Open Access Article
Open Access ArticleConvenient detection of metal–DNA, metal–RNA, and metal–protein adducts with a click-modified Pt(II) complex†
Alan D.
Moghaddam‡
a,
Jonathan D.
White‡
a,
Rachael M.
Cunningham
ab,
Andrea N.
Loes
ab,
Michael M.
Haley
a and
Victoria J.
DeRose
*ab
aDepartment of Chemistry & Biochemistry, University of Oregon, Eugene, OR 97403-1253, USA
bInstitute of Molecular Biology, University of Oregon, Eugene, OR 97403-1253, USA. E-mail: derose@uoregon.edu; Tel: +1-541-346-3568
First published on 8th October 2014
Abstract
cis-[Pt(2-azido-1,3-propanediamine)Cl2] is a reagent for high-yield post-treatment fluorescent labelling of Pt(II) biomolecular targets using click chemistry and exhibits a bias in conformational isomers in the context of duplex DNA. Pt–protein adducts are detected using BSA as a model. Following in vivo treatment, long-lived Pt–RNA adducts are detected on ribosomal RNA.
Platinum(II)-based drugs are administered in many anticancer treatment regimes;1 however, a broad identification of Pt-specific cellular targets is still lacking.2 As Pt(II) compounds are effective nucleic acid crosslinking agents, they also have potential as structure probes in complex RNAs.3 Click-functionalized Pt reagents provide a reactive handle for post-treatment labelling that would enable broad detection, isolation and identification of Pt-bound species. We recently described a new pyridine-based Pt click reagent, picazoplatin (1, Fig. 1).4 Despite the wide use of click chemistry and importance of platinum therapeutics, only one other recently reported compound, an extended Pt-acridine therapeutic, is available for post-treatment click analysis.5 In the case of picazoplatin, relatively slow reaction kinetics are built into the design of the parent compound picoplatin wherein the ring methyl shields Pt from substitution reactions.6 Moreover, although in-cell use was demonstrated, analysis of cellular targets may be complicated by the potential for trans-labilization of the azide-functionalized picoline ring. New click-functionalized Pt(II) reagents are of great interest in order to take advantage of bioorthogonal approaches7 for high-throughput identification of Pt-bound species in vivo.
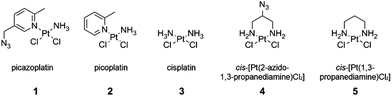 | ||
| Fig. 1 Click-functionalized picazoplatin 1 and its parent picoplatin 2, with cisplatin 3 and the 1,3-propanediamine derivatives 4 and 5. | ||
To provide a convenient and fast-reacting Pt(II)-azide click reagent, we have explored cis-[Pt(2-azido-1,3-propanediamine)Cl2] (4) in nucleic acid binding, protein binding, and click chemistry. While 4 has been previously used to create libraries of diazenecarboxamide–carboplatin conjugates,8 to our knowledge it has not been applied as an agent for post-treatment labelling by click modification. Herein, we demonstrate efficient click reactivity of the DNA-bound compound and an unusual isomeric differentiation of 4 when bound to duplex DNA. Click fluorescent labelling of Pt-bound proteins is demonstrated in bovine serum albumin (BSA). The utility of this compound for in vivo studies is demonstrated by tracking Pt-bound ribosomes from cells treated with 4.
DNA binding studies of 4 on a 13-nucleotide DNA containing a single GG site were analysed by dPAGE (Fig. 2). A higher yield of 4-bound DNA is observed in comparison with the sterically-hindered 1. Subsequent click reactivity with a dansyl alkyne fluorophore is seen in both cases.
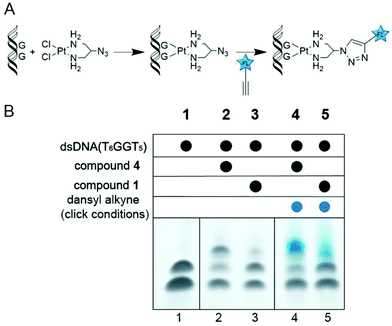 | ||
| Fig. 2 (A) Reaction scheme for Pt binding and subsequent click to an alkyne fluorophore. (B) dPAGE analysis of dsDNA bound by picazoplatin (1) and cis-[Pt(2-azido-1,3-propanediamine)Cl2] (4) and subsequently clicked to a dansyl alkyne fluorophore. The denatured DNA strands are visible in lane 1, while higher molecular weight Pt-bound species are visible in lanes 2 and 3. Click reaction results in fluorescence of the Pt–DNA band under UV light in lanes 4 and 5 (see ESI†). | ||
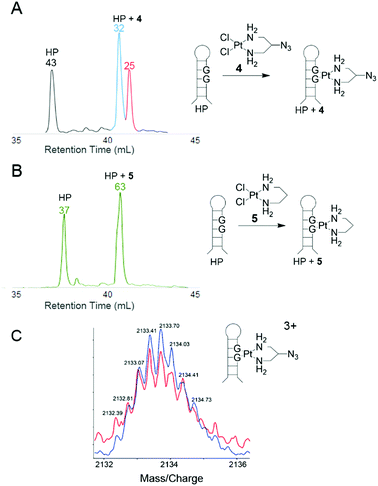
To further quantify Pt binding and click reactivity of 4, HPLC analysis was used with a short DNA hairpin sequence (TATGGTATTTTTATACCATA, HP). Upon Pt binding to the single GG site, two product peaks emerge that are distinct from the parent DNA peak. Both new peaks were confirmed by dPAGE to contain hairpin DNA bound by Pt (see ESI†). Both products contain one bound Pt(2-azido-1,3-propanediamine) as confirmed by identical 3+ charged ions in ESI-MS analysis (Fig. 3C). When reacted with dansyl alkyne, material in both product peaks yields clicked products as resolved by HPLC and identified by the fluorophore absorbance at 340 nm (>80% yield, see ESI†). Thus, this bimodal product distribution is hypothesized to originate from two different structural isomers of 4 bound to the asymmetric DNA strand. This hypothesis was tested with the analogous azide-free complex 5 (see ESI†).9 As expected, the HPLC trace of DNA-bound 5 exhibits only one product peak (Fig. 3B), indicating that the 2-azido group of 4 creates structural isomers.
The two isomer products of 4-treated hairpin DNA consistently yield non-equivalent peak areas, indicating preference of one conformation that is likely being dictated by the orientation of the azide relative to the helix. Supporting this, treatment of single-stranded DNA shows equivalent populations of both isomers (Fig. 4C), whereas the same sequence in duplex returns to an unequal population (Fig. 4D, see also ESI†).
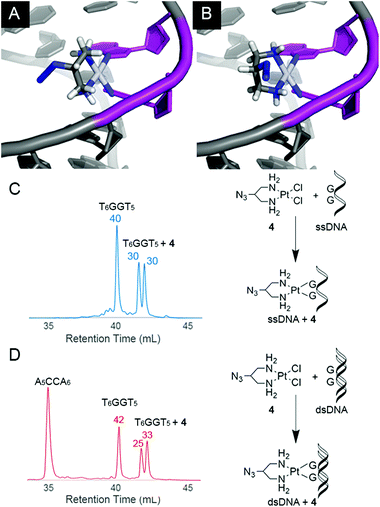 | ||
| Fig. 4 Molecular modelling (MMFF) of the two possible isomers of 4-bound double-stranded DNA with the azide oriented in towards (A) or out from (B) the centre of the helix. In the helical conformation of A as shown, the azide moiety is less solvent exposed than in B. Models were generated by substituting 4 into the crystal structure of cisplatin-bound DNA (PDB: 3LPV, see ESI†). The adjacent Pt-bound guanines of the intrastrand crosslink are shaded purple. HPLC absorbance traces at 260 nm for 4 reacted with (T6GGT5) as a single-strand (C, 55–65% yield) or in duplex (D, 55–65% yield). Normalized areas are indicated above peaks of interest. When reacted with the single-strand sequence, isomers of 4-bound DNA show equal population (ratio = 0.96 ± 0.05; see ESI† for data in triplicate) whereas they are unequal for the duplex (ratio = 1.3 ± 0.09; see also ESI†). | ||
Possible conformations of DNA-bound 4 are visualized (Fig. 4) in molecular models built by substituting 4 into the crystal structure of cisplatin-bound 12-mer DNA duplex (pdb: 3LPV).10 Bond lengths and angles around the Pt(II) centre were fixed and the position of the DNA was locked (see ESI†), and equilibrium geometries of the two structural isomers were determined using molecular mechanics (MMFF). The resulting models show distinct geometric properties, with the azide oriented either toward the major groove or towards the phosphate backbone. In the denatured strands analysed by HPLC, these isomers must give rise to slight differences in hydrophobicity; resolution of such minor differences in oligonucleotides has precedent.11,12 The inequivalent populations of isomers suggest that the helical structure of DNA has a direct impact on the preferred orientation of the azide substituent due to steric and electronic effects. Like the hairpin DNA, upon reaction with the dansyl alkyne fluorophore, 4-bound T6GGT5 is converted into clicked products with yields that range from 80–90% (see ESI†).
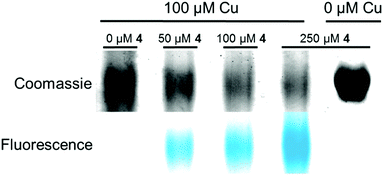
In furthering the analysis of potential targets for Pt(II) compounds, bovine serum albumin (BSA) was also investigated as a model compound for the ability of 4 to bind and participate in post-treatment labelling of proteins. Previous studies have shown Pt-binding to Cys 3413 following treatment of BSA with cisplatin. Here BSA is incubated with increasing equivalents of 4 followed by click to an alkyne-containing dansyl fluorophore (Fig. 5). Labelled protein was visualized by Coomassie staining and fluorescence imaging. Fluorescence is seen only in the 4-treated samples and increases in a dose-dependent manner (Fig. 5, lanes 2–4). Fluorescence is also only observed in reactions containing both 4 and the Cu catalyst.
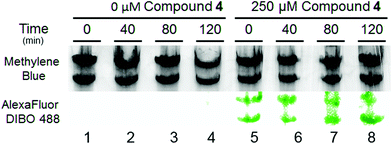
Ultimately, the power of clickable Pt compounds is to identify and analyze in vivo targets. In contrast to cisplatin, Pt(II) compounds with small chelating amine ligands such as ethylenediamine and 1,3-propanediamine are better tolerated by cells, making them particularly attractive as potential in vivo crosslinking reagents.14 We have previously shown significant accumulation of Pt on cellular RNA following treatment with cisplatin, with major accumulation being on ribosomal RNA.15,16 Here, we tracked the lifetime of Pt–RNA adducts using 4, and find surprising longevity of Pt adducts on ribosomal RNA. S. cerevisiae was treated with 0 μM and 250 μM 4 for 6 h and then re-suspended in fresh media. Cells resuspended in drug-free media showed growth similar to untreated control cells, consistent with the relatively low toxicity of 4 and in contrast to treatment with cisplatin.15 To determine whether Pt–rRNA adducts were quickly degraded or long-lived, RNA was extracted from cells at various timepoints and accumulated Pt detected following click labelling with Alexa Fluor 488 DIBO (Fig. 6). Lanes 1–4 contain rRNA isolated from untreated cells subjected to the same click labelling protocol. The absence of fluorescence in these lanes indicates the absence of nonspecific labelling during the DIBO/click procedure. Lanes 5–8 contain rRNA isolated from treated cells after up to 2 h in 4-free media. The normalized fluorescence per rRNA band shows no significant loss in Pt–RNA adducts, meaning that platinated ribosomes persist in doubling cells and that the majority of them are not targeted for fast degradation. The persistence of the fluorescent labelling even after 120 min post-treatment demonstrates the efficacy of 4 in post-labelling strategies to ascertain possible Pt(II) cellular targets.
Conclusions
This work demonstrates the ability of azide-modified Pt(II) compound 4 to readily bind oligonucleotides and proteins and subsequently undergo click reactions for post-treatment fluorescent labelling. This demonstrated reactivity of 4 expands the array of available azide-modified Pt(II) reagents to be used in post-treatment target analysis. In comparison with recently reported 1, 4 exhibits greatly increased efficiency in target binding. HPLC analysis and molecular modelling suggest formation of two different isomers of 4-bound DNA adducts, hypothesized to be biased by the orientation of the azide moiety of 4 in relation to the helical structure of DNA. Efforts to discern differences in click reactivity between the two isomers are currently underway in our laboratory, as well as efforts utilizing clickable Pt(II) reagents as RNA crosslinking agents to quantify and isolate platinum-bound species in cellulo.Acknowledgements
We thank the National Science Foundation (CHE-1153147) for support of this research.Notes and references
- (a) B. W. Harper, A. M. Krause-Heuer, M. P. Grant, M. Manohar, K. B. Garbutcheon-Singh and J. R. Aldrich-Wright, Chem. – Eur. J., 2010, 16, 7064–7077 CrossRef CAS PubMed; (b) P. J. Dyson and G. Sava, Dalton Trans., 2006, 1929–1933 RSC; (c) D. Wang and S. J. Lippard, Nat. Rev. Drug Discovery, 2005, 4, 307–320 CrossRef CAS PubMed.
- (a) G. Sava, G. Jaouen, E. A. Hillard and A. Bergamo, Dalton Trans., 2012, 41, 8226–8234 RSC; (b) A. Casini and J. Reedijk, Chem. Sci., 2012, 3, 3135–3144 RSC; (c) E. R. Guggenheim, D. Xu, C. X. Zhang, P. V. Chang and S. J. Lippard, ChemBioChem, 2009, 10, 141–157 CrossRef CAS PubMed; (d) E. Wexselblatt, E. Yavin and D. Gibson, Inorg. Chim. Acta, 2012, 393, 75–83 CrossRef CAS PubMed.
- (a) E. G. Chapman and V. J. DeRose, J. Am. Chem. Soc., 2012, 134, 256–262 CrossRef CAS PubMed; (b) E. G. Chapman and V. J. DeRose, J. Am. Chem. Soc., 2010, 132, 1946–1952 CrossRef CAS PubMed; (c) A. A. Hostetter, E. G. Chapman and V. J. DeRose, J. Am. Chem. Soc., 2009, 131, 9250–9257 CrossRef CAS PubMed.
- J. D. White, M. F. Osborn, A. D. Moghaddam, L. E. Guzman, M. M. Haley and V. J. DeRose, J. Am. Chem. Soc., 2013, 135, 11680–11683 CrossRef CAS PubMed.
- (a) S. Ding, X. Qiao, J. Suryadi, G. S. Marrs, G. L. Kucera and U. Bierbach, Angew. Chem. Int. Ed., 2013, 52, 3350–3354 CrossRef CAS PubMed; (b) X. Qiao, S. Ding, F. Liu, G. L. Kucera and U. Bierbach, J. Biol. Inorg. Chem., 2014, 19, 415–426 CrossRef CAS PubMed.
- (a) L. Kelland, Nat. Rev. Cancer, 2007, 7, 573–584 CrossRef CAS PubMed; (b) F. I. Raynaud, F. E. Boxall, P. M. Goddard, M. Valenti, M. Jones, B. A. Murrer, M. Abrams and L. R. Kelland, Clin. Cancer Res., 1997, 3, 2063–2074 CAS; (c) Y. Chen, Z. Guo, S. Parsons and P. J. Sadler, Chem. – Eur. J., 1998, 4, 672–676 CrossRef CAS; (d) Y. Chen, Y. Z. Guo, J. A. Parkinson and P. J. Sadler, Dalton Trans., 1998, 3577–3586 RSC; (e) L. R. Kelland, S. Y. Sharp, C. F. O'Neill, F. I. Raynaud, P. J. Beale and I. R. Judson, J. Inorg. Biochem., 1999, 77, 111–115 CrossRef CAS.
- (a) J. C. Jewett and C. R. Bertozzi, Chem. Soc. Rev., 2010, 39, 1272–1279 RSC; (b) H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem. Int. Ed., 2001, 40, 2004–2021 CrossRef CAS.
- (a) N. Stojanovic, D. Urankar, A. Brozovic, A. Ambriović-Ritsov, M. Osmak and J. Košmrlj, Acta Chim. Slov., 2013, 60, 368–374 CAS; (b) D. Urankar and J. Košmrlj, Inorg. Chim. Acta, 2010, 363, 3817–3822 CrossRef CAS PubMed.
- J. Zhang, L. Ma, H. Lu, Y. Wang, S. Li, S. Wang and G. Zhou, Eur. J. Med. Chem., 2012, 58, 281–286 CrossRef CAS PubMed.
- R. C. Todd and S. J. Lippard, J. Inorg. Biochem., 2010, 104, 902–908 CrossRef CAS PubMed.
- (a) W. L. Ward and V. J. DeRose, RNA, 2012, 18, 16–23 CrossRef CAS PubMed; (b) J. K. Frederiksen and J. A. Piccirilli, Methods in Enzymology, ed. D. Herschlag, Academic Press, 2009, ch. 14, vol. 468, pp. 289–309 Search PubMed.
- M. J. Bloemink, R. J. Heetebrij, K. Inagaki, Y. Kidani and J. Reedijk, Inorg. Chem., 1992, 31, 4656–4661 CrossRef CAS.
- A. Sugii, K. Nishimura, K. Harada, M. Nakayama and S. Masuda, Chem. Pharm. Bull., 1991, 39, 408–410 CrossRef CAS.
- N. H. Kuroda, Y. Hayami, O. K. Ekimoto and H. K. Takahashi, Chem. Pharm. Bull., 1989, 37, 2406–2409 CrossRef.
- A. A. Hostetter, M. F. Osborn and V. J. DeRose, ACS Chem. Biol., 2012, 7, 218–225 CrossRef CAS PubMed.
- M. F. Osborn, J. D. White, M. M. Haley and V. J. DeRose, ACS Chem. Biol., 2014, 9, 2404–2411 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, additional HPLC and dPAGE characterization, 1H, 13C, and 195Pt NMR spectra of 5. See DOI: 10.1039/c4dt02649g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |