3, 1H, Arphenolate–H), 7.13 (dd, 3JHH 8.6, 4JHH 1.3, 1H, Arphenolate–H), 7.26 (d, 3JHH 8.5, 2H, Armipp–H), 7.40 (ddd, 3JHH 8.5, 3JHH 6.8, 4JHH 1.5, 1H, Arphenolate–H), 7.49 (d, 3JHH 8.5, 2H, Armipp–H), 8.08–8.13 (2H, m, Ar–H), 8.39 (dd, 3JHH 8.7, 3JHH 7.5, 1H, Py–H), 8.67 (d, 3JHH 8.8, 1H, Py–H), the coordinated CH3CN ligand was not observed due to rapid exchange with bulk CD3CN. 13C{1H} NMR (CD3CN, 100 MHz): δ 17.4 (CH3C
N), 3.05 (sept, 3JHH 7.0, 1H, CHMe2), 6.93 (ddd, 3JHH 8.4, 3JHH 7.0, 4JHH 1.3, 1H, Arphenolate–H), 7.13 (dd, 3JHH 8.6, 4JHH 1.3, 1H, Arphenolate–H), 7.26 (d, 3JHH 8.5, 2H, Armipp–H), 7.40 (ddd, 3JHH 8.5, 3JHH 6.8, 4JHH 1.5, 1H, Arphenolate–H), 7.49 (d, 3JHH 8.5, 2H, Armipp–H), 8.08–8.13 (2H, m, Ar–H), 8.39 (dd, 3JHH 8.7, 3JHH 7.5, 1H, Py–H), 8.67 (d, 3JHH 8.8, 1H, Py–H), the coordinated CH3CN ligand was not observed due to rapid exchange with bulk CD3CN. 13C{1H} NMR (CD3CN, 100 MHz): δ 17.4 (CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 22.8 (CHMe2), 33.3 (CHMe2), 116.9 (CH), 118.5 (C), 120.7 (CH), 122.4 (CH), 125.6 (CH), 126.8 (CH), 127.2 (CH), 129.5 (CH), 132.9 (CH), 139.4 (CH), 142.9 (C), 149.5 (C), 150.3 (C), 155.2 (C), 160.0 (C), 177.8 (C
N), 22.8 (CHMe2), 33.3 (CHMe2), 116.9 (CH), 118.5 (C), 120.7 (CH), 122.4 (CH), 125.6 (CH), 126.8 (CH), 127.2 (CH), 129.5 (CH), 132.9 (CH), 139.4 (CH), 142.9 (C), 149.5 (C), 150.3 (C), 155.2 (C), 160.0 (C), 177.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Nimine), CF3SO3− not observed. 19F NMR (CD3CN, 376 MHz): δ −79.3 (s, 3F, CF3SO3). IR (cm−1): ν(C
Nimine), CF3SO3− not observed. 19F NMR (CD3CN, 376 MHz): δ −79.3 (s, 3F, CF3SO3). IR (cm−1): ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)imine 1597. ESIMS (+ve): m/z 476 [M − CF3SO3]+. ESIMS (−ve): m/z 149 [CF3SO3]−. HRMS (ASAP): m/z Calc. for C23H21N2O4SF3Pd [M − MeCN]+ 584.0218 Found 584.0482.
N)imine 1597. ESIMS (+ve): m/z 476 [M − CF3SO3]+. ESIMS (−ve): m/z 149 [CF3SO3]−. HRMS (ASAP): m/z Calc. for C23H21N2O4SF3Pd [M − MeCN]+ 584.0218 Found 584.0482.
General procedure for reactions of Pd–Cl complexes with the iodonium salt
A microwave vessel equipped with stirrer bar and open to the air was loaded with 3 or 4 (0.05 mmol) and di-p-tolyliodonium triflate (0.10 mmol, 2 eq.) and the contents suspended in toluene (4.5 mL) and MeCN (0.5 mL) before the system was sealed. The mixture was then stirred and heated to 100 °C for the specified time period. On cooling to room temperature the internal standard naphthalene (1 eq.) was added in hexane (2 mL). 1 mL of this reaction mixture was removed, diluted with a further 2 mL of hexane and the solids removed by filtration through a silica plug. The plug was washed with hexane (1 mL) and the filtrate was subject to analysis by GC. GC conditions: Hold oven temperature at 40 °C for 2 min; ramp 10 °C min−1 for 10 min; hold oven temperature at 180 °C for 12 min; injection temperature 250 °C; injection volume 1 μL; split ratio: 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. All reactions were repeated in triplicate.
1. All reactions were repeated in triplicate.
Crystallographic studies
Data for HL1a, HL2a, 1a, 1b, 2a, 2b, 3a and 5a were collected on a Bruker APEX 2000 CCD diffractometer. Details of data collection, refinement and crystal data are listed in Table 6. The data were corrected for Lorentz and polarisation effects and empirical absorption corrections applied. Structure solution by direct methods and structure refinement based on full-matrix least-squares on F2 employed SHELXTL version 6.10.22 Hydrogen atoms were included in calculated positions (C–H = 0.93–1.00 Å) riding on the bonded atom with isotropic displacement parameters set to 1.5Ueq(C) for methyl H atoms and 1.2Ueq(C) for all other H atoms. All non-H atoms were refined with anisotropic displacement parameters. Disordered solvent was omitted using the SQUEEZE option in PLATON for 1b and 2a.23| Complex | HL1a | HL2a | 1a | 1b |
|---|---|---|---|---|
| Formula | C22H22N2O | C23H26N2O | C96H96N8O12Pd4·7CHCl3·H2O | C27H30N2O3Pd·0.75C6H14 |
| M | 330.42 | 346.46 | 2833.00 | 623.10 |
| Crystal size (mm3) | 0.41 × 0.35 × 0.20 | 0.35 × 0.30 × 0.26 | 0.43 × 0.24 × 0.15 | 0.31 × 0.24 × 0.13 |
| Temperature (K) | 150(2) | 150(2) | 150(2) | 150(2) |
| Crystal system | Monoclinic | Monoclinic | Monoclinic | Monoclinic |
| Space group | P2(1)/c | P2(1)/c | P2(1)/c | C2/c |
| a (Å) | 7.6425(19) | 9.166(6) | 27.533(6) | 26.910(8) |
| b (Å) | 11.027(3) | 16.955(11) | 19.525(4) | 14.159(4) |
| c (Å) | 20.590(5) | 13.033(9) | 23.435(5) | 15.463(5) |
| α (°) | 90 | 90 | 90 | 90 |
| β (°) | 93.528(5) | 102.965(11) | 111.63(3) | 110.788(6) |
| γ (°) | 90 | 90 | 90 | 90 |
| U (Å3) | 1731.8(7) | 1974(2) | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 711(4) 711(4) |
5508(3) |
| Z | 4 | 4 | 4 | 8 |
| D c (Mg m−3) | 1.267 | 1.166 | 1.607 | 1.503 |
| F(000) | 704 | 744 | 5696 | 2608 |
| μ(Mo-Kα)(mm−1) | 0.078 | 0.071 | 1.144 | 0.712 |
| Reflections collected | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 311 311 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 931 931 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 937 937 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 197 197 |
| Independent reflections | 3410 | 3471 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 937 937 |
5402 |
| R int | 0.0574 | 0.0595 | 0.000 | 0.0834 |
| Restraints/parameters | 0/229 | 0/239 | 1134/1358 | 0/304 |
| Final R indices (I > 2σ(I)) | R 1 = 0.0489 | R 1 = 0.0528 | R 1 = 0.0958 | R 1 = 0.0490 |
| wR2 = 0.1054 | wR2 = 0.1289 | wR2 = 0.1419 | wR2 = 0.1019 | |
| All data | R 1 = 0.0705 | R 1 = 0.0708 | R 1 = 0.2822 | R 1 = 0.0713 |
| wR2 = 0.1147 | wR2 = 0.1385 | wR2 = 0.1947 | wR2 = 0.1084 | |
| Goodness of fit on F2 (all data) | 0.981 | 1.030 | 0.822 | 0.959 |
| Complex | 2a | 2b | 3a | 5a |
|---|---|---|---|---|
| a Data in common: graphite-monochromated Mo-Kα radiation, λ = 0.71073 Å; R1 = ∑||Fo| − |Fc||/∑|Fo|, wR2 = [∑w(Fo2 − Fc2)2/∑w(Fo2)2]1/2, w−1 = [σ2(Fo)2 + (aP)2], P = [max(Fo2, 0) + 2(Fc2)]/3, where a is a constant adjusted by the program; goodness of fit = [∑(Fo2 − Fc2)2/(n − p)]1/2 where n is the number of reflections and p the number of parameters. | ||||
| Formula | C25H28N2O3Pd·1.5CHCl3 | C28H34N2O3Pd·CH2Cl2 | C22H21ClN2OPd·CHCl3 | C25H24F3N3O4PdS·MeCN |
| M | 6889.95 | 637.90 | 590.63 | 666.99 |
| Crystal size (mm3) | 0.23 × 0.15 × 0.04 | 0.37 × 0.24 × 0.20 | 0.35 × 0.29 × 0.07 | 0.45 × 0.43 × 0.04 |
| Temperature (K) | 150(2) | 150(2) | 150(2) | 150(2) |
| Crystal system | Monoclinic | Monoclinic | Monoclinic | Triclinic |
| Space group | P2(1)/c | P2(1)/c | P2(1)/c |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a (Å) | 16.155(4) | 16.640(6) | 17.785(4) | 13.264(11) |
| b (Å) | 13.910(3) | 10.960(4) | 8.6156(19) | 13.822(11) |
| c (Å) | 13.360(3) | 17.137(6) | 16.469(4) | 17.160(14) |
| α (°) | 90 | 90 | 90 | 80.989(15) |
| β (°) | 109.643(5) | 116.252(5) | 110.168(4) | 78.907(15) |
| γ (°) | 90 | 90 | 90 | 64.369(13) |
| U (Å3) | 2827.5(11) | 2803.1(16) | 2368.8(9) | 2774(4) |
| Z | 4 | 4 | 4 | 4 |
| D c (Mg m−3) | 1.621 | 1.512 | 1.656 | 1.597 |
| F(000) | 1396 | 1312 | 1184 | 1352 |
| μ(Mo-Kα)(mm−1) | 1.113 | 0.866 | 1.253 | 0.805 |
| Reflections collected | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 017 017 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 332 332 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 002 002 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 653 653 |
| Independent reflections | 5551 | 5505 | 4659 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 758 758 |
| R int | 0.1884 | 0.0497 | 0.1264 | 0.1073 |
| Restraints/parameters | 277/285 | 0/341 | 0/283 | 36/740 |
| Final R indices (I > 2σ(I)) | R 1 = 0.0695 | R 1 = 0.0373 | R 1 = 0.0554 | R 1 = 0.0999 |
| wR2 = 0.1575 | wR2 = 0.0935 | wR2 = 0.0830 | wR2 = 0.2351 | |
| All data | R 1 = 0.1940 | R 1 = 0.0439 | R 1 = 0.1026 | R 1 = 0.1570 |
| wR2 = 0.1815 | wR2 = 0.0966 | wR2 = 0.0938 | wR2 = 0.2619 | |
| Goodness of fit on F2 (all data) | 0.725 | 1.059 | 0.897 | 1.033 |
CCDC reference numbers 1040521–1040528.
Acknowledgements
We thank the University of Leicester for financial assistance. Johnson Matthey PLC are thanked for their generous loan of palladium salts.References
- For reviews see: (a) N. R. Deprez and M. S. Sanford, Inorg. Chem., 2007, 46, 1924–1935 CrossRef CAS PubMed; (b) P. J. Stang and V. V. Zhdankin, Chem. Rev., 1996, 96, 1123–1178 CrossRef CAS PubMed; (c) A. Varvoglis, Tetrahedron, 1997, 53, 1179–1255 CrossRef CAS; (d) T. Wirth and U. H. Hirt, Synthesis, 1999, 1271–1287 CrossRef CAS PubMed; (e) T. Okuyama, Acc. Chem. Res., 2002, 35, 12–18 CrossRef CAS PubMed; (f) V. V. Zhdankin and P. Stang, Chem. Rev., 2002, 102, 2523–2584 CrossRef CAS PubMed; (g) Hypervalent Iodine Chemistry, Modern Developments in Organic Synthesis, in Topics in Current Chemistry, ed. T. Wirth, Springer, New York, 2003, vol. 224 Search PubMed; (h) A. J. Canty, T. Rodemann and J. H. Ryan, Adv. Organomet. Chem., 2008, 55, 279–313 CrossRef CAS; (i) K. Muñiz, Angew. Chem., Int. Ed., 2009, 48, 9412–9423 CrossRef PubMed.
- (a) J. Aydin, J. M. Larsson, N. Selander and K. J. Szabo, Org. Lett., 2009, 11, 2852–2854 CrossRef CAS PubMed; (b) E. A. Marritt and B. Olofsson, Angew. Chem., Int. Ed., 2009, 48, 9052–9070 CrossRef PubMed; (c) L.-M. Xu, B.-J. Li, Z. Yang and Z.-J. Shi, Chem. Soc. Rev., 2010, 39, 712–733 RSC; (d) P. D. Chaudhuri, R Guo and H. C. Malinakova, J. Organomet. Chem., 2007, 693, 567–573 CrossRef PubMed; (e) H. C. Malinakova, Top. Organomet. Chem., 2011, 35, 85–110 CrossRef CAS; (f) Y. Ye, N. D. Ball, J. W. Kampf and M. S. Sanford, J. Am. Chem. Soc., 2010, 132, 14682–14687 CrossRef CAS PubMed; (g) K. M. Engle, T.-S. Mei, M. Wasa and J.-Q. Yu, Acc. Chem. Res., 2012, 45, 788–802 CrossRef CAS PubMed; (h) X.-G. Zhang, H.-X. Dai, M. Wasa and J.-Q. Yu, J. Am. Chem. Soc., 2012, 134, 11948–11951 CrossRef CAS PubMed; (i) H. Zhang and A. Lei, Dalton Trans., 2011, 40, 8745–8754 RSC.
- (a) D. Kalyanai, N. R. Deprez, L. V. Desai and M. S. Sanford, J. Am. Chem. Soc., 2005, 127, 7330–7331 CrossRef PubMed; (b) E. W. Kalberer, S. R. Whifield and M. S. Sanford, J. Mol. Catal. A: Chem., 2006, 251, 108–113 CrossRef PubMed; (c) N. R. Deprez and M. S. Sanford, J. Am. Chem. Soc., 2009, 131, 11234–11241 CrossRef CAS PubMed; (d) A. J. Canty, A. Ariafard, M. S. Sanford and B. F. Yates, Organometallics, 2013, 32, 544–555 CrossRef CAS; (e) A. J. Canty, Dalton Trans., 2009, 10409–10417 RSC.
- (a) A. J. Canty, J. Patel, T. Rodemann, J. H. Ryan, B. W. Skelton and A. H. White, Organometallics, 2004, 23, 3466–3473 CrossRef CAS; (b) A. Bayler, A. J. Canty, J. H. Ryan, B. W. Skelton and A. H. White, Inorg. Chem. Commun., 2000, 3, 575–578 CrossRef CAS.
- K. J. Szabó, J. Mol. Catal. A: Chem., 2010, 324, 56–63 CrossRef PubMed.
- (a) J. Vicente, M. T. Chicote, J. Martin, M. Artigao, X. Solans, M. Font-Altaba and M. Aguilo, J. Chem. Soc., Dalton Trans., 1988, 141–147 RSC; (b) A. J. Canty, S. D. Fritshe, H. Jin, B. W. Skelton and A. H. White, J. Organomet. Chem., 1995, 490, C18–C19 CrossRef CAS; (c) A. J. Canty, H. Jin, A. S. Roberts, B. W. Skelton and A. H. White, Organometallics, 1996, 15, 5713–5722 CrossRef CAS; (d) R. van Belzen, C. J. Elsevier, A. Dedieu, N. Veldman and A. L. Spek, Organometallics, 2003, 22, 722–736 CrossRef CAS; (e) S. R. Whitfield and M. S. Sanford, J. Am. Chem. Soc., 2007, 129, 15141–15143 CrossRef PubMed.
- C–Cl bond forming reductive elimination from Pd(IV) is considered highly thermodynamically favourable when compared with the corresponding elimination from Pd(II): See ref. 1a and A. H. Hoy and J. F. Hartwig, Organometallics, 2004, 23, 1533–1541 CrossRef.
- (a) M.-C. Lagunas, R. A. Gossage, A. L. Spek and G. van Koten, Organometallics, 1998, 17, 731–741 CrossRef CAS; (b) A. J. Canty, M. C. Denney, G. van Koten, B. W. Skelton and A. H. White, Organometallics, 2004, 23, 5432–5439 CrossRef CAS; (c) L. T. Pilarski, N. Selander, D. Boese and K. J. Szabó, Org. Lett., 2009, 11, 5518–5521 CrossRef CAS PubMed; (d) N. Selander, B. Willy and K. J. Szabó, Angew. Chem., Int. Ed., 2010, 49, 4051–4053 CrossRef CAS PubMed.
- P. L. Alsters, P. F. Engel, M. P. Hogerheide, M. Copijn, A. L. Spek and G. van Koten, Organometallics, 1993, 12, 1831–1844 CrossRef CAS.
- (a) J. Vicente, A. Arcas, F. Julia-Hernández and D. Bautista, Chem. Commun., 2010, 46, 7253–7255 RSC; (b) J. Vicente, A. Arcas, F. Julia-Hernández and D. Bautista, Inorg. Chem., 2011, 50, 5339–5341 CrossRef CAS PubMed; (c) J. Vicente, A. Arcas, F. Julia-Hernández and D. Bautista, Angew. Chem., Int. Ed., 2011, 50, 6896–6899 CrossRef CAS PubMed.
- (a) T. Furuya and T. Ritter, J. Am. Chem. Soc., 2008, 130, 10060–10061 CrossRef CAS PubMed; (b) T. Furuya, D. Benitez, E. Tkatchouk, A. E. Strom, P. Tang, W. A. Goddard III and T. Ritter, J. Am. Chem. Soc., 2010, 132, 3793–3807 CrossRef CAS PubMed; (c) M. G. Campbell and T. Ritter, Org. Process Res. Dev., 2014, 18, 474–480 CrossRef CAS.
- W. Alkarekshi, A. P. Armitage, O. Boyron, C. J. Davies, M. Govere, A. Gregory, K. Singh and G. A. Solan, Organometallics, 2013, 32, 249–259 CrossRef CAS.
- C. J. Davies, A. Gregory, P. Griffith, T. Perkins, K. Singh and G. A. Solan, Tetrahedron, 2008, 64, 9857–9864 CrossRef CAS PubMed.
- O. Adeyi, W. B. Cross, G. Forrest, L. Godfrey, E. G. Hope, A. McLeod, A. Singh, K. Singh, G. A. Solan, Y. Wang and L. A. Wright, Dalton Trans., 2013, 42, 7710–7723 RSC.
- W. B. Cross, E. G. Hope, G. Forrest, K. Singh and G. A. Solan, Polyhedron, 2013, 59, 124–132 CrossRef CAS PubMed.
- (a) M. Lopez-Torres, P. Juanatey, J. J. Fernandez, A. Fernandez, A. Suarez, D. Vazquez-Garcia and J. M. Vila, Polyhedron, 2002, 21, 2063–2069 CrossRef CAS; (b) H. Onoue and I. Moritani, J. Organomet. Chem., 1972, 43, 431–436 CrossRef CAS; (c) H. Onoue, K. Minami and K. Nakagawa, Bull. Chem. Soc. Jpn., 1970, 43, 3480–3485 CrossRef CAS.
- K. Nakamoto, IR and Raman Spectra of Inorganic and Coordination Compounds, Wiley, New York, 5th edn, 1997, Part B, p. 271 Search PubMed.
- (a) E. T. J. Strong, J. T. Price and N. D. Jones, Dalton Trans., 2009, 9123–9125 RSC; (b) D. M. Pearson, N. R. Conley and R. M. Waymouth, Organometallics, 2011, 30, 1445–1453 CrossRef CAS.
- W. L. F. Armarego and D. D. Perrin, in Purification of Laboratory Chemicals, Butterworth Heinemann, 4th edn, 1996 Search PubMed.
- J. E. Parks, B. E. Wagner, R. H. Holm and J. E. Parks, J. Organomet. Chem., 1974, 56, 53–66 CrossRef.
- M. Bielawski and B. Olofsson, Chem. Commun., 2007, 2521–2523 RSC.
- G. M. Sheldrick, SHELXTL Version 6.10, Bruker AXS, Inc., Madison, Wisconsin, USA, 2000 Search PubMed.
- A. L. Spek, Acta Crystallogr., Sect. A: Fundam. Crystallogr., 1990, 46, C34 Search PubMed.
Footnote |
| † CCDC 1040521–1040528. For crystallographic data in CIF or other electronic format see DOI: 10.1039/c5dt00062a |
| This journal is © The Royal Society of Chemistry 2015 |