Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
RNA and DNA binding of inert oligonuclear ruthenium(II) complexes in live eukaryotic cells
Xin
Li
a,
Anil K.
Gorle
a,
Tracy D.
Ainsworth
b,
Kirsten
Heimann
*cd,
Clifford E.
Woodward
a,
J.
Grant Collins
*a and
F.
Richard Keene
*def
aSchool of Physical, Environmental and Mathematical Sciences, University of New South Wales, Australian Defence Force Academy, Canberra, ACT 2600, Australia. E-mail: g.collins@adfa.edu.au
bARC Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, QLD 4811, Australia
cCollege of Marine & Environmental Sciences, James Cook University, Townsville, QLD 4811, Australia. E-mail: kirsten.heimann@jcu.edu.au
dCentre for Biodiscovery and Molecular Development of Therapeutics, James Cook University, Townsville, QLD 4811, Australia
eDepartment of Matter & Materials, College of Science, Technology & Engineering, James Cook University, Townsville, QLD 4811, Australia
fSchool of Chemistry and Physics, University of Adelaide, Adelaide, SA 5005, Australia. E-mail: richard.keene@adelaide.edu.au
First published on 21st October 2014
Abstract
Confocal microscopy was used to study the intracellular localisation of a series of inert polypyridylruthenium(II) complexes with three eukaryotic cells lines – baby hamster kidney (BHK), human embryonic kidney (HEK-293) and liver carcinoma (Hep-G2). Co-staining experiments with the DNA-selective dye DAPI demonstrated that the di-, tri- and tetra-nuclear polypyridylruthenium(II) complexes that are linked by the bis[4(4′-methyl-2,2′-bipyridyl)]-1,12-dodecane bridging ligand (“bb12”) showed a high degree of selectivity for the nucleus of the eukaryotic cells. Additional co-localisation experiments with the general nucleic acid stain SYTO 9 indicated that the ruthenium complexes showed a considerable preference for the RNA-rich nucleolus, rather than chromosomal DNA. No significant differences were observed in the intracellular localisation between the ΔΔ and ΛΛ enantiomers of the dinuclear complex. Cytotoxicity assays carried out over 72 hours indicated that the ruthenium complexes, particularly the tri- and tetra-nuclear species, were significantly toxic to the eukaryotic cells. However, when the activity of the least cytotoxic compound (the ΔΔ enantiomer of the dinuclear species) was determined over a 24 hour period, the results indicated that the ruthenium complex was approximately a 100-fold less toxic to liver and kidney cells than to Gram positive bacteria. Circular dichroism (CD) spectroscopy was used to examine the effect of the ΔΔ and ΛΛ enantiomers of the dinuclear complex on the solution conformations of RNA and DNA. The CD experiments indicated that the RNA maintained the A-type conformation, and the DNA the B-type structure, upon binding by the ruthenium complexes.
Introduction
There has been significant interest over the last forty years in the non-covalent interactions of inert transition metal complexes with DNA and RNA.1–3 In particular, the nucleic acid binding properties of ruthenium(II) complexes containing polypyridyl ligands have been extensively studied.4–8 These metal complexes have a rigid octahedral framework and can interact with nucleic acids through a variety of different modes, with the particular mode of binding being predictably governed by the metal complex structure. Furthermore, the structure of a ruthenium(II) complex can be readily modified – e.g. shape, charge, or the addition of specific recognition elements – to “tune” nucleic acid binding. Additionally, and if applicable, the chirality of the ruthenium complex can also be used to gain even greater control over the specificity or selectivity of the binding.More recently, due to the nucleic binding properties of inert polypyridylruthenium(II) complexes, there has been increasing interest in their biological properties.9–16 A variety of mononuclear and dinuclear complexes have shown good in vitro anticancer activity, which is generally considered to be due to DNA binding. However, in some cases other mechanisms of action have been proposed – e.g. interactions with membranes or mitochondrial-mediated apoptosis.15 In addition to the established anticancer properties of inert polypyridylruthenium(II) complexes, there is now growing recognition of their potential as antimicrobial agents. Antimicrobial resistance is an increasingly serious threat to global public health: infections caused by antibiotic-resistant bacteria are associated with increased morbidity and mortality.17 The lack of new antimicrobials in the pipeline to replace those in current use which are becoming ineffective has fostered research into the development of new types of drugs.
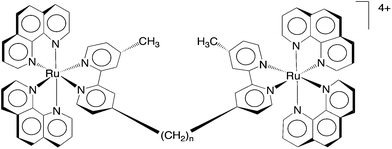
Dwyer and co-workers initially demonstrated the antimicrobial activity of mononuclear polypyridylruthenium(II) complexes against both Gram negative and Gram positive bacteria.18,19 We have subsequently shown that dinuclear analogues have even greater antimicrobial potential: [{Ru(phen)2}2{μ-bbn}]4+ {“Rubbn”; where phen = 1,10-phenanthroline; bbn = bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane for n = 5, 7, 10, 12 and 16 – see Fig. 1} showed excellent activity, and they maintained the activity against drug-resistant strains such as methicillin-resistant Staphylococcus aureus (MRSA).20 Furthermore, preliminary toxicity assays against human red blood cells and a human white blood leukemia cell line (THP-1) demonstrated that the Rubbn complexes were not toxic to human cells at the concentrations required to kill the bacteria.20
While the affinity of polypyridylruthenium(II) complexes for nucleic acids can be readily demonstrated in vitro, it is more important to establish nucleic acid binding in live cells at concentrations similar to those required for anticancer or antimicrobial activities. Although there have been relatively few cellular localisation studies of polypyridylruthenium(II) complexes, the results reported to date have demonstrated a surprisingly diverse range of binding sites in eukaryotic cells. For example, Svensson et al. showed that the cellular localisation of a series of ruthenium dipyridophenazine (dppz) complexes in Chinese hamster ovarian cells was dependent upon the relative lipophilicity.21 The least lipophilic complex was predominantly found in the nucleus and the most lipophilic accumulated outside of the nucleus and probably in the endoplasmic reticulum. Furthermore, Gill et al. demonstrated that the DNA groove-binding dinuclear complex [{Ru(phen)2}2{μ-tpphz}]4+ (where tpphz = tetrapyridophenazine) could be used to image nuclear DNA in eukaryotic cells.11 Alternatively, the 4,7-diphenyl-1,10-phenanthroline analogue [{Ru(DIP)2}2{μ-tpphz}]4+ localised in the endoplasmic reticulum.22 By contrast, the Rubbn complexes were shown to localise in the mitochondria of L1210 white blood cells.12 Mitochondrial targeting has also been observed for other ruthenium complexes.15
As preliminary pharmacokinetic studies indicated that the Rubbn complexes accumulate in the liver and kidney of mice,23 we sought to confirm that the ruthenium complexes localised in the mitochondria of liver and kidney cells as we had previously demonstrated with the L1210 cells.12 In the present study, we examined the localisation of Rubb12 and its tri- and tetra-nuclear analogues in liver and kidney cells by confocal microscopy. In order to examine the effect of the ruthenium complexes on large DNA and RNA molecules, we also studied the binding of the ruthenium complexes to calf thymus DNA and baker's yeast RNA by CD spectroscopy. Interestingly, Rubb12 was found to selectively accumulate in the nucleolus, the RNA-rich component of the nucleus, rather than in the mitochondria.
Experimental
Synthesis of ruthenium(II) complexes
The ruthenium complexes used in this study were synthesised and characterised as previously described.24,25Cell culture
The BHK (baby hamster kidney) cell line and two human cell lines – HEK-293 (embryonic kidney) and Hep-G2 (liver carcinoma) – were used in this study. All cell lines were generously supplied by Australian Army Malaria Institute (AMI, Enoggera, QLD, Australia), and originated from the American Type Culture Collection (ATCC, Manassas, VA). All cell lines were cultured in 75 mL culture flasks in RPMI-1640 (Roswell Park Memorial Institute 1640; Sigma-Aldrich) culture media supplemented with 10% fetal bovine serum (Sigma-Aldrich), 4 mM L-glutamine (Sigma-Aldrich) and 1.5 g L−1 sodium bicarbonate (Sigma-Aldrich) at 37 °C in an atmosphere of 5% humidified CO2. Cells used in the study were in the logarithmic growth phase. Cells were grown to 70% confluence, and then trypsinised with 0.25% trypsin–0.02% EDTA (Sigma-Aldrich).Cytotoxicity
The cytotoxicities of the ruthenium complexes were determined using the Alamar Blue cytotoxicity assay as previously described.26 All data were from at least three independent experiments and the IC50 determined using GraphPad Prism 6.0 (GraphPad Software, San Diego, USA).Incubation of cells with ruthenium(II) complexes and organelle stains
The trypsinised cells were seeded on the coverslips in petri dishes. The ruthenium complexes were applied to the cells in RPMI-1640 media to make the desired concentration (arranged from 5 to 50 μM) and incubated at 37 °C with 5% CO2 for 4 h or overnight as described. During the final 30 min of incubation, 100 nM Mitotracker® Green FM (Invitrogen) was added for mitochondrial staining, 100 nM DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) for nuclear staining and 50 nM SYTO 9 for nucleolus staining, and the cells incubated for a further 5 min. The coverslips were rinsed gently with phosphate buffer solution and mounted on bridged slides for imaging.Cellular localisation studies
The cellular localisation of the ruthenium(II) complexes was determined using a Zeiss laser scanning confocal microscope (LSM 700). Samples were viewed under a 40× or 63× oil immersion lens. Metal complexes (λex = 450 nm, λem = 610 nm) and Mitotracker Green FM (λex = 490 nm, λem = 516 nm) were excited using blue argon laser (λex = 488 nm), and emissions were collected at 570–650 nm and 470–550 nm, respectively. For DAPI excitation, diode laser (λex = 405 nm) was used and the emission detected at 430–500 nm. SYTO9 was excited with λex = 488 nm, and the emission collected at 495–510 nm. Image data acquisition and processing were performed using Zen software 2009 (Carl Zeiss).CD spectroscopy
Solutions of CT-DNA (Sigma Aldrich) in phosphate buffer (650 μL, 1 mM Na2EDTA, 10 mM Na2HPO4, 20 mM NaCl, pH 7.0) gave a ratio of UV absorbances at 260 and 280 nm, A260/A280, of 1.80–1.90, indicating that the DNA was sufficiently free from protein.27 The concentration of CT-DNA stock solution was determined from UV absorption at 260 nm. The circular dichroism spectral titration experiments were performed by keeping the CT-DNA concentration constant (2.7 mM bases) while varying the concentration of metal complexes from 0 to 39 μM. The CD spectra were measured in 1 mm path length quartz cuvettes on a JASCO J-815 circular dichroism spectrometer. Two scans were accumulated at a scan speed of 100 nm min−1. All CD spectra were recorded at every 0.5 nm from 200 to 350 nm. Sample temperature was maintained at 35 °C using a JASCO MCB-100 mini-circulation bath. Spectra were corrected for buffer signal.The baker's yeast RNA (Sigma Aldrich) stock solution was also prepared in phosphate buffer in DEPC (Sigma Aldrich) treated water. The A260/A280 value was 2.10, indicating the RNA was pure.28 The initial RNA concentration was 2.3 mM (bases). The titration of ruthenium complexes and the data collection were the same as indicated for the DNA experiments.
Results
Based upon our previous studies on the antimicrobial activities and the corresponding toxicities to eukaryotic cell lines,20 Rubb12 appears to have the best therapeutic window (antimicrobial activity compared to toxicity) of the dinuclear complexes. Furthermore, the tetranuclear analogue Rubb12-tetra (see Fig. 2) has the best antimicrobial activity of all the oligonuclear ruthenium complexes we examined.25 Consequently, this study focused on the Rubb12, Rubb12-tri and Rubb12-tetra complexes, and in order to examine the effect of the chirality of the complexes, we examined the toxicity, cellular localisation and DNA/RNA binding of the ΔΔ and ΛΛ enantiomers of Rubb12.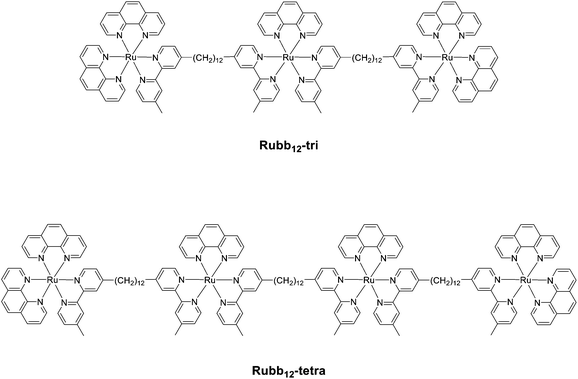 | ||
| Fig. 2 Structures of the trinuclear (Rubb12-tri) and tetranuclear (Rubb12-tetra) ruthenium(II) complexes. | ||
In vitro toxicity against kidney and liver cells
In order to determine the biologically relevant concentrations of the ruthenium complexes, and to ascertain the toxicity of the tri- and tetra-nuclear species against eukaryotic cells for the first time, the IC50 values of Rubb12, Rubb12-tri and Rubb12-tetra were determined against three cell lines (BHK, HEK-293 and Hep-G2). The results are summarised in Table 1. All ruthenium complexes were toxic against the three cell lines, particularly the cancer cell line Hep-G2. The dinuclear complexes ΔΔ/ΛΛ-Rubb12 were less toxic than Rubb12-tri, which was slightly less toxic than Rubb12-tetra. For Rubb12, there were only small differences in the IC50 values for the ΔΔ and ΛΛ enantiomers.Table 2 shows the comparison of the IC50 values of the ruthenium complexes against the eukaryotic cells to the corresponding MIC values against the Gram positive bacterium S. aureus and the Gram negative species E. coli. Compared to the healthy eukaryotic BHK and HEK-293 cell lines, the ruthenium complexes exhibited a selectivity index (SI = IC50/MIC) of between 12 and 91 when compared to the Gram positive bacterium S. aureus, but only between 5 and 22 for the Gram negative E. coli. Interestingly, all ruthenium complexes were more toxic to the cancer cell line Hep-G2, and consequently they exhibited a lower SI value. Of the ruthenium complexes, ΔΔ-Rubb12 exhibited the best SI when compared to the healthy eukaryotic cell lines.
| BHK | HEK-293 | Hep-G2 | ||||
|---|---|---|---|---|---|---|
| S. aureus | E. coli | S. aureus | E. coli | S. aureus | E. coli | |
| ΔΔ-Rubb12 | 91 | 22 | 25 | 6 | 9 | 2 |
| ΛΛ-Rubb12 | 40 | 19 | 12 | 6 | 8 | 4 |
| Rubb12-tri | 53 | 13 | 22 | 6 | 19 | 5 |
| Rubb12-tetra | 44 | 11 | 21 | 5 | 17 | 4 |
Time-course cytotoxicity assays
As the MIC values for the antimicrobial activities were determined over 16–18 hours and the incubation times for the confocal microscopy were also much shorter than the standard 72 hour incubation used for the cytotoxicity assays, the IC50 of the complex exhibiting the best SI (ΔΔ-Rubb12) was determined as a function of time. The results are summarised in Table 3. As would be expected, the IC50 values significantly increased with decreasing incubation time. The SI values for the ΔΔ-Rubb12 complex based upon the 16–18 hour incubation against the bacteria and the 24 hour IC50 values for the eukaryotic cell lines are 85 to 117 for S. aureus and 20 to 28 for E. coli.| 4 hour | 8 hour | 24 hour | 48 hour | 72 hour | |
|---|---|---|---|---|---|
| BHK | 190.9 ± 36.5 | 103.8 ± 8.5 | 70.5 ± 26.4 | 57.5 ± 7.1 | 54.3 ± 3.2 |
| HEK-293 | 90.8 ± 17.9 | 90.48 ± 34.3 | 50.9 ± 19.9 | 24.9 ± 1.1 | 15.1 ± 2.8 |
| Hep-G2 | 103.2 ± 3.8 | 109.7 ± 29.3 | 61.7 ± 5.5 | 15.8 ± 10.4 | 5.2 ± 2.0 |
Cellular localisation study
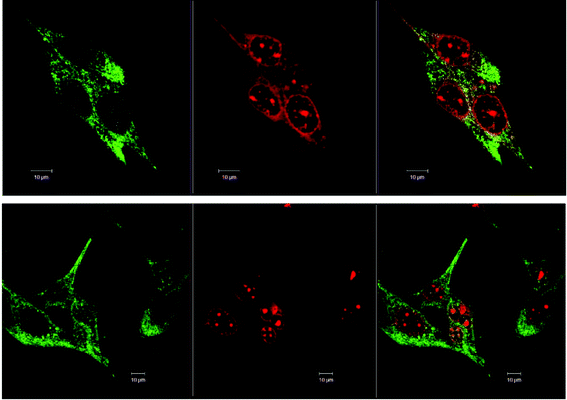
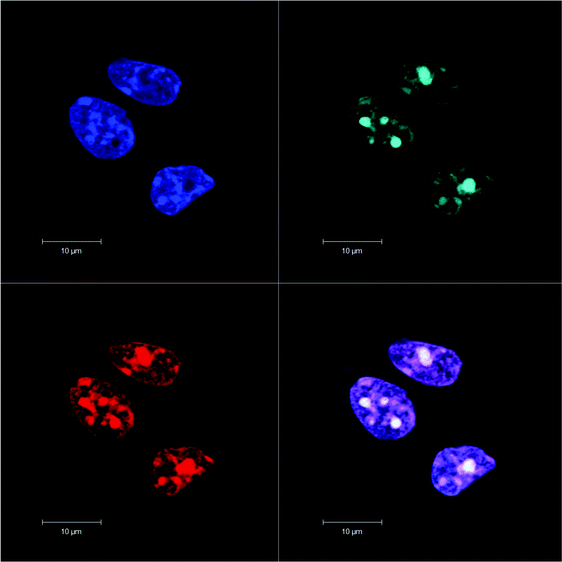
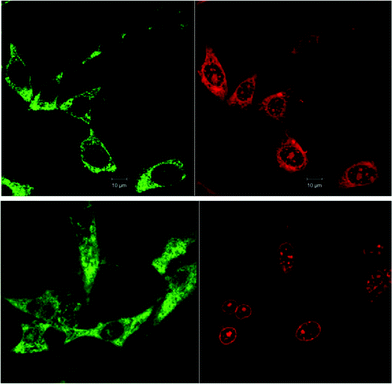
Fig. 3 shows the comparison of the localisation of ΔΔ-Rubb12 in BHK cells with the selective mitochondrial stain Mitotracker Green (20 hour incubation). It is clearly observed that ΔΔ-Rubb12 does not localise in the mitochondria, but appears to preferentially accumulate in the cell nucleus, as shown by the results at 5 μM. Similar results were obtained with the other cell lines and the other ruthenium complexes (data not shown).The localisation in the nucleus was confirmed through co-staining with DAPI. DAPI is considered to be a DNA-selective stain, as it binds DNA 100-fold more strongly than RNA and has a 3-fold higher fluorescence quantum yield when bound to DNA than to RNA.29 In Fig. 4 we show the results of the DAPI co-staining experiments with ΔΔ-Rubb12 and BHK cells. The ΔΔ-Rubb12 concentration was 50 μM (approx. IC50) and the incubation time was 20 hours. These results confirm the preferential accumulation of the ruthenium complexes in the nucleus, however the localisation pattern was not identical. While significant DNA binding of the complex was observed at this concentration (as evidenced by the overlap with DAPI staining), there is also intense ΔΔ-Rubb12 red fluorescence in areas of the nucleus where there is little or no DAPI fluorescence. These so-called “DAPI holes” are generally recognised as nucleoli.30 The nucleolus is the site within the nucleus where ribosomal-RNA (r-RNA) is synthesised, and consequently is rich in r-RNA. The nucleoli can be highlighted through staining with SYTO 9. This general nucleic acid stain binds both DNA and RNA but binds RNA with greater affinity. The results of SYTO-9 co-staining experiments (also shown in Fig. 4) confirmed that ΔΔ-Rubb12 does accumulate in the nucleoli.
As is observed in Fig. 4, there is considerable DNA co-staining at 50 μM; however, at 10 μM there appears to be predominant RNA binding, and almost exclusive RNA binding at 5 μM (see Fig. 3).
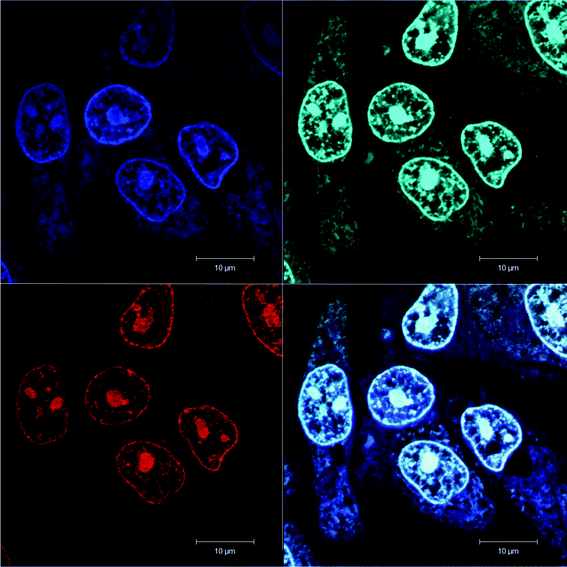
Similar results were obtained with the other eukaryotic cells. For example, Fig. 5 shows the preferential accumulation of ΔΔ-Rubb12 in the nucleoli of Hep-G2 cells.
No significant difference in the localisation of the ΔΔ-Rubb12 and ΛΛ-Rubb12 enantiomers was observed. Similarly, for the Rubb12-tri and Rubb12-tetra complexes the same pattern of localisation was observed; however, the total accumulation appeared to be greater with more DNA binding observed for the Rubb12-tri and Rubb12-tetra complexes. Furthermore, increased accumulation was also observed outside of the nucleus (Fig. 6).
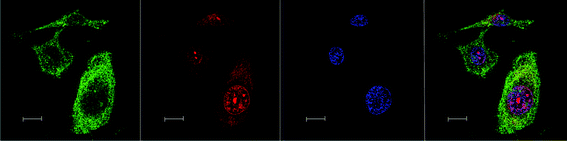 | ||
| Fig. 6 Left to right – Rubb12-tetra localisation in BHK cells at 10 μM, stained by Mitotracker (green), Rubb12-tetra (red), DAPI (blue) and merged image. Scale bar = 10 μm. | ||
To examine the effect of time on the localisation of ΔΔ-Rubb12, BHK cells were incubated with ΔΔ-Rubb12 at 55 μM for both 4 and 20 hours. The resultant images are shown in Fig. 7. After a 4 hour incubation, ΔΔ-Rubb12 was localised to a greater extent in the cytoplasm compared to nucleolus. Subsequently, after the longer incubation time, the ΔΔ-Rubb12 was predominantly observed in the nuclear region, particularly in the nucleolus and nuclear envelope. These observations suggest that the ruthenium complexes will accumulate in the endoplasmic reticulum after passing through the cell membrane, but finally accumulate in the nucleolus. Similar results were obtained with ΛΛ-Rubb12 (data not shown).
CT-DNA binding
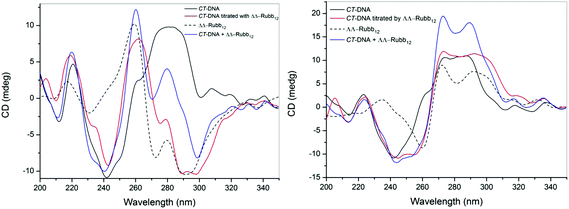
While the confocal microscopy experiments demonstrated that the Rubbn complexes bound RNA and DNA in live cells, the effect of the ruthenium complexes on the solution conformation of the nucleic acids is unknown. In order to examine the effect of micro-molar concentrations of the ruthenium complexes on the solution conformation of large segments of DNA and RNA, an in vitro binding study with ΔΔ- and ΛΛ-Rubb12 was conducted by circular dichroism spectroscopy (CD). In a CD spectrum, B-form CT-DNA is characterised by a positive band at 260–280 nm due to base stacking and a negative band around 245 nm due to the helicity of the structure.31 However, the enantiomers of Rubb12 also have strong CD signals in the 200–300 nm range. Consequently, DNA binding was determined by comparing the observed signal upon titration of ΔΔ- or ΛΛ-Rubb12 into the CT-DNA sample with the arithmetic sum of the individual CD spectra of the metal complex and DNA.Addition of both ΔΔ- and ΛΛ-Rubb12 induced significant decreases in the CD signal for the CT-DNA with added ruthenium complex in the 260–300 nm range at concentrations below the IC50 values (Fig. 8). However, and most clearly seen for the titration with ΛΛ-Rubb12, the basic B-type conformation is maintained (negative peak at 245 nm and positive peak at 260–280 nm). The decrease in intensity of the CD signal between 260 and 300 nm is consistent with the changes noted for the addition of high concentrations (5 M) of NaCl to CT-DNA32 – a decrease in the CD signal between 260 and 300 nm caused by high salt concentration is generally interpreted as the DNA structure becoming more tightly wound, but remaining in the B conformation.
RNA binding
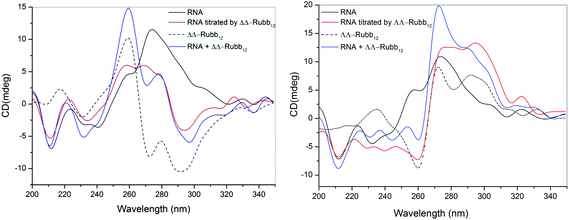
The binding of ΔΔ- and ΛΛ-Rubb12 to RNA was examined by CD spectroscopy in an analogous manner to the study described for DNA. The CD spectrum of free RNA at 35 °C has two positive bands at 225 and 270 nm and two negative bands at 210 and 230 nm, which is consistent with the double-stranded A-conformation.33,34 The large reduction in the CD signal at 260–280 nm upon addition of either ΔΔ- or ΛΛ-Rubb12 indicates that both enantiomers interact strongly with RNA at concentrations below the IC50 values (see Fig. 9). This band is sensitive to base-stacking;35 consequently, the decrease in its intensity can be interpreted as a modification of base-stacking that potentially partially destabilises the A-type conformation. Consistent with this interpretation is the decrease in the base-stacking band observed for RNA oligonucleotides upon lowering the ionic strength of the solution from 1 M to 0.01 M NaCl.36 Importantly, the CD results indicate that the ruthenium complex-bound RNA maintains the A-type structure.Discussion
The results of this study indicate that the oligonuclear inert ruthenium complexes linked by the bbn ligand are toxic to kidney and liver cells. However, even when comparing the 72 hour cytotoxicity data with the 16–18 hour antimicrobial MIC values, it is clear that the ruthenium complexes are more toxic to bacteria than the eukaryotic cells examined in this study. For the BHK and HEK-293 cell lines, the dinuclear complex was less toxic than the tri- and tetra-nuclear species, and showed the largest relative (and absolute) difference between cytotoxicity and antimicrobial activity. Toxicity is related to the cellular uptake. The lower toxicity of the dinuclear complex is possibly due to its lower lipophilicity, with the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values for Rubb12, Rubb12-tri and Rubb12-tetra being −2.9, −1.0 and −1.6 respectively.25 Interestingly, even though the trinuclear species is more lipophilic than the tetranuclear complex, it was generally less toxic to the eukaryotic cells. This demonstrates the importance of the cationic charge of the ruthenium complex in the mechanism of the observed toxicity towards eukaryotic cells.
P values for Rubb12, Rubb12-tri and Rubb12-tetra being −2.9, −1.0 and −1.6 respectively.25 Interestingly, even though the trinuclear species is more lipophilic than the tetranuclear complex, it was generally less toxic to the eukaryotic cells. This demonstrates the importance of the cationic charge of the ruthenium complex in the mechanism of the observed toxicity towards eukaryotic cells.
Confocal microscopy was used to determine the cellular localisation of the ruthenium complexes in the three cell lines. By comparison with DAPI and SYTO 9 staining, it was concluded that Rubb12, Rubb12-tri and Rubb12-tetra preferentially accumulated in the nucleolus at low complex concentrations, while significant DNA binding is also observed at higher concentrations. The preference for RNA is consistent with our previous study on the localisation of ΔΔ-Rubb16 in the ribosomes of E. coli.37 The overall preference of these complexes to the nucleus is surprising given the mitochondrial selectivity we observed for the Rubbn complexes in L1210 cells.12 Also of note is the difference between the rigidly linked tetrapyridophenazine (tpphz) dinuclear ruthenium complexes studied by Thomas and co-workers11,22 and the flexibly-linked bbn complexes examined in this study. The less lipophilic [{Ru(phen)2}2{μ-tpphz}]4+ (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = −0.96) targeted the nucleus, but not the nucleolus, and showed little toxicity towards MCF-7 cancer cells IC50 = 138 μM).11 On the other hand, the more lipophilic analogue containing the 4,7-diphenyl-1,10-phenanthroline ligand [{Ru(DIP)2}2{μ-tpphz}]4+ (log
P = −0.96) targeted the nucleus, but not the nucleolus, and showed little toxicity towards MCF-7 cancer cells IC50 = 138 μM).11 On the other hand, the more lipophilic analogue containing the 4,7-diphenyl-1,10-phenanthroline ligand [{Ru(DIP)2}2{μ-tpphz}]4+ (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.52) targets the endoplasmic reticulum and is highly toxic to MCF-7 cells (IC50 = 7 μM).22 The bbn linked oligonuclear complexes are less lipophilic but all show greater toxicity to the cell lines studied than [{Ru(phen)2}2{μ-tpphz}]4+, and despite the differences in log
P = 1.52) targets the endoplasmic reticulum and is highly toxic to MCF-7 cells (IC50 = 7 μM).22 The bbn linked oligonuclear complexes are less lipophilic but all show greater toxicity to the cell lines studied than [{Ru(phen)2}2{μ-tpphz}]4+, and despite the differences in log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values, they all target the nucleolus. The results of this study suggest that in these cases log
P values, they all target the nucleolus. The results of this study suggest that in these cases log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values do not reflect the ease with which the ruthenium complexes can cross cell membranes. Although it is acknowledged that the [{Ru(phen)2}2{μ-tpphz}]4+ complexes enter cells by active transport,11 the results of this study suggest that the distance between the ruthenium centres (compared to the length of the highly non-polar section of a lipid bilayer) could be a more important factor for cellular uptake than lipophilicity, per se.
P values do not reflect the ease with which the ruthenium complexes can cross cell membranes. Although it is acknowledged that the [{Ru(phen)2}2{μ-tpphz}]4+ complexes enter cells by active transport,11 the results of this study suggest that the distance between the ruthenium centres (compared to the length of the highly non-polar section of a lipid bilayer) could be a more important factor for cellular uptake than lipophilicity, per se.
The CD spectroscopy experiments confirmed that Rubb12 can interact with DNA and RNA at biologically relevant concentrations. While the CD results suggested that Rubb12 affected the base-stacking of both DNA and RNA, there was no indication that the ruthenium complex condensed or aggregated either nucleic acid at ≤IC50 concentrations. Furthermore, both the RNA and DNA maintained their normal solution conformations upon ΔΔ/ΛΛ-Rubb12 binding. Given the preferential RNA binding exhibited by the ruthenium complexes, it is possible that RNA binding is responsible for the cellular toxicity. In support of this proposal is the observation that after 4 hours incubation with BHK cells, confocal microscopy indicated that a large proportion of the administered Rubb12 was located outside of the nucleus, but after 20 hours nearly all the ruthenium complex was inside the nucleus in the nucleolus. The IC50 value after a 4 hour incubation in the BHK cells was 190.9 μM, but this dropped to 70.5 μM after 24 hours and then only decreased to a small extent over the next 48 hours.
The results of this study indicate that the oligonuclear ruthenium complexes do bind nucleic acids in live cells, thereby supporting the proposed biological potential suggested in the many studies of nucleic acid binding by cationic transition metal complexes.1–8 However, it appears the ruthenium complexes target RNA rather than DNA. We have previously demonstrated that the bulky dinuclear ruthenium complexes bind in the DNA minor groove and preferentially target non-duplex features, such as bulges and hair-pin loops, compared to standard duplex structures.38,39 It could be argued that RNA contains a greater proportion of non-duplex structures than does DNA. However, generally only a slight difference was seen between the ΔΔ and ΛΛ enantiomers in terms of toxicity, intracellular localisation or in binding to long segments of DNA or RNA. In particular, a relatively larger enantiomeric effect is seen for Hep-G2 and S. aureus. As we have observed differences in the way the enantiomers interact with DNA oligonucleotides,7,38 it is possible that the effects observed in this study are primarily due to non-specific electrostatic interactions that cause sufficient structural modifications to inhibit RNA-driven transcription. The CD spectroscopy studies indicated that the ruthenium complex-bound DNA maintained the B-conformation, while the bound-RNA maintained the A-form. The A-form RNA has a shorter rise per base pair (≈2.8 Å) than B-DNA (≈3.4 Å).40 Consequently, A-RNA will have an increased linear negative charge density compared to B-form DNA. This should impact on the binding of polycations, and potentially when coupled to the increased proportion of more flexible non-duplex structures found in RNA, provides an explanation for the observed binding preference of the ruthenium complexes for RNA in the eukaryotic cells studied here.
In conclusion, the results of this study demonstrate that the Rubbn class of antimicrobial agents selectively accumulate in the nucleus of eukaryotic cells. However, the ruthenium complexes preferentially localise in the RNA-rich nucleoli, rather than with the chromosomal DNA. Although RNA and DNA binding is most likely responsible for the toxicity of the ruthenium complexes to the eukaryotic cells, the cytotoxicity assays indicated that the lead complex, ΔΔ-Rubb12, is approximately 100-fold less toxic to eukaryotic cells than to Gram positive bacteria.
Acknowledgements
We thank Professor Alan Baxter (Comparative Genomics Centre, James Cook University, Townsville, Australia) for the use of their laboratories and tissue culture facilities, and the Australian Army Malaria Institute (Enoggera, Queensland, Australia) for the donation of the cell lines.Notes and references
- S. J. Lippard, Acc. Chem. Res., 1978, 11, 211 CrossRef CAS.
- B. Nordén, P. Lincoln, B. Akerman and E. Tuite, Met. Ions Biol. Syst., 1996, 33, 177 Search PubMed.
- A. C. Komor and J. K. Barton, Chem. Commun., 2013, 49, 3617 RSC.
- K. E. Erkkila, D. T. Odom and J. K. Barton, Chem. Rev., 1999, 99, 2777 CrossRef CAS PubMed.
- C. Metcalfe and J. A. Thomas, Chem. Soc. Rev., 2003, 32, 215 RSC.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565 RSC.
- F. R. Keene, J. A. Smith and J. G. Collins, Coord. Chem. Rev., 2009, 253, 2021 CrossRef CAS PubMed.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179 RSC.
- C. A. Puckett, R. J. Ernst and J. K. Barton, Dalton Trans., 2010, 39, 1159 RSC.
- C. A. Puckett and J. K. Barton, Biochemistry, 2008, 47, 11711 CrossRef CAS PubMed.
- M. R. Gill, J. Garcia-Lara, S. J. Foster, C. Smythe, G. Battaglia and J. A. Thomas, Nat. Chem., 2009, 1, 662 CrossRef CAS PubMed.
- M. J. Pisani, P. D. Fromm, Y. Mulyana, R. J. Clarke, H. Korner, K. Heimann, J. G. Collins and F. R. Keene, ChemMedChem, 2011, 6, 848 CrossRef CAS PubMed.
- M. Matson, F. R. Svensson, B. Nordén and P. Lincoln, J. Phys. Chem. B, 2011, 115, 1706 CrossRef CAS PubMed.
- F. R. Svensson, J. Andersson, H. L. Åmand and P. Lincoln, J. Biol. Inorg. Chem., 2012, 17, 565 CrossRef CAS PubMed.
- T. Chen, Y. Liu, W.-J. Zheng, J. Liu and Y.-S. Wong, Inorg. Chem., 2010, 49, 6366 CrossRef CAS PubMed.
- M. R. Gill, H. Derrat, C. G. W. Smythe, G. Battaglia and J. A. Thomas, ChemBioChem, 2011, 12, 877 CrossRef CAS PubMed.
- H. W. Boucher, G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert, L. B. Rice, M. Scheld, B. Spellberg and J. Bartlett, Clin. Infect. Dis., 2009, 48, 1 CrossRef PubMed.
- F. P. Dwyer, E. C. Gyarfas, W. P. Rogers and J. H. Koch, Nature, 1952, 170, 190 CrossRef CAS.
- F. P. Dwyer, I. K. Reid, A. Shulman, G. M. Laycock and S. Dixson, Aust. J. Exp. Biol. Med. Sci., 1969, 47, 203 CrossRef CAS.
- F. Li, Y. Mulyana, M. Feterl, J. M. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032 RSC.
- F. R. Svensson, M. Matson, M. Li and P. Lincoln, Biophys. Chem., 2010, 149, 102 CrossRef CAS PubMed.
- M. R. Gill, D. Cecchin, M. G. Walker, R. S. Mulla, G. Battaglia, C. Smythe and J. A. Thomas, Chem. Sci., 2013, 4, 4512 RSC.
- F. Li, Inert dinuclear polypyridylruthenium(II) complexes as antimicrobial agents, PhD thesis, University of New South Wales, Australia, 2013 Search PubMed.
- Y. Mulyana, D. K. Weber, D. P. Buck, C. A. Motti, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 1510 RSC.
- A. K. Gorle, M. Feterl, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2014 10.1039/c4dt02139h.
- J. O'Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 267, 5421 CrossRef.
- S. Prakash, V. P. Vaidya, K. M. Mahadevan, M. K. Shivananda, P. A. Suchetan, B. Nirmala and M. Sunitha, J. Chem. Pharm. Res., 2012, 4, 1179 Search PubMed.
- W. W. Wilfinger, K. Mackey and P. Chomczynski, BioTechniques, 1997, 22, 474 CAS.
- F. A. Tanious, J. M. Veal, H. Buczak, L. S. Ratmeyer and W. D. Wilson, Biochemistry, 1992, 31, 3103 CrossRef CAS.
- D. Hernandez-Verdun, in The Nucleolus, ed. M. O. J. Olson, Springer, New York, 2011, Ch. 1, pp. 3–28 Search PubMed.
- W. C. Johnson, CD of Nucleic Acids, in Circular dichroism: principles and applications, ed. K. Nakanishi, N. Berova and R. W. Woody, VCH, New York, 1994, pp. 523–540 Search PubMed.
- J. Kypr, I. Kejnovská, D. Renciuk and M. Vorlícková, Nucleic Acids Res., 2009, 37, 1713 CrossRef CAS PubMed.
- M. Vorlícková, Biophys. J., 1995, 69, 2033 CrossRef.
- J. Kypr and M. Vorlícková, Biopolymers, 2002, 67, 275 CrossRef PubMed.
- E. P. Loret, P. Georgel, W. C. Johnson Jr. and P. S. Ho, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 9734 CrossRef CAS.
- T. Kirihata, S. Nakano and N. Sugimoto, Nucleic Acids Symp. Ser., 2004, 48, 109 CrossRef PubMed.
- F. Li, E. J. Harry, A. L. Bottomley, M. D. Edstein, G. W. Birrell, C. E. Woodward, F. R. Keene and J. G. Collins, Chem. Sci., 2014, 5, 685 RSC.
- J. L. Morgan, C. B. Spillane, J. A. Smith, P. D. Buck, J. G. Collins and F. R. Keene, Dalton Trans., 2007, 4333 RSC.
- F. Li, D. K. Weber, J. L. Morgan, J. G. Collins and F. R. Keene, Dalton Trans., 2012, 41, 6528 RSC.
- S. A. Pabit, X. Qiu, J. S. Lamb, L. Li and S. P. Meisburger, Nucleic Acids Res., 2009, 37, 3887 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |