Open Access Article
Open Access ArticleThe nanoparticle biomolecule corona: lessons learned – challenge accepted?
D.
Docter
*a,
D.
Westmeier
a,
M.
Markiewicz
b,
S.
Stolte
bc,
S. K.
Knauer†
d and
R. H.
Stauber†
*a
aDepartment of Nanobiomedicine/ENT, University Medical Center of Mainz, Langenbeckstr. 1, 55101 Mainz, Germany. E-mail: docter@uni-mainz.de; roland.stauber@unimedizin-mainz.de
bDepartment Sustainable Chemistry, Center for Environmental Research and Sustainable Technology (UFT), University of Bremen, Leobener Str. 1, D-28359, Bremen
cDepartment of Environmental Analytics, Faculty of Chemistry, University of Gdańsk, Wita Stwosza 63, 80-308 Gdańsk, Poland
dInstitute for Molecular Biology, CENIDE, Mainz Scientific Screening Center UG&Co. KG, University Duisburg-Essen, Universitätsstr. 5, 45117 Essen, Germany
First published on 11th June 2015
Abstract
Besides the wide use of engineered nanomaterials (NMs) in technical products, their applications are not only increasing in biotechnology and biomedicine, but also in the environmental field. While the physico-chemical properties and behaviour of NMs can be characterized accurately under idealized conditions, this is no longer the case in complex physiological or natural environments. Herein, proteins and other biomolecules rapidly bind to NMs, forming a protein/biomolecule corona that critically affects the NMs' (patho)biological and technical identities. As the corona impacts the in vitro and/or in vivo NM applications in humans and ecosystems, a mechanistic understanding of its relevance and of the biophysical forces regulating corona formation is mandatory. Based on recent insights, we here critically review and present an updated concept of corona formation and evolution. We comment on how corona signatures may be linked to effects at the nano–bio interface in physiological and environmental systems. In order to comprehensively analyse corona profiles and to mechanistically understand the coronas' biological/ecological impact, we present a tiered multidisciplinary approach. To stimulate progress in this field, we introduce the potential impact of the corona for NM–microbiome–(human)host interactions and the novel concept of ‘nanologicals’, i.e., the nanomaterial-specific targeting of molecular machines. We conclude by discussing the relevant challenges that still need to be resolved in this field.
Introduction
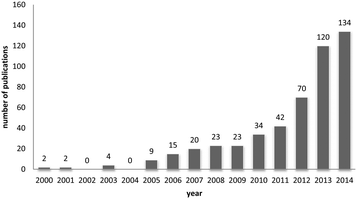
One way of swiftly judging the relevance and up-to-dateness of a scientific topic is to examine the available databases. As illustrated in Fig. 1, querying PubMed by using the search criteria ‘nanoparticles’ and ‘corona’ reveals that more than 430 reports have been published on this topic, and the number has increased almost sevenfold over the last 5 years. Albeit numbers do not always equalize scientific quality, it is obvious that the ‘nanoparticle corona’ represents a still unresolved hot topic, with high scientific and economic relevance.This trend is almost expected, since the technical applications of engineered nanomaterials (NMs) and also their (intended) use in biotechnology and biomedicine is steadily growing.1,2 Such developments will also lead to an increasing exposure of humans and the environment to NMs. NMs can be produced in almost unlimited combinations to take advantage of properties related to their chemistry, shape, size and surface characteristics.3 Due to their high surface free energy, NMs adsorb (bio)molecules upon contact with biological and/or abiotic environments, forming the so-called (bio)molecule corona.4–6
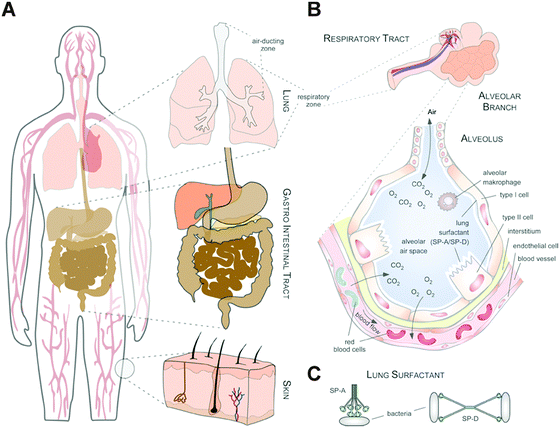
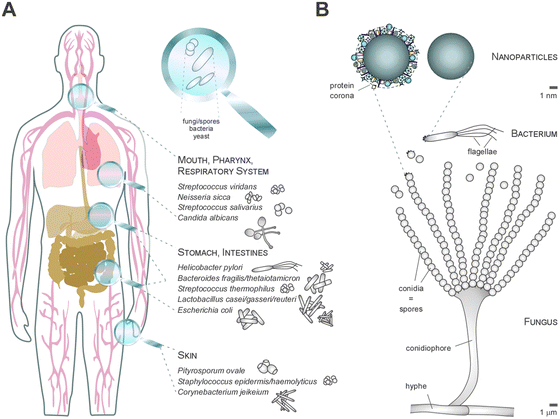
Thus, whereas the physico-chemical properties and behaviour of NMs can be engineered and controlled in technically stable, protected environments, such as technical products, this is no longer the case in complex physiological or natural environments. Such ‘complex environments’ are not only represented by simple and higher organisms, including humans, but also by complex solid and liquid interfaces to which NMs are exposed during their technical application and/or following the intended/unintended exposure of humans and following their release into the ecosystem. Moreover, complex organisms are further composed of various additional ‘complex microenvironments’, such as organs and cells, which also differ quite dramatically in their physico-chemical composition (Fig. 2A and B).
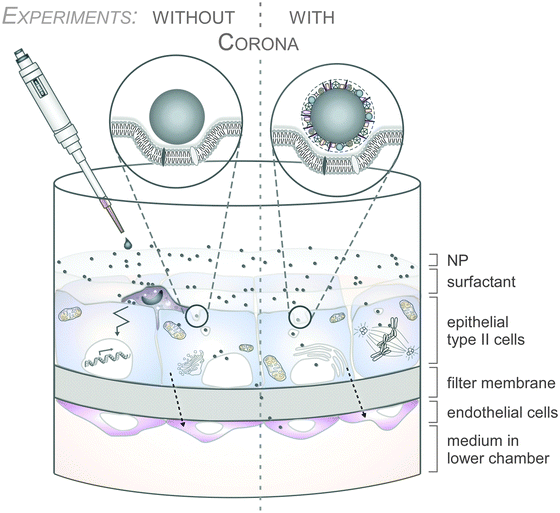
As an example, the human lung is depicted in Fig. 2. The respiratory system is not only involved in breathing, but also represents a main target for inhaled or deliberately applied nanoparticles (NPs). In humans and other mammals, the lung contains the respiratory bronchioles, the alveolar ducts and the alveoli, in which gas exchange takes place (Fig. 2B). Oxygen and carbon dioxide are passively exchanged by diffusion between the gaseous external environment and the blood. There are several reasons why the alveolar-capillary barrier can be considered as one of the main targets of inhaled NPs, leading to local or systemic effects: (i) the location of the alveolar epithelium beyond the mucociliary escalator clearance mechanism of the conducting airways; (ii) the impaired or less effective recognition of NPs by alveolar macrophages, the first line of defence against particulates that have escaped the mucociliary escalator; (iii) the relatively large alveolar epithelial surface area estimated at 100 to 140 m2; and (iv) only a thin cellular barrier from the airspace to the capillary blood is presented by the alveolar epithelial (type I (ATI) and type II (ATII)) and pulmonary capillary endothelial cells (Fig. 2B). Moreover, the lung contains the pulmonary surfactant, a thin lipid–protein film coating the cellular surface of the mammalian lung (Fig. 2B). Its major function is the reduction of surface tension, thereby reducing the work of breathing. The main constituents of the lung surfactant are lipids. Although present only in small amounts, surfactant spsecific proteins (SP-A, SP-B, SP-C and SP-D) are highly important for the proper function of the lung surfactant. In particular, the proteins SP-A and SP-D can bind to carbohydrates on the surface of pathogens and thereby opsonize them for uptake by phagocytes (Fig. 2C). Surfactant degradation or inactivation may thus contribute to enhanced susceptibility to lung inflammation and infection. However, whether the interaction with nanosized objects has similar (patho)physiological consequences remains to be investigated. Clearly, the lung surfactant, epithelial, endothelial and immune cells represent quite different microenvironments that the NPs encounter and have to overcome prior to potentially entering the bloodstream.7,8 It is expected, though not yet fully proven, that (all) of these microenvironments affect the formation and fate of a nanoparticle corona.
So far, the focus in ‘corona research’ has mostly been on blood plasma- or/and bovine serum containing cell culture media-induced protein corona formation.4–6 Whereas studies of the NP corona recovered from physiological fluids from other organs, such as the lung, the brain or gut, are emerging,7,8 comprehensive reports on the composition and relevance of the corona in ecosystems are still missing. Indeed, querying PubMed reveals only a few studies investigating the composition of the corona in ‘natural environments’, such as complex aqueous media or soil systems.9–12 Even though physiological environments, such as plasma, intestinal fluid or the lung surfactant, are complex enough, environmental systems, such as different types of water or soil, are even more complex, being composed of many different chemical and physical molecules, and biotic and abiotic matter, which, in addition, are experimentally hard to control. Also, very little is (mechanistically) known about the relevance and impact of the biomolecule corona on the interaction of NPs with environmental soil or water organisms, or on the NMs' environmental fate, their biotransformation and/or their potential adverse effects. Hence, this topic requires particular scientific and regulatory attention and, thus, will be further discussed in our review.
Die hard – hard corona, soft corona, protein clouds – what do we really need?
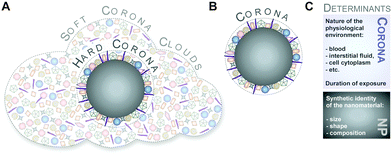
Although protein adsorption on to surfaces has long been known about since the pioneering work by Vroman in 1962,13 the term ‘protein corona’ was only introduced to the nanoparticle community much later in the study of Cedervall and colleagues,14 which stimulated a whole field of new investigations.Albeit somehow loosely defined, the term ‘hard protein corona’ was coined to describe the long-lived equilibrium state, representing an analytically accessible protein/biomolecule signature of a nanoparticle/NM in a certain environment.5,6,15,16 In addition, some models further suggest that on top of this ‘hard corona’ with a rather low complexity, a ‘soft protein corona’ or even ‘protein clouds’ exist, consisting of loosely associated and rapidly exchanging highly complex layers of biomolecules (Fig. 3A).4,14,16–18 However, since this ‘soft corona’ desorbs during current purification processes, its existence, analytical dissection and, particularly, its relevance for effects at the nano–bio interface remains to be fully confirmed. Moreover, these terms are not used when describing other corona types, such as those formed by lipids.7
Thus, transferring knowledge from the field of cell biology, natural or chemical ligands can stimulate (patho)physiological reactions by interacting with receptors.19 However, albeit low affinity binding to other receptors or non-receptor proteins may exist as well, only efficient ligand–target protein interactions will ultimately trigger biological responses (Fig. 15).19 Thus, in the context of the life sciences, such as biology or medicine, unspecific ligand–receptor interactions are hardly discussed and no discrimination between ‘soft’ or ‘hard’ ligand–receptor interactions are made.
Hence, as inspection of the current literature revealed that the term ‘soft’ versus ‘hard’ corona seems to mostly create more confusion than helping to describe and resolve scientific questions, we suggest to generally refer to analytically accessible NP–protein complexes simply as the ‘protein corona’ (Fig. 3B). As such, we will use this term throughout the review.
Crown of thorns – factors influencing the biomolecule–corona's composition
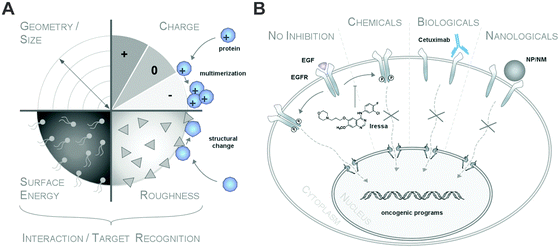
Clearly, it is the biomolecular corona (herein termed the biomolecule corona) that primarily interacts with biological systems and thereby constitutes a major element of the nanoparticles' biological identity.5,6,16,20,21 The bio-physical properties of such a corona-covered NP often differ significantly from those of the formulated pristine particle during manufacturing.5,6,16,22–24 Albeit ‘chemically speaking’, there are no ‘naked’ or ‘pristine’ NPs, we here refer to such NPs in biomolecule-free environments. From a regulatory aspect, (bio)molecule-coated NMs may therefore be even considered as novel materials with different properties compared to pristine NMs during production.6,23Typically, corona profiles differ significantly from the protein composition of the (biological) fluid investigated.4,6,16,25–27 Distinct proteins will be either enriched or will display only a weak affinity for the nanoparticle surface. Furthermore, not only have the particle material, size and the surface properties been shown to play a role in determining the composition of the protein corona but so have the exposure time and the relative ratio of the physiological fluid to the nanoparticle dispersion (Table 1).4–6,16,25–29
| Factors | Ref. |
|---|---|
| Exposure temperature | 30 |
| Exposure time | 6, 18 and 31–36 |
| Nanoparticle hydrophobicity | 14 and 37–42 |
| Nanoparticle size/surface curvature | 14, 15, 18, 40 and 43–49 |
| Nanoparticle surface charge | 6, 36, 37, 42, 48, 50 and 51 |
| Nanoparticle surface functionalization | 14, 15, 37 and 50 |
| Relative ratio of physiological media to nanoparticle concentration | 29 and 41 |
| Topology | 51 |
Although some studies are emerging on the regulation of different types of coronas, such as lipids or natural organic matter (NOM), the physico-chemical factors regulating these types of non-protein coronas are currently even less understood.4,7,52
Though, some studies still tend to suggest having identified ‘the major factor’ controlling protein/biomolecule corona formation; however, as convincingly shown by recent comprehensive studies, none of the above-mentioned factors, such as the NPs' physico-chemical properties or exposure time, alone is able to be used to determine the formation and composition of the protein/biomolecule corona.6,16 In the study by Tenzer and colleagues,6 bioinformatic unsupervised hierarchical cluster analysis revealed that protein corona profiles could be correctly grouped according to exposure time, as well as to the NPs' physico-chemical properties (Fig. 4).
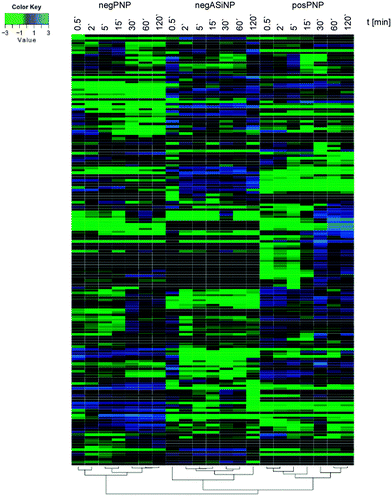 | ||
| Fig. 4 Plasma protein corona profiles are affected by the incubation time as well as by the NPs' physico-chemical characteristics. Unsupervised hierarchical cluster analysis of the relative abundance of plasma proteins bound to negatively charged amorphous silica (aSiNP) and negatively (negPS) or positively (posPS) charged polystyrene NPs. The dendrograms illustrate the sample similarities. Colour scheme is based on log2 of the ratio (protein amount at the respective time point/average protein amount across all time points). Blue indicates proteins with higher than average abundance; green indicates proteins with lower than average abundance. The exposure times to human plasma are indicated. For further details see ref. 6. | ||
A recent study also showed that protein signatures alone without the kinetic information do not seem to allow prediction of the hematocompatibility of NPs.28 However, the relation between the pristine NPs' characteristics and the nature of the corona is far from trivial, and currently still remains impossible to predict in complex physiological environments.4,6,16,25–28 Despite the complexity and analytical challenges of the biomolecule corona during its ex situ characterization, researchers face additional challenges during its in situ analysis in both physiological and environmental systems.
As outlined above, organisms and ecosystems are composed of various additional ‘complex microenvironments’, which may differ dramatically in their physico-chemical composition, such as pH, ion strength and/or temperature. Moreover, when NPs move from one biological environment to another, e.g. from the lung surfactant through epithelial/endothelial cells to the blood system and subsequent organs, such as the liver or spleen (Fig. 2), a key question is whether the original corona remains stable or is subjected to substantial changes.4,30 The current model assumes that after passing through several ‘biological environments’, the final corona still contains a fingerprint of its history and keeps a memory of its prior journey through the body.4,34 Indeed, recent quantitative kinetic data demonstrate that, for example, the plasma protein corona is surprisingly stable and matures only quantitatively rather than qualitatively ex situ.6 Hence, although previous studies suggested a dynamic corona evolution, one might hypothesize that the corona may not be subjected to significant changes, even when passing through several ‘biological environments’ ex situ, unless processing is performed by enzymatic cellular machineries.4,34,53 In particular for metal and metal oxide NPs, dissolution processes have been recognized as being essential for the NPs' fate, biodistribution and also toxicity.16,54–58
Close inspection of the literature indeed reveals that the detailed fate of the original corona in situ, as it travels through different organs and ecosystems, thereby interacting with various environments, changes in physical parameters, and that the enzymatic machineries of this remain to be resolved.
Summarizing our current knowledge, a multi-parameter classifier will be required to generally model and predict nanoparticle–protein/biomolecule interaction profiles in complex physiological and environmental systems in the future.
Go with the flow – dynamics of corona formation, evolution and complexity under physiological conditions
Protein/biomolecule adsorption to any surface, including to nanomaterials, is a dynamic process, with protein/biomolecules continually adsorbing and desorbing.13,33,36,59,60 The time-dependence of these processes, i.e. the rates of association and dissociation of a protein/biomolecule, are described by the parameters kon and koff, respectively. The balance between kon and koff determines the affinity of a protein/biomolecule to the (nano)material, and is defined by the dissociation constant (Kd).13,33,36,59,60 The value of kon depends on the contact frequency between the protein/biomolecule and the (nano)material, along with the probability that such a contact results in an adsorption event.13,33,36,59,60 Protein/biomolecules that are present at high concentrations and that diffuse rapidly mostly show high kon values. The value of koff depends on the binding energy of the protein/biomolecule–(nano)material complex. Protein/biomolecules that adsorb with a large binding energy thus have a lower koff value. In complex environments, protein/biomolecules compete for binding sites on the (nano)material surface during adsorption, with their characteristic individual Kd.13,33,36,59,60 Notably, Kds determined for the adsorption of certain proteins in isolation to NPs were reported to range between 10−4 and 10−9 M36,61 – very similar to what is measured for antibody–antigen interactions.In contrast to nanoscale objects, the formation and evolution of protein layers on flat surfaces was first analysed by Vroman.13 The so-called ‘Vroman-effect’ postulates that the identities of the adsorbed proteins can change over time even though the total amount of adsorbed protein remains roughly constant. The Vroman-effect describes a time-dependent composition of the biocoating, in which highly abundant proteins adsorbing only weakly dominate the early state. Subsequently, these adsorbed proteins are replaced by less abundant proteins, which, however, bind with a higher affinity, resulting in a complex series of adsorption and displacement steps.13,33,36,59,60 However, one has to keep in mind that the Vroman-effect has been demonstrated only for a mixture of a few proteins and is unable to predict binding kinetics to NPs in complex protein mixtures.13,59 Nevertheless, several models have directly used the Vroman-effect to explain the evolution of the protein corona around NPs, even in complex environments such as FCS or human serum, resulting in the concept of a ‘dynamic protein corona evolution’.4,27,32,34 However, in complex physiological liquids, such as blood, containing more than two thousand different proteins, or in ecosystems, a high-resolution time-resolved knowledge of NP-specific protein/biomolecules adsorption is required, as various protein/biomolecules are expected to display increased or reduced binding over time. Indeed, snapshot kinetic proteomic profiling recently demonstrated for silica and polystyrene NPs of various sizes and surface functionalization, the existence of complex protein adsorption kinetics.6 As predicted from the Vroman-effect, protein groups displaying increased or reduced binding over time were observed also in the complexity of human plasma (Fig. 5).6
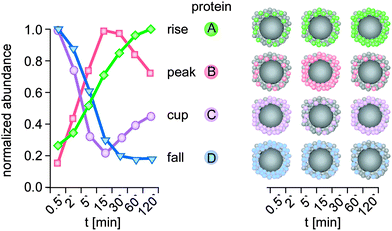 | ||
| Fig. 5 Kinetic protein-binding modalities during the temporal evolution of the plasma protein corona found for negatively charged polystyrene NPs:6 protein groups showed increasing (A, rise) or decreasing (D, fall) binding over time, respectively. ‘Peak’ proteins (B), display low abundance at the beginning of plasma exposure and at later time points, but higher abundance at intermediate time points. ‘Cup’ proteins (C) show the opposite behaviour, with a high abundance at early and late time points, but low abundance at intermediate time points. Kinetic of a representative protein (colour) is shown for each binding profile. t: plasma exposure time. | ||
Interestingly, the novel protein binding kinetics discovered cannot be solely explained by the Vroman-effect.6 Classification of the protein-binding modalities identified proteins characterized by low abundance at the beginning of plasma exposure and at later time points, but that displayed ‘peak’ abundance at intermediate time points (Fig. 5, peak), while other proteins showed exactly the opposite behaviour. These proteins are characterized by a ‘cup-shaped’ binding kinetics, i.e. being highly abundant at early and late exposure time points, but not at intermediate exposure time points (Fig. 5, cup).
A potential reason why such complex binding kinetics have not been noticed so far is the fact that most kinetic studies to date have not employed sensitive quantitative LC-MS-based proteomics. Thus, a protein that was no ‘longer detectable’ at a certain time point, was then classified as ‘absent’ or ‘disappeared’ in previous studies, thereby contributing to the model of a highly dynamic protein corona.4,27,32,34 Hence, we again want to emphasize that it is highly important to use the highest technological standards and SOPs for the determination of protein corona binding kinetics,62 also allowing inter-laboratory comparison and model building. Although the study examined 11 different types of NPs,6 further analyses are required to determine whether such adsorption kinetics, including Vroman-effect type binding kinetics, indeed exist for all NPs and nanomaterials in general. Moreover, we still lack experimental data to decide whether similar corona binding kinetics are also observed for other biomolecules, such as lipids, sugars, NOM or metabolites.
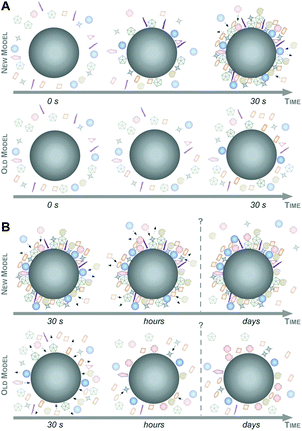
Intriguingly, a quantitative snapshot of the proteomics demonstrated that the plasma protein corona is highly complex, containing over 200 different proteins, and is established in less than one minute.6 In contrast, previous studies suggested that the protein corona has a rather low complexity, consisting of only a few tens of proteins, even when NPs are introduced into highly complex environments, such as the human blood, and evolved rather slowly.4,27,32,41 Moreover, the study by Tenzer and colleagues also showed that the corona composition changed almost exclusively quantitatively but not qualitatively over time.6 Previous models, however, proposed a highly dynamic protein corona, changing its composition over time due to continuous protein association and dissociation events, predominantly controlled by the Vroman-effect.4,27,32
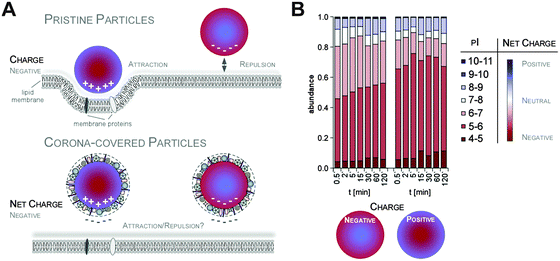
Taking these recent insights and comprehensive data into account,6,16,47 we here would like to suggest a new model of protein corona formation and evolution (Fig. 6), and which is significantly different from the old model proposed so far.4,27,32,34
As illustrated in Fig. 6, this mainly relies on the speed of corona formation, its complexity, and the predominant quantitative instead of qualitative maturation with complex binding kinetics, which cannot be explained by the Vroman-effect alone. Hence, it is suffice to assume that pristine NPs in general exist only for a short period of time in complex physiological or ecological environments. As also depicted in Fig. 6, the number of different corona proteins exceeds in most cases the number of proteins that theoretically could be accommodated on a single NP surface.6 These results give the first indication that the corona exists most likely not as a simple monolayer in situ but possibly composed of multiple core–shell structures or higher order ‘Christmas tree-like’ structures, also involving protein–protein interactions.
Additionally, it was suggested that negatively charged NPs attract primarily positively charged proteins and vice versa. Classifying corona proteins according to their predicted isoelectric point (pI) showed that proteins displaying a negative charge (pI < 7) at physiological pH 7.3 were preferentially bound by all the investigated silica and polystyrene NPs, irrespective of the particles' negative or positive surface functionalization.6,47 Indeed, zeta-potential measurements confirmed that the plasma-corona-covered NPs were overall negatively charged.6 We are not aware of any reports convincingly demonstrating the existence of positively charged plasma-corona-covered NPs so far. Hence, the hypothesis that positively charged NPs preferentially interact with the negatively charged cell membrane, resulting in improved cellular uptake, seems too simplified and may only be relevant for pristine NPs in environments with a low concentration of proteins/biomolecules (Fig. 7).63 However, whether and what type of supramolecular interactions with charged microdomains on corona proteins, NPs and/or cellular structures occur and if these are biologically relevant remains to be resolved. Consequently, it is interesting to correlate NP uptake not only to properties of the bare NPs, but also to properties of the NP–protein corona complex, as the interaction with cell membranes and the mechanism of cellular uptake is expected to be (partially) controlled by the adsorbed proteins.6,41,64–66
Blood is thicker than water – the impact of corona formation on blood system physiology
As the perturbation of physiological systems by nanoparticles has recently been reviewed,27,67,68 we here focus on novel insights into the role of the protein corona for the blood system.When designed for nanomedical applications, such as drug delivery or imaging, nanoparticle administration often requires intravenous injections.2,69 Hence, detailed knowledge about the physical and chemical aspects associated with the behaviour of NPs in the complex protein-rich environment of the blood system is thus key for (re)shaping the future of nanobiomedicine. As the complete plasma proteome reference set contains more than two thousand different proteins,70 such knowledge is particularly relevant for plasma proteins.4,6,26,28,71 Indeed, the plasma protein corona has recently been shown to be indeed highly complex as well.6
Proteins involved in physiological as well as toxicological relevant processes in the blood system, such as to complement activation and coagulation, have been identified in the coronas of various NPs.4,6,26,28,44,71,72 The identified proteins span about three to four orders of magnitude dynamic range, most likely covering most biologically relevant corona proteins.6 Notably, the respective abundance of all of these proteins was affected by the plasma exposure time and the characteristics of the NPs, such as size and surface functionalization.
As NP uptake is also an important determinant for nanopathology and targeted delivery,4,65,73 several reports have shown that the protein corona has a major impact on the NPs' cellular uptake. In particular, the recent study by Walkey and colleagues indicates that distinct protein corona signatures are indeed able to predict the cellular uptake of gold and silver NPs.16 Hence, covering NPs with a ‘physiological coating’ can indeed promote or inhibit their interaction with the cellular uptake machinery, whereas the surface charge of the bare NPs appears to be less important.6,63,65,66
Consequently, one may use this information to rationally engineer the uptake-properties of NPs by modulating the corona fingerprints. Different apolipoproteins have been described to promote transport across the blood–brain-barrier74 and different immunoglobulins and complement factors, known as opsonins, enable uptake into monocytes69 while dysopsonins like albumin and again apolipoproteins14 inhibit uptake. However, these findings are mostly based on the prior knowledge of selected proteins based on their biological function in isolation. As also the functionalization of NPs with proteins seems not to (completely) prevent corona formation, the complexity of the protein corona with more than a hundred different proteins, makes it difficult to predict the impact of individual proteins in vivo.4,6,16,75 Hence, the engineering of modified coronas by depletion or enrichment of the protein groups is required as the next step to identify corona components causally involved in (cell type specific) NP uptake.
Cleary, obtaining comprehensive quantitative and qualitative protein corona signatures, and ideally the implementation of an international standardized corona profile database resource, will finally allow the bioinformatic analysis and exploitation of signatures to guide a subsequent rational in vitro/in vivo investigation of the potential impact of corona proteins in physiological systems.4,6,16,26
Although several studies have reported the impacts of corona formation on NP exposure of the blood system, most of these effects were described as occurring at rather late exposure time points.44,76,77 By employing primary human cell models of the blood system, it was recently demonstrated that the early corona formation indeed affected processes at the nano–bio interface.6 The study showed that although the studied pristine NPs existed only for a short period in the blood system, they were still able to affect the vitality of endothelial cells, trigger thrombocyte activation and induce hemolysis.6 Hence, it was concluded that the formation of the biomolecule corona rapidly modulated the NPs' decoration with bioactive proteins, thereby protecting the cellular components of the blood system against nanoparticle-induced (patho)biological processes, as well as also influencing cellular uptake of the NPs (Fig. 8).6 However, whether this general statement is indeed valid for all existing and future NP formulations, as well as for every biomedical relevant environment in humans or in the ecosystem, remains to be experimentally confirmed.
The protein and also other biomolecule coronas are currently still unpredictable complex factors, potentially triggering not only nanomedical desired reactions but also undesired toxicological biological responses.4,6,16,28 Hence, there are currently numerous attempts to chemically completely prevent protein adsorption, which have also been reviewed previously.4,27,35,78–81 NPs functionalized with certain polymer chains, such as by the addition of various polyethylene glycol-based chains (‘PEGylation’) onto the NP surface, are often referred to as highly ‘biocompatible’, as unspecific interactions with biological components are minimized.35,78,79 NPs functionalized with PEG confers colloidal stability, even under physiological salinity conditions caused by interparticular repulsion.35,78,79 However, even complex ‘PEGylation’ is unable to completely prevent protein/biomolecule corona formation, albeit the extent of protein adsorption is clearly reduced.4,27,35,78–81 As protein adsorption is reduced, it is assumed that numerous cellular responses are affected, including opsonization by cells of the RES.4,27,82 Thus, the circulation time in the blood system, as well as the biodistribution of NPs, may be modulated via PEGylation, although the detailed mechanisms are not yet resolved.44,76,77 Again, standardized corona profiling combined with data mining and subsequent experimental verification is required to answer these questions.
Also, extensive surface functionalization with biomolecules, such as antibodies tailored to achieve the specific targeting of cell types and/or organs, or ‘cellular uptake proteins’ does not completely prevent corona formation, albeit such modifications clearly affect corona profiles. In some cases, protein corona formation was even considered a major factor in significantly reducing cell-targeting efficacy in vitro, as well as in vivo.74,83–85
Collectively, although sophisticated surface modifications reduce the adsorption of biomolecules on NMs, an association with biomolecules does still occur. Indeed, we are currently unaware of any nanomaterial functionalization that can completely prevent the formation of a biomolecule corona in complex environments. The design and synthesis of such nanodevices represents one of the key challenges required not only to finally understand the regulation and impact of the biomolecule corona but also to allow novel nanomaterial applications.
The call of nature – the biological and ecological impact of the NP–biomolecule corona
Physiological environments, such as plasma, intestinal fluid or the lung surfactant, are already complex enough; nevertheless, the natural aquatic or terrestrial environment with all their biotic and abiotic matter form far more compound mixtures. Clearly, besides the known formation of protein–lipid coronas, it is certain that heterogeneous, mixed coronas will form when NPs are released into ecosystems.9–12NPs' released into the environment
Recent increases in using NPs in medical applications, cosmetics, textiles, paints, coatings and food packaging, etc. results in their presence in the natural environment. In general the predicted aqueous concentrations of NPs range from fractions of ng L−1 to tenths of μg L−1, depending on the type of particles and the extent of their usage.86,87 Recently, fullerenes and TiO2 of urban origin were detected in the μg L−1 range in natural water.88 The importance of examining the influence of NPs on the environment has been recently recognized and many environmental agencies now have NM-dedicated projects (e.g. EPA, OECD, etc.).The environmental risk assessment of chemicals is based on an evaluation of the indigenous dangers posed by a chemical (e.g. hazard identification, dose–response assessment) and an assessment of the likelihood of these adverse effects occurring (exposure assessment). In the case of NPs, both ‘hazard’ and ‘exposure’ are quite difficult to define. For NPs, the toxic effect is not only a function of ‘the material’ but also size, surface area, redox activity, doping, surface modifiers and stability/aggregation. It has been generally observed that the smaller the NPs are, the higher is their aquatic toxicity, mostly because smaller particles are more stable (and therefore more available to free swimming organisms).89,90 In the case of redox-active NPs (e.g. Ag, Zn or Cu), smaller particles with a higher surface area release more of the free ions that are largely responsible for their toxicity. In the case of photoactive NM (e.g. TiO2 or quantum dots), the toxicity is significantly enhanced in the presence of light.87,91 Therefore, clearly defining the material and testing conditions is crucial in allowing for any kind of comparison or standardization.
Also, defining exposure is not an easy task since, like in the case of biological media, the NPs can be stabilized or destabilized and their affinity to biological surfaces can change depending on the conditions in the surrounding medium, such as the pH, ionic strength or presence of colloids. Natural colloids play a particularly important role in forming the corona and determining the fate of NPs. Apart from an inorganic fraction (e.g. ions, natural minerals), they encompass a much more complex organic fraction, collectively termed as the natural organic matter (NOM).92 The NOM content in aqueous environments ranges from 0.1 mg L−1 for groundwater, 2–10 mg L−1 in lake water to 50 mg L−1 in bogs.93,94 In aqueous environments, NOM can be very diverse and is often divided into allochthonous and autochthonous. Allochthonous organic matter – mostly humic (HA) or fulvic acids (FA) of terrestrial origin formed in the process of plant residue decomposition – is characterized by a higher molecular mass, higher flexibility and higher degree of hydrophobicity and aromaticity (all of these parameters are usually more pronounced for humic acids than fulvic acids). Autochthonous organic matter is composed of substances produced by organisms inhabiting a biotope (polysaccharides, proteins, lipids, nucleic acids, etc.) and is usually lower in molecular mass and has a less aromatic, more rigid nature.94–97 Although data regarding the influence of natural corona on NPs stability are scarce and largely focused on humic or fulvic substances, it is reasonable to expect that the influence of allochthonous and autochthonous organic matter on NPs will be quite different.
The formation of the corona depends on the characteristics of the NPs and the characteristics of the medium, like pH, ionic strength and the properties of molecules participating in corona formation.98 It was found that the adsorption of NOM occurs on ZnO, TiO2, Al2O3, NZVI, CuO Ag, Au, quantum dots, fullerenes and carbon nanotubes.11,86,87,98–105
Influence of pH and ionic strength on corona formation
The pH and presence of other ions influence the stability of NPs, especially metals, metal oxides or engineered NPs with ionisable surface modifiers, by affecting the surface charge properties. At pH values close to the isoelectric point (IEP), the stability of NPs is largely decreased. At pH values lower than the IEP, the NPs will be positively charged, while above it they will carry negative surface charge if other so-called potential determining ions are not present; therefore, the aggregation can be brought about by changes in pH. The most readily pronounced influence of pH on corona formation is also a result of surface charge modification. Since NOM and the surface of prokaryotic or eukaryotic cells are often negatively charged,106 their electrostatic interaction with NPs is favoured when they themselves are positively charged. This implies two things: (i) the pH will influence the corona formation if the NPs' surface or the adsorbing corona molecules possess chargeable sites (e.g. hydroxyl groups on the surface of metal oxides), and (ii) the pH causing charge reversal falls within environmental limits (i.e. the IEP clearly occurs in the pH range 5–9). Indeed the adsorption of bacterial exopolysaccharides on the surface of Ag NPs or humic acid on TiO2 decreased significantly when the pH exceeded the IEP.107,108 On the other hand, it is hardly possible to reverse the surface charge of e.g. silica to favour the electrostatic interaction with HA, and so here, the pH value is less important. The electrostatic attraction is not the only mechanism of corona formation, ligand exchange or hydrophobic interactions are equally important, especially for non-metallic NPs, e.g. fullerenes adsorbing NOM by hydrophobic interactions.86,109Another factor that influences the corona formation is the ionic strength of the medium. The presence of high amounts of monovalent salt can decrease the amount of organic matter adsorbed on NPs;110 whereas the presence of Ca2+ or Mg2+ ions promotes the adsorption of NOM by cation bridging between the negative NPs' surface and the negative functional groups of NOM.111,112 It was shown that in the presence of equally charged ions (e.g. phosphates or carbonates), the adsorption of NOM is lower, due to competition for the charged sides on the surface of the NPs.112 Conversely, the adsorption of NOM on carbon nanotubes, which occurs mostly through hydrophobic interactions and that correlates with the aromatic carbon content, increases with the increasing amount of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 electrolyte. The reason behind this is the tighter coiling on NOM in higher ionic strength solutions, which allows a tighter packing of molecules on the surface of the adsorbent.113
1 electrolyte. The reason behind this is the tighter coiling on NOM in higher ionic strength solutions, which allows a tighter packing of molecules on the surface of the adsorbent.113
Influence of the environmental corona on the properties of NPs
Natural or anthropogenic colloids might participate in the formation of a corona that affects the behaviour of NPs in both ways: stabilizing and destabilizing. Generally, bare NPs usually have a decreased stability with increasing ionic strength (especially when multivalent cations are present) or with pH values approaching the IEP. This is caused by the reduced repulsive forces between NPs, which allows a closer approach between them and thus increases their chances of collision.87,114–116 Stabilization by the corona is attained by electrostatic repulsion, which is achieved by rendering the surface charge negative (since NOM is mostly negatively charged) or by providing a steric barrier.117 It was shown that the presence of NOM increases the stability of Ag, TiO2, Al2O3, ZnO, Fe0 NPs, fullerenes and carbon nanotubes11,109,113,115,118 and that NOM-coated NPs are much more resistant against destabilization by multivalent cations.103,104,115 Nevertheless, formation of the corona can also destabilize NPs by cross-linking NOM molecules (especially the rigid autochthonous ones), which is particularly pronounced in the presence of multivalent cations.117,119 Nevertheless, the comparison of the TiO2 stable fraction in natural waters (lake water, tap water, seawater, and groundwater) with EPA artificial DOC-free test waters of different hardnesses showed no significant differences in stability. This is because all of these waters are similar regarding the parameters influencing TiO2 stability (i.e. organic matter content up to 2 mg L−1, pH between 6.5 and 8.5, moderate ionic strength). Therefore, at least in the case of TiO2, it seems that unless the natural environment of interest is characterized by extreme values of those parameters, the natural and synthetic waters will give comparable results.120Another way of achieving stabilization by NOM is observed in the case of redox-active NPs e.g. Fe0, which usually undergo corrosion in natural waters, so that the Fe0 is transformed to higher oxidation state species. The corona can screen them from oxidation, thereby retaining their redox activity, which is often responsible for their toxic action.121 There are, however, examples where NOM can increase oxidation, which would potentially decrease at least the oxidative-stress-based toxicity.122 Even though polyaspartate-modified Fe0 particles retained their redox activity to a high extent during aging, their toxicity actually decreased compared to fresh bare NPs. Interestingly enough, the decreased toxicity was more pronounced in cells that internalize NPs by phagocytosis – supposedly because surface modification prevents the aggregation of particles to reach a size optimal for phagocytosis (1–3 μm). For cells not capable of phagocytosis, the toxicity was solely dependent on the Fe0 content, which was slightly higher for fresh bare NZVI particles but lower for e.g. aged bare-NPs.121 This suggests that the mitigation of toxicity by NOM is mostly caused by a decreased contact with living cells.
Influence on toxicity
There are many examples where the presence of an organic matter corona mitigates the toxicity. The lower toxicity of Fe0, CuO or Ag NPs towards bacteria, crustaceans and fish has been observed in the presence of organic matter.102,123,124 The bacterial toxicity of CuO and Fe0 NPs was mitigated in the presence of FA, mostly as a result of hindering the direct contact with cells.125 Also, the effect of CuO NPs and Cu2+ ions on Daphnia magna (water flea) was one to two orders of magnitude higher when measured in natural river water as compared to artificial fresh water, showing the positive correlation between the organic matter content of river water and EC50 values. Interestingly no such correlation was observed for Daphnia magna and ZnO NPs or Zn2+ ions. Similarly, the toxicity of CuO, Cu2+, ZnO, Zn2+ towards another crustacean Thamnocephalus platyurus varied, depending on the type of water used as the test medium, but did not show a clear dependence on the organic matter content, except for the fact that the EC50 values in fresh waters were lower.126 In some cases, the test medium does not need to contain NOM for the corona-driven destabilization of NPs to take place, as some bacteria and algae have been shown to produce exopolysaccharides as a protection mechanism against NPs. Those compounds – representatives of autochthonous organic matter – are known to induce flocculation due to their rigid structure.108The lack of a corona or destabilization by a corona causing NPs aggregation decreases the bioavailability for free swimming organisms, but makes them more available to sediment dwelling organisms. It was shown that juvenile, demersal zebrafish accumulated three times as much TiO2 when no NOM was present, since bare NPs sediment more readily and were accordingly present near the bottom. Nevertheless, the mortality rate was higher in the presence of NOM due to NOM enhancing the ROS formation and oxidative DNA damage.127
There are also examples where the presence of a corona increases the toxicity for free swimming organisms. The toxicity of CuO NPs covered by FA towards the green algae Microcystis aeruginosa was higher (EC50 value three times lower). In the presence of FA, the release of Cu2+ from CuO NPs and the amount of copper bound to the cell (both CuO NPs and Cu2+ ions) was six times higher than in the case of bare particles, which contributed to the higher toxicity. Additionally, the presence of FA promoted the internalization of NPs by endocytosis, leading to DNA damage and the formation of ROS.105
Implications
To achieve the standardization of ecotoxicity testing procedures for NPs, the tested material (size distribution, surface area, oxidative state, release of ions), its stability and testing conditions (including pH, ionic strength of medium, presence of colloids, presence of activators e.g. light, redox active species) must be scrutinized. It seems that including evaluation of the NOM corona in the assessment of the risk of NPs could deliver environmentally relevant insights. Nevertheless, some kind of standard procedure, specifying what kind of model NOM and in what quantity it should be used, needs to be developed for that purpose.Despite the fact that this complexity has so far prevented progress being made in investigating the corona in the environment, it is now accepted that understanding also the impact of the corona in environmental systems is needed, but this requires new tools for its analytical dissection and the identification of corona-structure-function relationships. Hence, also in the field of environmental research, the biomolecule corona can no longer be ignored as an ‘unknown factor’, but may now represent a yet unexplored opportunity to understand and predict the impact of NMs in the environment. Generating such knowledge may also lead to more sustainable and greener nanotechnology.85,128,129
Must haves – requirements for a comprehensive analysis and functional understanding of the protein corona
Given the now (almost) accepted relevance and potentially predictive power of the NP corona, it is sufficient to propose a hypothetical research programme, called ‘systems biology of the NP corona’, or in short: CoronaSystems. In order to comprehensively analyse corona profiles and to mechanistically understand the coronas' biological/ecological impact, a tiered multidisciplinary approach is clearly required. In order to stimulate the realization of such a hypothetical research programme, we will subsequently present some of the work packages we feel are essential to successfully execute CoronaSystems and thus, to improve our current knowledge of mechanisms at the nano–bio interface.Although it is impossible to discuss in detail all the potential experimental systems applicable to address corona-related questions, we will describe and discuss the state-of-the-art requirements, as well as the challenges and caveats of such key work packages. As the majority of previous studies were performed on the protein corona, we will focus on this aspect of corona research, but will also mention the challenges of environmental coronas, such as those formed by natural organic matter (NOM). Still, we feel that the principles and comments mentioned are also transferable to other types of coronas, such as those formed by lipids or metabolites, although the methodologies will certainty need to be adapted to obtain optimal results.
Analytical challenges and strategies for a comprehensive analysis of protein or NOM corona profiles
Upon contact with a biological environment, NPs interact strongly with proteins and other biomolecules, which dramatically changes their physico-chemical and biological identities. To date, numerous studies have been conducted to identify biomolecules associating with NPs and to attempt to correlate their binding with the NPs' physico-chemical and biological properties.4,6,16,18,25–28,47,130 Albeit some common principles have emerged from these studies, the heterogeneity in their analytical approaches have so far resulted not only in a still incoherent picture of how the composition and evolution of the protein corona is affected by various factors but has sometimes even led to conflicting conclusions.Therefore, it is essential to apply and develop analytical methods with standardized protocols to investigate and characterize NP–biomolecule interactions, allowing researchers to understand the mechanistic basis for the possible biological activity of these complexes and to achieve a safety assessment of the NMs used for biological applications. As several such methods have been reviewed and described in detail before, we merely summarize such approaches in Table 2 and refer the reader to the respective reviews or original publications in which the different analytical procedures were described. In more detail, we discuss recent developments, which particularly allow one to achieve a comprehensive, kinetic snapshot analysis of the evolution of the protein corona.
| Analytical method | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| Analytical ultracentrifugation (UC) | For cell components isolation and protein thermodynamics exploration; density gradient UC for protein–NP complex purification and isolation; low impact on complex structure; high resolution in separation compared to conventional centrifugation; complexes can be further analysed; feasible for separation of NOM | Mere separation of protein–NP complexes possible, further analytical methods to study protein–NP interaction needed | 88, 92, 132–134 and 204 |
| Circular dichroism (CD) | Conformational changes can be detected on a single protein level | Problems in absolute secondary structure determination; information about structural alterations at the level of individual amino acids cannot be provided; cannot be applied on complex protein mixtures; CD signal only reflects an average of the total molecular population | 61 and 151–155 |
| Differential sedimentation centrifugation (DCS) | For isolation of protein–NP complexes; gives reliable/reproducible results | Multiple purification steps needed which alter the equilibrium of the system, leading to corona modifications of small and low density NPs (diameters about 5–20 nm; close to 1 g cm−3); does not lead to a good separation between unbound proteins and protein–NP complexes; no recovery of protein–NP complexes for further studies | 18 and 41 |
| Dynamic light scattering (DLS) | Method to measure size distribution of NPs and NOM under physiological conditions; fast and accurate | All the particles present in a biological or environmental sample contribute to the scattered light; challenging characterization of heterogeneous biological samples; sizes measured normally larger than those obtained by EM; not suitable for polydispersed samples; no element analysis available | 88, 92 and 139 |
| Electron microscopy (Scanning electron microscopy (SEM) transmission electron microscopy (TEM) and environmental scanning electron microscopy) | Frequently applied to measure size and morphology of colloidal samples (NPs and NOM) | Drying at room temperature can produce severe artefacts; large numbers of particles need to be imaged and analysed to obtain sufficient statistics; high ion (and biomolecule) concentrations in physiological samples may generate significant background levels; only used for elements with high electron density | 20, 88 and 92 |
| Fluorescence correlation spectroscopy (FCS) | Low particle concentration; hydrodynamic radius can be calculated with sub-nanometre precision | Fluorescently labelled particles needed; sensitive to the presence of aggregates; can only be applied to samples with good colloidal stability | 36 and 141–144 |
| Fourier Transform Infrared (FTIR) | Cheap; easy to identify functional groups; sensitive to protein conformation; not constrained by size and material; characterization of NOM possible | Characterization not possible in complex media; time consuming; sample preparation destroys sample | 159, 160, 168, 170 and 205 |
| Gel electrophoresis (1D/2D) | Separation of protein mixtures; suitable for quantitative and qualitative analysis; easy to perform; cheap; widely-used; inter-lab comparison possible; reproducible; separation of NOM possible | Low detection sensitivity; elution of proteins from NP necessary; harsh buffers needed; tedious; reveals only a partial view of the protein corona; propensity to form artefacts | 6, 15, 34, 47, 74, 169 and 206 |
| Isothermal titration calorimetry (ITC) | Quantitative measurement of binding affinity constant, enthalpy changes and binding stoichiometry; not limited by ligand or protein size; not affected by optical properties of the sample | Requires high sample concentration; difficult data interpretation | 14, 40, 146, 147 and 207 |
| Liquid chromatography mass spectrometry (LC-MS) | High resolution; unique to obtain protein identities; quantitative and qualitative; less user bias; characterization of NOM possible | Time consuming; expensive; requires experimental and theoretical expertise | 6, 47 and 74 |
| Nuclear magnetic resonance (NMR) spectroscopy | High-resolution information on protein structural changes; quantitative and qualitative; organic and inorganic materials; versatile; characterization of NOM possible | Time consuming; expensive; experimental and theoretical expertise is required | 156–158, 167 and 168 |
| Raman spectroscopy (SERS) | Usable with solids and liquids; no sample preparation needed; fast; spectra can be collected from small volume; no interference from water | Weak Raman effect; sensitive and highly optimized instrumentation; cannot be used for metals and alloys; impurities can hide Raman spectrum; sample heating through laser can destroy the sample | 20 and 162 |
| Surface plasmon resonance (SPR) | Study conformation of immobilized proteins and binding kinetics to NP surfaces; sensitive to changes in refractive index of medium surrounding sensor | Detection limit of the system | 14 and 148 |
| UV/Vis spectroscopy | Cheap; fast; simple; small amount of sample | Solution (pH, electrolyte, presence of interfering substances) can influence the adsorption spectrum; variables like effective bandwidth must be controlled; quantitative results difficult to achieve; needs to be used in combination with complementary spectroscopic and structural investigations | 32, 163 and 164 |
| Zeta potential | Measures surface charge in different biological environments; indicator of NP- and NOM-stability | NPs need to be monodisperse; pH-dependent surface charge and aggregation behaviour | 6, 36, 37, 42, 48, 50, 51, 88 and 92 |
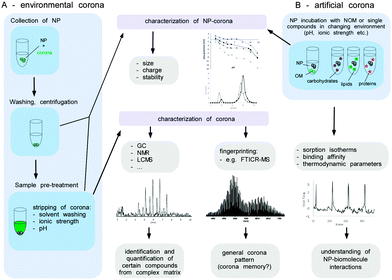
Until today, there has been no single gold standard for analysing the interaction of proteins/biomolecules with NPs, but the combination of different characterization methods is highly recommended as a strategy to compensate the drawbacks and limitations of individual methods. In general, the methods and techniques used to study the formation, evolution, composition and structure of the corona can be roughly divided into ex situ or in situ methods. Ex situ approaches seek the isolation of biomolecule–NP-complexes from the biological/ecological environment, followed by their subsequent analysis; whereas in situ methods try to measure the influence of the biomolecule corona on the NPs while the material is still dispersed in the physiological/environmental surrounding.
Notably, most physiological and environmental systems are highly dynamic.4,6 Hence, beside the full recovery of all the biomolecule–NP complexes, the development of methods allowing a snapshot time-resolved knowledge of nanoparticle-specific fingerprints and their evolution is required to fully understand their interaction with biological environments. Albeit biomolecule coronas can in principle be obtained by standard centrifugation methods, these involve mostly relatively long centrifugation steps.4 Therefore, while the ‘intended exposure time’ of NPs to physiological or environmental fluids have been reported to be relatively short, the centrifugation times required to pellet the nanoparticle–biomolecule complexes represent an additional ‘unintended exposure time’, during which the NPs are still in contact with the physiological/environmental fluids, potentially resulting in the further binding and/or dissociation of corona proteins.4,16,33 Thus, most previous studies did not precisely control the (short) exposure times of NPs to the physiological/environmental fluid of interest. These limitations have so far precluded a rapid and high-resolution kinetic analysis of corona profiles, thus necessitating the development of standardized protocols to quantitatively and qualitatively analyse the biomolecule corona of NPs.6,14,60
To minimize the contact time of NPs with the biological fluids of interest, a sucrose cushion-based centrifugation method was recently introduced and applied to resolve plasma corona formation (Fig. 9).6,62
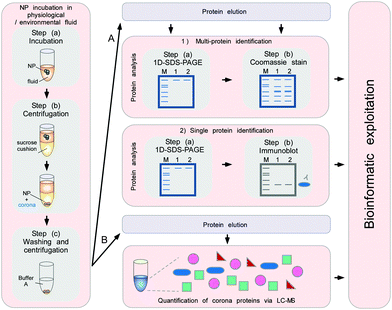 | ||
| Fig. 9 Schematic to obtain the quantitative and qualitative protein corona signatures. Following nanoparticle incubation in a physiological or environmental fluid, nanoparticle–protein complexes are rapidly separated from unbound proteins by sedimentation through a sucrose cushion, and then washed. Corona component analysis can subsequently be achieved by different methods. (A) Protein elution and separation via 1D SDS-PAGE allows directly visualizing and comparing stained protein patterns. Immunoblot analysis (semi)quantify the presence of specific corona components. (B) Protein elution and analysis via label-free quantitative LC-MS allow obtaining qualitative as well as quantitative comprehensive corona protein signatures. Further bioinformatic analysis and exploitation of the data facilitates a rational in vitro/in vivo analysis of the potential impact of corona proteins in physiological systems. In principle, similar approaches are applicable to analyse other types of biomolecule coronas, i.e. containing lipids or small metabolites, by using adequate LC-MS-based methods combined with bioinformatic exploitation strategies.7 | ||
The cushion-based centrifugation features two key advancements: first, by limiting the contact time of NPs with the biological fluid of interest, it renders the analysis of short time periods feasible. and secondly, it reduces the unintended exposure time. The isolation of biomolecule–NP complexes from excess biological/environmental fluids allows identifying and quantifying individual proteins within the corona by subsequent analytical procedures (Fig. 9).
Following protein detachment, 1D and/or 2D polyacrylamide gel electrophoresis followed by in-gel tryptic digestion and mass spectrometry have been mostly used for the identification and relative quantification of corona proteins.4,60 Albeit these methods are relatively easy procedures to directly visualize and to qualitatively compare corona protein patterns, they suffer from low throughput, reveal only a partial view of the protein corona profiles and do not allow a global quantification of the corona constituents. To overcome these limitations, label-free liquid chromatography mass spectrometry (LC-MS) has recently been introduced as the ‘gold standard’ for the qualitative and quantitative profiling of the protein corona.6,16,62 Compared to other techniques, requiring time-consuming and the expensive introduction of a label into proteins and peptides, these steps can be omitted. Although some approaches using stable isotope labelling allow multiplexing, typically only up to eight experiments can be directly compared,138 which does not seems ideal for the detailed time-resolved analyses of multiple corona profiles. In contrast, label-free techniques have been shown to achieve a reliable and highly reproducible comprehensive quantification of plasma protein corona components.4,6
Fig. 9 illustrates a proteomic workflow for the quantitative and qualitative characterization of protein corona profiles. The basic steps shown are suitable for various biotic and environmental protein-containing fluids, such as ichor, CSF-fluid, cell/organ lysates and NOM, as well as for exposure with different nanoparticle formulations.
The isolation of the biomolecule–NP complexes in ex situ approaches often requires sequential cycles of enrichment/washing steps to recover the NP–corona complexes.4,16,26,27,62 Albeit these approaches are suitable and give reliable results, one might keep in mind that multiple purification steps may in principle affect the ‘true corona’, which is formed in situ.6,31,32 Also, the separation of NPs with small diameters (less than 20 nm) and low densities (close to 1 g cm−3) is not trivial and high speed and long centrifugation times are often necessary, which also may affect the in situ corona composition and the in situ colloidal status of the NP–corona complexes. Hence, if applicable, methodologies that minimize the number of steps for the ex situ isolation/recovery of the biomolecule–NPs complexes should be used.
Another ‘complication’, which most likely occurs, is that biomolecule–NP complexes may be heterogeneous but are presented as a mixture of monomers, dimers, trimers, etc. (x-mers), for which the actual corona composition is unknown. Hence, a statistical fluctuation in corona composition could not only arise from particle to particle, but also between heterogeneous NP–corona complexes within the same sample.4,18 These different structures of co-existing complexes will have different sizes, may carry different proteins and the biological impact of these complexes might be different. Despite these complications, which are likely to exist, the biological/ecological relevance of such complexes is presently unknown. As several studies have shown that protein coronas in general are quite complex, potentially consisting of more than 200 different proteins, one might first try to dissect and understand the biological relevance of such ‘heterogeneous biomolecule–NP complexes’. Then, in a second step one should try to investigate whether and what type of relevance different nanoparticle protein complexes indeed have for the in vivo situation. Hence, albeit the study of NP–corona monomers, dimers and x-mers is certainly of academic interest, its implication for actual exposure scenarios remains to be clarified. Clearly, accidental or deliberate exposure of physiological or ecological environments to NMs will always result in an exposure to a heterogeneous population of biomolecule–NP complexes.
NPs are usually stabilized by electrostatic repulsion, steric hindrance or depletion forces.20 Charged functional groups or surfactant molecules on the NP surface produce a coulombic potential and, thus, give rise to electrostatic repulsion between individual particles carrying charges of the same polarity. The zeta potential gives information on the surface charge of the NPs and can be used to detect protein binding to the NP surface, as this will change the overall surface charge. By binding biomolecules to the NP surface, the zeta potential will change, which may also lead to changes in the NPs' colloidal stability.6,145
Another method to study the strength of protein interactions is isothermal titration calorimetry (ITC). ITC provides additional information about the thermodynamics of the protein adsorption in a quantitative and qualitative manner (e.g. binding affinity constant, enthalpy changes, binding stoichiometry between NPs and proteins).146,147 The interpretation of ITC data should be treated with caution, since in complex protein mixtures many concurrent thermodynamic processes, like ligand exchange or protein structure changes, are constantly happening. Similar to ITC, the strength of protein interactions with NPs can also be investigated by surface plasmon resonance (SPR).14,148 SPR provides information about the adsorption kinetics of the proteins to the NP' surface. The SPR technology is based on the change of oscillation of surface plasmon waves caused by the adsorption of molecules onto a metal (nanoparticle) surface (typically gold and silver). It is the fundamental principle behind many colour-based biosensor applications and different lab-on-a-chip sensors.149,150
A powerful tool to investigate the conformation or the conformational changes of an adsorbed protein is circular dichroism (CD).61,151–155 The secondary structures of proteins have their own characteristic CD spectra in the UV region. The disadvantage of CD is that it cannot be applied to complex protein mixtures and, like all spectroscopic methods, the CD signal only reflects an average of the total molecular population. However, single protein conformational changes due to NP binding can be detected using CD.
A method to quantify the extent and nature of nanoparticle surfaces is to use nuclear magnetic resonance (NMR). NMR can provide high-resolution information on protein structural changes in biomolecule–NP complexes in aqueous solution, i.e. in an environment very close to that found in physiological conditions. Also, NMR is particularly well suited to detecting detailed information in the case of weak protein–target interactions.156–158 Two other (vibrational) spectroscopic methods to detect protein binding on a NP surface by investigating the vibration modes are Fourier transform infrared (FTIR) and Raman spectroscopy.20,159–162 Compared to FTIR, Raman spectroscopy can measure the biomolecule–NP complexes in aqueous solution; furthermore, the Raman spectra of electron-rich groups or double or triple bonds are easier to interpret, because they produce more intense Raman bands. UV/vis spectroscopy can be used to analyse biomolecule binding, although quantitative and conclusive results are difficult to achieve, as binding to the NPs induces changes in the absorption spectra of the NP.32,163,164 UV/vis spectroscopy needs to be performed in association with other complementary spectroscopic and structural investigations, as NP dimer or trimer conjugates have different size distributions to bare NPs and different absorption spectra, respectively.
Collectively, the current ‘corona analysis toolbox’ contains many powerful techniques, allowing a comprehensive characterization of pristine and corona-covered NPs, as well as allowing delivery qualitative and quantitative biomolecule corona profiles. Clearly, to compensate for the drawbacks and limitations of individual methods, the combination of different characterization methods is highly recommended but should be adapted according to the scientific question investigated. Although one may consider the fact that many methods measure an ensemble average of all the adsorbed proteins, and thus cannot map the position or determine the structure of individual proteins within the corona in situ or in vivo, there are many still unresolved corona formation mechanisms that can be addressed by systematically applying the ‘toolbox’. Such integrative efforts are clearly necessary since, despite the fact that the relationship between nanomaterial design, corona formation and the responses at the nano–bio interface has been studied intensely for more than two decades, only some general principles have emerged so far.
As not much is (mechanistically) known about the relevance and impact of the biomolecule corona on the interaction of NPs with environmental systems, its comprehensive analytical characterization is highly important. Albeit several of the methods mentioned above are in principle applicable to also characterize the environmental biomolecule corona, several key steps need to be considered that are normally not found in physiological environments, such as blood. Herein, we give a brief description of the methods, as well as the challenges and analytical caveats, which should be considered when investigating the NP corona in ecosystems.
A broad range of advanced instrumental analytics such as size-exclusion chromatography,166 multidimensional NMR,167 GC,168 capillary electrophoresis169 or mass spectrometry e.g. ultrahigh-resolution Fourier transform ion cyclotron resonance (FTICR-MS)170 have been applied to characterize the NOM. The qualitative and quantitative determination of the NOM components in low concentrations from a highly complex matrix, however, represents an enormous challenge. In this context, the analysis of the NP corona appears even more complex, since most of the stripped molecules presumably occur in much lower concentrations than present in the environmental fluids.
Before the analysis of the composition of the environmental corona, the biomolecules have to be stripped from the surface of the NPs by e.g. extraction with solvents or via changes of pH or ionic strength. Such a remobilization of corona molecules represents a major obstacle, since it might not be quantitative or may alter the structure of the substances involved.
The isolation of the biomolecules allows the characterisation of the corona components by subsequent analytical procedures (Fig. 11). Thereby, the identification and quantification of certain individual components within the corona might be achieved by various analytical techniques. The knowledge about the detailed NP corona composition would surely be desirable, but this can hardly be resolved at a molecular level. Also, for an explanation and understanding of the ecotoxicological effects this might not be necessary. A general understanding of how particle characteristics such as size, surface area, redox activity, doping, surface modifiers, etc. affect the sorption behaviour of biomolecules in dependency of their molecular interaction potentials or environmental conditions (ionic strength, pH, redox potential and temperature) seems to be more important (Fig. 11B). To achieve this, it is not mandatory to experiment with highly complex mixtures, instead the sorption isotherms of single representatives of different types of biomolecules (e.g. proteins, lipids, polysaccharides) forming an artificial corona could be measured to elucidate each structural or molecular parameter involved in the sorption process. To determine the amount of biomolecules bound at the surface as a function of the molecules present in the solution compound, specific instrumental analysis can be applied. ITC can provide additional information about the thermodynamics of the sorption process (Fig. 11B).
Moreover, the NP properties (including ecotoxicity) of ‘naked’ and single-compound biomolecule ‘coated’-NPs could be compared and used for interpretation of the toxicological data. Even though a detailed qualitative and quantitative characterization of the NP corona appears to be beyond realization, a peak pattern analysis might reveal highly interesting information. The composition of organic matter can differ strongly depending on its origin (terrestrial, fresh or marine water, atmospheric organic carbon). The qualitative analysis of a NP corona peak pattern via e.g. FTICR-MS could give a NP corona ‘fingerprint’. If the NP was isolated from the environmental media, this ‘eco-fingerprint’ might allow tracing back the chemical fate-and-transport of a NP within the environment (corona memory).
The power of the NP–corona in in silico modelling
Driven by recent increases in computational power, mathematical calculation and the in silico modelling of the molecular details about how chemical/protein ligands interact with proteins, have stimulated the field to also characterize and predict protein–nanomaterial interactions. Indeed, powerful tools are available to calculate, simulate and predict protein–chemical/NM interactions, including quantum mechanical, atom empirical force field and coarse-grained strategies.20,27,52,171 Albeit such methods allow, in principle, determining the binding orientations and elucidating the specific interactions between amino acids and the nanomaterial surface, these methods are mostly restricted to studying and predicting the interactions of individual proteins with NMs.Nevertheless, in silico simulation is gaining popularity in the field as a complement or even as a replacement of experimental techniques. As such, several studies have used in silico methods to study and predict protein and lipid corona formation as a function of the NMs' physico-chemical properties, such as size, charge and surface structure.27,33,49,52,171–176 For example, plasma protein adsorption to NIPAM/BAM copolymer NPs was modelled using a biexponential function.60
This research area is highly important for the development of accurate in silico prediction tools and models in the future. However, we feel that the relevance of most of these predictions for the formation and impact of coronas in realistic complex physiological or ecological environments is still limited. For example, we have learned that protein coronas are highly complex, consisting of more than a hundred different proteins, which adsorb rapidly following exposure to human plasma.6 Clearly, these experimental data demonstrate that the binding patterns observed in plasma cannot be explained by current mathematical protein corona evolution models, as these are derived from simplified experimental systems.4,27,32–34,52,171–176 As reviewed by Vogler,59 it is currently impossible to model and predict the regulation of proteins binding to human-sized objects in such complex environments. Fundamentally, the challenge of deciphering these detailed relationships is the complexity inherent in the system.59 As the simulations' reliability critically depend on the quality of the experimental data used for the model development and validation, it is highly important to generate data to the highest level of accuracy. As mentioned above, a variety of techniques are available in the current (protein) corona ‘toolbox’. However, in order to generate high quality data for model building, verification and optimization, standard operating procedures and methods giving reliable qualitative and quantitative data should be applied across laboratories worldwide. Collecting such high-quality data in a CoroNano database would allow establishing a valuable data resource for model building, verification and training. Combined with improvements in software development and computational power, the in silico simulation and prediction of nanomaterial–protein interactions could certainly become a powerful tool to support the prediction of the (patho)biological relevance and biomedical impact of the nanomaterial–(protein) corona in general.
Resolving the impact of the NP–corona for processes at the nano–bio interface – from in vitro to in vivo
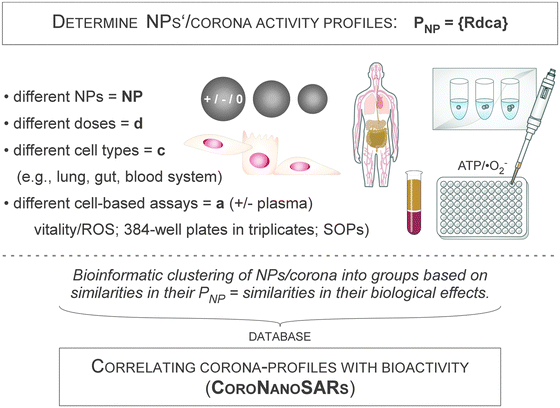
Clearly, determination of the qualitative and quantitative (kinetic) corona profiles is a prime and critical step in NanoSystems in order to understand the biological impact of the nanoparticle corona on living systems. However, although such comprehensive information is key for the subsequent interpretation and prediction of potential corona-mediated biological effects, these data have to be complemented by biological assay systems of varying complexity. Here, a tiered approach should be applied in order to reduce the complexity of biological systems and finally to understand the biological impact of the NP corona on living systems.Currently, cell models are exposed to pristine or corona-covered NPs often in low throughput applications. Assays are performed either by precoating the NPs with biomolecules or by performing the experiments in biomolecule-containing liquids, such as NOM- or FCS-containing cell culture media. Albeit such studies have provided basic insights into corona-mediated effects, their low throughput and lack of standardization make interlaboratory comparisons often difficult, sometimes even leading to contradictory results.4,16,82
To overcome these problems, high-throughput screening (HTS)/high-content screening (HCS) experimental systems should be used, exploiting technological knowledge from the drug development pipelines and toxicity testing in industry and academia gained over the last ten years. Cell-based HCS has evolved dramatically, allowing HTS applications to measure the responses of cells to chemicals and, as recently described, also to NMs.6,54,183–185 The concept and technological execution of such assay systems have been review by Nel and colleagues,183,184,186 albeit the impact of the biomolecule corona has not been specifically investigated so far.
The workflows used predominantly for such HCS/HTS screening approaches mostly focussed on classical plate reader-based assays using biochemical readouts.183,184,186 However, the ability to automate the capture and analysis of the fluorescent images of thousands of cells has made fluorescence microscopy an additional tool for systematic cell biology investigations, and also applicable for investigating nanobiology. Importantly, HTS assays have been developed that can be automatically performed in microtiter plates, and thus, the analysis is highly economical for cells and NPs, and, furthermore, can be executed under stable standardized experimental procedures (SOPs).
An example for such a high-throughput microscopy-based screening workflow to investigate the impact of the plasma protein corona is illustrated in Fig. 12. Here, the biological activity of well-characterized NPs will be assessed by high content cell-based assays, in multiple cell types and multiple doses, in the absence and presence of the plasma protein corona. Experimentally, pristine and protein-coated NPs can be profiled using an automated high-throughput microscopy platform combined with a pipetting robot, data storage and bioinformatic analysis hardware and software.62,82,183,184,186 Each NP can then be characterized by a reactivity profile PNP = {Rdca}, in which each feature is the normalized assay result Rdca that emerges when the NP is added at dose d to cell type c, and its effect is measured using assay a. As each NP profile is composed of (d c a) features, it samples much more biological activity information than is accessible by characterizing NPs in a single cell type and using only a single phenotypic assay. Bioinformatic clustering methods can then be applied to classify the NPs into groups based on similarities in their PNP, i.e. based on similarities in their patterns of biological effects in different cellular contexts.62,82,183,184,186 Subsequently, data can be bioinformatically exploited to correlate the nano-structure activity relationships (NanoSARs) with the corona profiles.
Combining systematic analytical and cell-based technologies with recent advances in bioinformatics is accepted as a powerful approach to rationally dissect and understand the cause-effect relationships of NMs in living systems. Thus, analytical and experimental data delivered by proteomic and in vitro HTS profiling experiments, can be collected in a data repository. This data repository can be used for the identification of CoroNanoSARs linking protein coronas with the NPs' characteristics and biological effects. Here, hierarchical clustering (unsupervised and consensus clustering), as well as the class prediction of NPs based on biological pathways, can be performed by using state-of-the-art software tools.62,82,183,184,186–188 Importantly, recent developments have aimed to combine and mine data obtained by methodological divergent ‘omics-technologies’, such as proteomics, transcriptomics, metabolomics, cellomics and epigenomics, allowing researchers to build a multi-classifier score for each type of NP studied.187,189 Moreover, biological pathway exploitation will provide rational information about which type of NP affects the corona-dependent cellular responses to aid the subsequent identification of key corona proteins causally involved in the observed biological effects.
In principle, the outlined workflow is also applicable to other exposure routes, such as the gastrointestinal tract, the lung or other organs, as well as to investigate other types of biomolecule coronas. Albeit comprehensive reports investigating the impact of NMs on ecosystems by using appropriate cell-based assays are still lacking, the technologies for developing such assays are in place, and would be useful for resolving a variety of corona-related and corona-unrelated questions in nano-ecology.
Nevertheless, one should keep in mind that the design and execution of such HTS assays is demanding, especially concerning technology platforms, experimental workflow and experience in data acquisition and handling. Also, in contrast to soluble chemicals, one will certainly encounter additional problems that need to be resolved when studying pristine and corona-covered NMs, such as the aggregation of certain NMs upon contact with salt and/or protein/biomolecule-containing liquids. Such a type of aggregation may be problematic when using automatic pipetting stations and may need appropriate pre-treatment, such as sonication or other dissolution techniques.12,62,82,183,184,186,190
It takes three to tango – impact of the NP corona on the microbiome–host interaction
The human microbiome is defined as the sum of all microbial organisms that reside in the body, including bacteria, fungi and archaea. Scientific and medical interest in the human microbiome has increased dramatically in recent years, following reports that the type and number of microorganisms seem to play important roles in the onset of several medical conditions, including obesity, cancer and diabetes.208–210 In the healthy state, the microbiome actually helps with basic physiological processes ranging from digestion to growth to self-defense against hostile microorganisms. Hence, it is now accepted that the microbial communities that colonize different human organs play important roles in many aspects of health and disease.211–213Notably, different types of microorganisms are present in all the major exposure and entry sites for NPs in the human body, as well as in environmental organisms (Fig. 14A). As xenobiotics are involved in actively shaping the human microbiome,214 the field has just now started to explore the impact of NPs on endogenous microbial communities and the subsequent relevance for nanotoxicology and an organism's health in mammalian, as well as in environmental, model systems.211–213 Indeed, recent studies in rats, as well as in zebrafish, have provided the first evidence that exposure of the gastrointestinal system with Ag- or Cu-NPs affects the populations of intestinal microbiota and intestinal-mucosal gene expression patterns.211–213 These results indicate that at least the oral exposure route to NPs may alter mucosa-associated microbiota, and modulate the gut-associated immune response and the overall homeostasis of the intestinal tract. It is estimated that the human body contains over 10 times more microbial cells than human cells.208–210 Hence, albeit we are faced with more than a thousand reports on the impact of the NP corona on human cells, it is more than surprising that no studies on the relevance of the NP corona on the microbiome, as well as on microbiome–host interactions, have been reported so far.
As discussed above, it is expected that microbial organisms, as well as the microbiome–host cell microenvironments, face mostly biomolecule corona-covered rather than pristine NPs. Albeit experimental data are still lacking, the impact of the biomolecule corona can be manifold. Here, not only the characteristics of the NP itself but also the type and profile of the corona may directly affect the interaction of the microbial cell with the NPs, thereby influencing the vitality and activity of microbial populations (Fig. 14A). In addition, the decoration of microorganisms with pristine or biomolecule corona-covered NPs may determine whether a NP–microorganism complex is more or less effective when interacting with host cells. For example, the recognition, uptake and destruction of microorganisms by macrophages or other immune cells might be influenced by this ménage à trois (Fig. 14B). In the different organs, such as the lung, mouth or the gastrointestinal tract, not only the protein corona but also other types of biomolecule coronas will certainly play a major role. As such interactions will particularly be of high importance for the physiological in vivo situation, including in ecosystems, the field is now challenged with the design of experimental systems and methods to address these questions.
Fact or fiction – nanologicals: NP-based targeting of molecular machines
The applications of nanotechnology in life sciences imply the development of materials designed to interact with biomolecules at the molecular scale with a high degree of specificity. In this review, so far we have predominantly focused on what happens to the technical and biological identity of NPs once they are covered by a biomolecule corona. However, let's flip the coin and vice versa discuss the consequences of the binding of proteins or biomolecules to NPs, with regards to their structure and function, as well as the overall impact of these interactions for biological or ecological systems.Protein adsorption occurs spontaneously if it is thermodynamically favourable. During adsorption, conformational changes may be induced in proteins, particularly when hydrophobic or charged protein domains interact with hydrophobic or charged NP surfaces. Clearly, the degree of conformational change upon binding depends on both the structure and chemistry of the protein, as well as on the physico-chemical characteristics of the NP. Highly stable proteins, i.e. proteins with strong internal stabilizing forces such as salt bridges or disulfide bonds, are expected to be less affected upon binding. In contrast, proteins with highly flexible domains, like loop structures or protruding helices, will most likely be influenced more severely, which may even lead to the loss of protein activity. Conformational changes may also alter the way in which an adsorbed protein is presented to its environment by exposing normally inaccessible domains or by rearranging critical binding or catalytic domains.17,67
Indeed, there are several examples that NPs are capable of inactivating proteins such as proteolytic enzymes, cytochrome c, fibrinogen, LDH and others.27,60,61,215,216 Circular dichroism spectroscopy has suggested that the structure of some of the examined proteins, although not all of them, was significantly altered by adsorption to the nanoparticle surface. Also, the charge was found to be an important determinant in some cases, partially explaining the NPs' inhibitory activity.27,60,61,215 In contrast, a range of NMs with different sizes and shapes has been reported to show positive effects on enzymes and molecular machines, such as the proteasome.215,217
Collectively, these data indicate that, in contrast to bulk materials, a certain binding specificity of nano-sized objects for a specific (class of) protein(s), and also other biomolecules may exist. Notably, the binding constants, Kds, determined for the adsorption of proteins to NPs were reported to range between 10−4 and 10−9 M, very similar to the highly selective and biologically relevant antibody–antigen interactions.36,61
Although we are still at the beginning of understanding the rules and underlying mechanisms of these effects, we hypothesize that such ‘targeted’ nanomaterial–biomolecule interactions may be of high translational relevance for biotechnology and nanomedicine applications.
Current nanomedical applications are mostly restricted to imaging, implant functionalization or drug delivery, often relying on additional laborious (bio)molecular functionalization of the NM (of chemical and/or biological origin) to obtain specificity and the desired activity.
In contrast, we propose the new concept of ‘nanologicals’. Here, target-specificity and activity are exclusively conferred by the physico-chemical characteristics of the respective nanomaterial, such as size, geometry, charge, roughness and/or surface energy (Fig. 15A).
A potential modus operandi of nanologicals in an extracellular environment is exemplified in Fig. 15B for the cancer relevant epidermal growth factor receptor (EGFR) signalling pathway.
Besides surgery or irradiation, current therapies are based on the application of ‘chemicals’ and/or ‘biologicals’. As depicted in Fig. 15, treatment with chemicals, such as the tyrosine kinase inhibitor Iressa, or biologicals, such as the monoclonal antibody Cetuximab, impede the intra- or extracellular activation of the EGFR pathway, and thus ultimately the tumour progression.218,219 The activity of both, chemicals and biologicals is, however, mediated by specific chemical (supra)molecular interactions between the agents and the target domains of the EGFR. Herein, we now propose that also certain NPs may be capable of (specifically) binding to the EGFR, thereby blocking/inducing conformational changes in domains critical for natural ligand binding or the receptor's catalytic activity. Consequently, EGFR signalling is inhibited, potentially resulting in the termination of oncogenic programmes in cancer cells. As most NPs are also taken up by many cell types,220 the inhibition or even activation of multi-protein complexes may also occur intracellularly. Nanologicals thus challenge the current paradigm that drugs have to be exclusively based on chemicals or biologicals.
As exemplified by the EGFR, proteins and enzymes are large biomolecules regulating almost all chemical reactions in numerous (patho)biological processes, including signal transduction, gene expression, immune responses, metastasis and metabolism. As enzymes are also widely used in the pharmaceutical and medical fields, and food and environmental industries, the rational regulation of enzyme activity and stability by such novel devices could become an important research area.215
However, proof-of-concept reports, particularly in complex biological environments such as cancer cells or other disease models are lacking so far. In the majority of studies, the impact of the NPs on the target protein was shown in the absence of competing proteins. In some studies, target protein inhibition was prevented by the presence of other proteins, most likely through competition for direct target protein–NPs interaction.221
However, upon success this concept will open up a variety of unprecedented applications of nanologicals, not only as ‘nanoprobes’ to dissect (patho)biological processes but also as potential tools for the treatment and diagnosis of diseases. Compared to certain chemicals and particularly biologicals, nanologicals are expected to be more stable outside and inside physiological environments, and are also likely to be produced easier and cheaper under highly standardized (GLP) conditions. Clearly, the steadily increasing advances in the chemical/physical design and synthesis of NMs now allow the rational production of a huge variety of NMs, representing a so far unexplored valuable reservoir.
Still, we are facing a variety of challenges before nanologicals might become a reality. At the moment, the underlying physico-chemical mechanism and criteria on both the NPs and on the target side allowing one to predict a priori a positive, neutral or negative effect of a nanomaterial on a specific protein are far from understood. Hence, similar to global corona profiling, comprehensive (e.g. high-throughput) screens testing a variety of NMs against numerous targets need to be performed in order to identify and dissect these NanoSARs. Notably, the bioinformatic analysis of global proteomic corona binding profiles obtained from various environments, such as from the extracellular matrix and intracellular or organelle lysates, may allow a first selection of ‘matching pairs’ of target protein and nanomaterials for the rational planning of subsequent studies.
Whether nanologicals will indeed be active in the highly complex in vivo or ecosystem environments remains to be proven.
The future is now – conclusion and outlook
In this review, we have provided ample argumentation as to why the biomolecule corona is far from being an ‘out-dated’ and already resolved topic in basic, as well as in applied, nanoscience. Even regulatory agencies have begun to accept that corona signatures might be relevant for NMs' risk assessment and prediction. Although the relationship between nanomaterial design and physiological responses has been studied intensely for almost two decades, only some general principles have emerged. Even less is known in the complex area of nanoecology and ‘green chemistry’. However, even in the field of environmental research, the biomolecule corona is now no longer ignored as an unknown factor, but instead is acknowledged as a potential but yet unexplored opportunity to understand and predict the impact of NMs on the environment.In addition to unresolved old challenges, such as the reliable determination of the physical and chemical parameters of NPs in complex fluids, herein we introduced and discussed several new challenges that the field is facing. The complexity of environmental systems has so far prevented swift progress in investigating and understanding ‘coronas’ in the environment. Here, new tools for their analytical dissection and simplified model systems for the identification of corona-structure function relationships are needed.
Also, the corona's complexity increases once we are dealing with physiologically relevant complex microenvironments in biological systems, including the human body. Here, interactions between nanomaterials, cells and additional ‘inhabitants’ such as microbes or fungi have not been addressed in detail so far. However, we predict that the corona will also be a critical determinant rather than a bystander in regulating nanomaterial–microbiome–host interactions. Moreover, besides proteins, also other biomolecule coronas require intensified analytical as well as functional investigation.
Despite these still pending challenges, we also learned some lessons. It is now accepted that biomolecule coronas are established rapidly, and that their formation most likely occurs to varying degrees for all nano-sized materials in general. Hence, the (long term) existence of pristine NPs in complex physiological or ecological environments appears to be rather an exception to the rule. Corona profiles are highly complex, consisting of a variety of adsorbed proteins potentially capable of modulating (patho)biological responses, and are rather stable ex situ. Indeed, the first protein corona signatures seem to be now emerging, and are being used to predict responses at the nano–bio interface. However, there is still an unresolved debate about whether corona profiles change significantly over time, particularly in time frames in which biological responses are expected. Nevertheless, the long-term effects and the relevance of coronas on the fate and transformation of NPs deposited in organs or ecosystems require future attention.
The good news is that we do not have to reinvent the wheel. Our ‘corona analysis toolbox’ contains many powerful techniques, thus allowing a comprehensive characterization of pristine and corona-covered NPs. Furthermore, novel bioinformatic algorithms are under development for in silico modelling and prediction. Quantitative, high-resolution LC-MS/MS is now capable of dramatically reducing the biomolecule corona characterization time and can provide quantitative data from large libraries of nanomaterials. Nevertheless, moving forward clearly requires applying these methods under standardized operating procedures – detailed enough to ensure data quality and to allow inter-laboratory comparison. In order to comprehensively analyse corona profiles and to mechanistically understand the coronas' biological/ecological impact, a tiered multidisciplinary approach is clearly mandatory/requisite, including not only comprehensive analytical methods but also the involvement of high throughput/high content ‘omics’ technologies together with bioinformatic data mining to reach the next level. Despite the achievements of the past years, the establishment of nanostructure activity-relationships linking NP/corona properties to (patho)physiological or ecological responses remains a still distant goal. Such knowledge is ultimately needed not only to understand and minimize nanotoxicity but also to develop NMs for improved future applications.
However, we are not only facing challenges but also encountering novel opportunities. Will nanologicals remain fiction or potentially become a fact?
Clearly, to further translate the ‘lessons learned’ and to accept, mitigate and finally resolve the challenges, comprehensive interdisciplinary research programmes such as the drafted CoronaSystems are a must!
Acknowledgements
Grant support: BMBF-MRCyte/NanoBEL/DENANA, Zeiss-ChemBioMed, Stiftung Rheinland-Pfalz (NanoScreen), DFG-SPP1313, PTE-foundation. We thank Dr R. Qiao for helpful discussion. We apologize to all colleagues whose work could not be cited due to space limitations.Notes and references
- M. Reese, Health Matrix, 2013, 23, 537–572 Search PubMed.
- T. J. Webster, Nanomedicine, 2013, 8, 525–529 CrossRef CAS PubMed.
- H. Rauscher, B. Sokull-Kluttgen and H. Stamm, Nanotoxicology, 2013, 7, 1195–1197 CrossRef PubMed.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- M. P. Monopoli, F. B. Bombelli and K. A. Dawson, Nat. Nanotechnol., 2011, 6, 11–12 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- A. A. Kapralov, W. H. Feng, A. A. Amoscato, N. Yanamala, K. Balasubramanian, D. E. Winnica, E. R. Kisin, G. P. Kotchey, P. Gou, L. J. Sparvero, P. Ray, R. K. Mallampalli, J. Klein-Seetharaman, B. Fadeel, A. Star, A. A. Shvedova and V. E. Kagan, ACS Nano, 2012, 6, 4147–4156 CrossRef CAS PubMed.
- Y. Y. Tyurina, E. R. Kisin, A. Murray, V. A. Tyurin, V. I. Kapralova, L. J. Sparvero, A. A. Amoscato, A. K. Samhan-Arias, L. Swedin, R. Lahesmaa, B. Fadeel, A. A. Shvedova and V. E. Kagan, ACS Nano, 2011, 5, 7342–7353 CrossRef CAS PubMed.
- J. Gao, S. Youn, A. Hovsepyan, V. L. Llaneza, Y. Wang, G. Bitton and J. C. Bonzongo, Environ. Sci. Technol., 2009, 43, 3322–3328 CrossRef CAS.
- A. A. Keller, H. Wang, D. Zhou, H. S. Lenihan, G. Cherr, B. J. Cardinale, R. Miller and Z. Ji, Environ. Sci. Technol., 2010, 44, 1962–1967 CrossRef CAS PubMed.
- K. Yang, D. Lin and B. Xing, Langmuir, 2009, 25, 3571–3576 CrossRef CAS PubMed.
- G. V. Lowry, K. B. Gregory, S. C. Apte and J. R. Lead, Environ. Sci. Technol., 2012, 46, 6893–6899 CrossRef CAS PubMed.
- L. Vroman, Nature, 1962, 196, 476–477 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, F. Song, R. Liu, H. Guo, D. W. Olsen, Y. Cohen, A. Emili and W. C. Chan, ACS Nano, 2014, 8, 2439–2455 CrossRef CAS PubMed.
- I. Lynch, T. Cedervall, M. Lundqvist, C. Cabaleiro-Lago, S. Linse and K. A. Dawson, Adv. Colloid Interface Sci., 2007, 134–135, 167–174 CrossRef CAS PubMed.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed.
- A. Schweitzer, S. K. Knauer and R. H. Stauber, Int. J. Cancer, 2010, 15, 801–809 Search PubMed.
- L. Treuel, K. A. Eslahian, D. Docter, T. Lang, R. Zellner, K. Nienhaus, G. U. Nienhaus, R. H. Stauber and M. Maskos, Phys. Chem. Chem. Phys., 2014, 16, 15053–15067 RSC.
- Y. K. Lee, E. J. Choi, T. J. Webster, S. H. Kim and D. Khang, Int. J. Nanomed., 2015, 10, 97–113 Search PubMed.
- E. R. Toke, O. Lorincz, E. Somogyi and J. Lisziewicz, Int. J. Pharm., 2010, 392, 261–267 CrossRef CAS PubMed.
- A. M. Nystrom and B. Fadeel, J. Controlled Release, 2012, 161, 403–408 CrossRef PubMed.
- J. Wolfram, Y. Yang, J. Shen, A. Moten, C. Chen, H. Shen, M. Ferrari and Y. Zhao, Colloids Surf., 2014, 124, 17–24 CrossRef CAS PubMed.
- J. S. Gebauer, M. Malissek, S. Simon, S. K. Knauer, M. Maskos, R. H. Stauber, W. Peukert and L. Treuel, Langmuir, 2012, 28, 9673–9679 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- C. D. Walkey and W. C. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- M. A. Dobrovolskaia, B. W. Neun, S. Man, X. Ye, M. Hansen, A. K. Patri, R. M. Crist and S. E. McNeil, Nanomedicine, 2014, 10, 1453–1463 CrossRef CAS PubMed.
- G. Caracciolo, D. Pozzi, A. L. Capriotti, C. Cavaliere, P. Foglia, H. Amenitsch and A. Lagana, Langmuir, 2011, 27, 15048–15053 CrossRef CAS PubMed.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz, M. R. Ejtehadi, R. Hartmann, K. Kantner, U. Linne, P. Maffre, S. Metzler, M. K. Moghadam, C. Pfeiffer, M. Rezaei, P. Ruiz-Lozano, V. Serpooshan, M. A. Shokrgozar, G. U. Nienhaus and W. J. Parak, ACS Nano, 2013, 7, 6555–6562 CrossRef CAS PubMed.
- A. L. Barran-Berdon, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Lagana, Langmuir, 2013, 29, 6485–6494 CrossRef CAS PubMed.
- E. Casals, T. Pfaller, A. Duschl, G. J. Oostingh and V. Puntes, ACS Nano, 2010, 4, 3623–3632 CrossRef CAS PubMed.
- D. Dell'Orco, M. Lundqvist, C. Oslakovic, T. Cedervall and S. Linse, PLoS One, 2010, 5, e10949 Search PubMed.
- M. Lundqvist, J. Stigler, T. Cedervall, T. Berggard, M. B. Flanagan, I. Lynch, G. Elia and K. Dawson, ACS Nano, 2011, 5, 7503–7509 CrossRef CAS PubMed.
- K. Natte, J. F. Friedrich, S. Wohlrab, J. Lutzki, R. von Klitzing, W. Osterle and G. Orts-Gil, Colloids Surf., B, 2013, 104, 213–220 CrossRef CAS PubMed.
- C. Rocker, M. Potzl, F. Zhang, W. J. Parak and G. U. Nienhaus, Nat. Nanotechnol., 2009, 4, 577–580 CrossRef PubMed.
- M. S. Ehrenberg, A. E. Friedman, J. N. Finkelstein, G. Oberdorster and J. L. McGrath, Biomaterials, 2009, 30, 603–610 CrossRef CAS PubMed.
- T. M. Goppert and R. H. Muller, Eur. J. Pharm. Biopharm., 2005, 60, 361–372 CrossRef PubMed.
- D. Labarre, C. Vauthier, C. Chauvierre, B. Petri, R. Muller and M. M. Chehimi, Biomaterials, 2005, 26, 5075–5084 CrossRef CAS PubMed.
- S. Lindman, I. Lynch, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Nano Lett., 2007, 7, 914–920 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- D. E. Owens 3rd and N. A. Peppas, Int. J. Pharm., 2006, 307, 93–102 CrossRef PubMed.
- S. Chakraborty, P. Joshi, V. Shanker, Z. A. Ansari, S. P. Singh and P. Chakrabarti, Langmuir, 2011, 27, 7722–7731 CrossRef CAS PubMed.
- M. A. Dobrovolskaia, A. K. Patri, J. Zheng, J. D. Clogston, N. Ayub, P. Aggarwal, B. W. Neun, J. B. Hall and S. E. McNeil, Nanomedicine, 2009, 5, 106–117 CrossRef CAS PubMed.
- D. Dutta, S. K. Sundaram, J. G. Teeguarden, B. J. Riley, L. S. Fifield, J. M. Jacobs, S. R. Addleman, G. A. Kaysen, B. M. Moudgil and T. J. Weber, Toxicol. Sci., 2007, 100, 303–315 CrossRef CAS PubMed.
- S. H. Lacerda, J. J. Park, C. Meuse, D. Pristinski, M. L. Becker, A. Karim and J. F. Douglas, ACS Nano, 2010, 4, 365–379 CrossRef PubMed.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed.
- H. Zhang, K. E. Burnum, M. L. Luna, B. O. Petritis, J. S. Kim, W. J. Qian, R. J. Moore, A. Heredia-Langner, B. J. Webb-Robertson, B. D. Thrall, D. G. Camp 2nd, R. D. Smith, J. G. Pounds and T. Liu, Proteomics, 2011, 11, 4569–4577 CrossRef CAS PubMed.
- D. Dell'Orco, M. Lundqvist, T. Cedervall and S. Linse, Nanomedicine, 2012, 8, 1271–1281 CrossRef PubMed.
- A. Gessner, A. Lieske, B. Paulke and R. Muller, Eur. J. Pharm. Biopharm., 2002, 54, 165–170 CrossRef CAS.
- M. Mahmoudi, S. Sant, B. Wang, S. Laurent and T. Sen, Adv. Drug Delivery Rev., 2011, 63, 24–46 CrossRef CAS PubMed.
- G. Hu, B. Jiao, X. Shi, R. P. Valle, Q. Fan and Y. Y. Zuo, ACS Nano, 2013, 7, 10525–10533 CrossRef CAS PubMed.
- S. Lin, X. Wang, Z. Ji, C. H. Chang, Y. Dong, H. Meng, Y. P. Liao, M. Wang, T. B. Song, S. Kohan, T. Xia, J. I. Zink, S. Lin and A. E. Nel, ACS Nano, 2014, 8, 4450–4464 CrossRef CAS PubMed.
- H. Zhang, S. Pokhrel, Z. Ji, H. Meng, X. Wang, S. Lin, C. H. Chang, L. Li, R. Li, B. Sun, M. Wang, Y. P. Liao, R. Liu, T. Xia, L. Madler and A. E. Nel, J. Am. Chem. Soc., 2014, 136, 6406–6420 CrossRef CAS PubMed.
- X. Wang, Z. Ji, C. H. Chang, H. Zhang, M. Wang, Y. P. Liao, S. Lin, H. Meng, R. Li, B. Sun, L. V. Winkle, K. E. Pinkerton, J. I. Zink, T. Xia and A. E. Nel, Small, 2014, 10, 385–398 CrossRef CAS PubMed.
- B. Gilbert, S. C. Fakra, T. Xia, S. Pokhrel, L. Madler and A. E. Nel, ACS Nano, 2012, 6, 4921–4930 CrossRef CAS PubMed.
- N. Konduru, J. Keller, L. Ma-Hock, S. Groters, R. Landsiedel, T. C. Donaghey, J. D. Brain, W. Wohlleben and R. M. Molina, Part. Fibre Toxicol., 2014, 11, 55 CrossRef PubMed.
- R. Li, Z. Ji, C. H. Chang, D. R. Dunphy, X. Cai, H. Meng, H. Zhang, B. Sun, X. Wang, J. Dong, S. Lin, M. Wang, Y. P. Liao, C. J. Brinker, A. Nel and T. Xia, ACS Nano, 2014, 8, 1771–1783 CrossRef CAS PubMed.
- E. A. Vogler, Biomaterials, 2012, 33, 1201–1237 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, M. Foy, T. Berggard, S. C. Donnelly, G. Cagney, S. Linse and K. A. Dawson, Angew. Chem., Int. Ed., 2007, 46, 5754–5756 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- D. Docter, U. Distler, W. Storck, J. Kuharev, D. Wunsch, A. Hahlbrock, S. K. Knauer, S. Tenzer and R. H. Stauber, Nat. Protoc., 2014, 9, 2030–2044 CrossRef CAS PubMed.
- D. Huhn, K. Kantner, C. Geidel, S. Brandholt, I. De Cock, S. J. Soenen, P. Rivera Gil, J. M. Montenegro, K. Braeckmans, K. Mullen, G. U. Nienhaus, M. Klapper and W. J. Parak, ACS Nano, 2013, 7, 3253–3263 CrossRef PubMed.
- M. Mahmoudi, J. Meng, X. Xue, X. J. Liang, M. Rahman, C. Pfeiffer, R. Hartmann, P. R. Gil, B. Pelaz, W. J. Parak, P. Del Pino, S. Carregal-Romero, A. G. Kanaras and S. Tamil Selvan, Biotechnol. Adv., 2014, 32, 679–692 CrossRef CAS PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Aberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed.
- A. Lesniak, A. Salvati, M. J. Santos-Martinez, M. W. Radomski, K. A. Dawson and C. Aberg, J. Am. Chem. Soc., 2013, 135, 1438–1444 CrossRef CAS PubMed.
- Y. Zhang, Y. Bai, J. Jia, N. Gao, Y. Li, R. Zhang, G. Jiang and B. Yan, Chem. Soc. Rev., 2014, 43, 3762–3809 RSC.
- B. Pelaz, G. Charron, C. Pfeiffer, Y. Zhao, J. M. de la Fuente, X. J. Liang, W. J. Parak and P. Del Pino, Small, 2013, 9, 1573–1584 CrossRef CAS PubMed.
- K. Riehemann, S. W. Schneider, T. A. Luger, B. Godin, M. Ferrari and H. Fuchs, Angew. Chem., Int. Ed., 2009, 48, 872–897 CrossRef CAS PubMed.
- T. Farrah, E. W. Deutsch, G. S. Omenn, D. S. Campbell, Z. Sun, J. A. Bletz, P. Mallick, J. E. Katz, J. Malmstrom, R. Ossola, J. D. Watts, B. Lin, H. Zhang, R. L. Moritz and R. Aebersold, Mol. Cell. Proteomics, 2011, 10, M110 Search PubMed 006353.
- S. Barkam, S. Saraf and S. Seal, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 544–568 CrossRef CAS PubMed.
- S. M. Moghimi and Z. S. Farhangrazi, Nanomedicine, 2013, 9, 458–460 CrossRef CAS PubMed.
- O. Lunov, T. Syrovets, C. Rocker, K. Tron, G. U. Nienhaus, V. Rasche, V. Mailander, K. Landfester and T. Simmet, Biomaterials, 2010, 31, 9015–9022 CrossRef CAS PubMed.
- S. Meister, I. Zlatev, J. Stab, D. Docter, S. Baches, R. H. Stauber, M. Deutsch, R. Schmidt, S. Ropele, M. Windisch, K. Langer, S. Wagner, H. von Briesen, S. Weggen and C. U. Pietrzik, Alzheimer's Res. Ther., 2013, 5, 51 CrossRef PubMed.
- M. Zhu, G. Nie, H. Meng, T. Xia, A. Nel and Y. Zhao, Acc. Chem. Res., 2013, 46, 622–631 CrossRef CAS PubMed.
- P. Aggarwal, J. B. Hall, C. B. McLeland, M. A. Dobrovolskaia and S. E. McNeil, Adv. Drug Delivery Rev., 2009, 61, 428–437 CrossRef CAS PubMed.
- M. A. Dobrovolskaia, D. R. Germolec and J. L. Weaver, Nat. Nanotechnol., 2009, 4, 411–414 CrossRef CAS PubMed.
- C. Sacchetti, K. Motamedchaboki, A. Magrini, G. Palmieri, M. Mattei, S. Bernardini, N. Rosato, N. Bottini and M. Bottini, ACS Nano, 2013, 7, 1974–1989 CrossRef CAS PubMed.
- A. K. Murthy, R. J. Stover, A. U. Borwankar, G. D. Nie, S. Gourisankar, T. M. Truskett, K. V. Sokolov and K. P. Johnston, ACS Nano, 2013, 7, 239–251 CrossRef CAS PubMed.
- F. Q. Hu, L. Wei, Z. Zhou, Y. L. Ran, Z. Li and M. Y. Gao, Adv. Mater., 2006, 18, 2553–2556 CrossRef CAS PubMed.
- D. Pozzi, V. Colapicchioni, G. Caracciolo, S. Piovesana, A. L. Capriotti, S. Palchetti, S. De Grossi, A. Riccioli, H. Amenitsch and A. Lagana, Nanoscale, 2014, 6, 2782–2792 RSC.
- D. Docter, S. Strieth, D. Westmeier, O. Hayden, M. Gao, S. K. Knauer and R. H. Stauber, Nanomedicine, 2015, 10, 503–519 CrossRef CAS PubMed.
- A. Salvati, A. S. Pitek, M. P. Monopoli, K. Prapainop, F. B. Bombelli, D. R. Hristov, P. M. Kelly, C. Aberg, E. Mahon and K. A. Dawson, Nat. Nanotechnol., 2013, 8, 137–143 CrossRef CAS PubMed.
- G. von Maltzahn, J. H. Park, K. Y. Lin, N. Singh, C. Schwoppe, R. Mesters, W. E. Berdel, E. Ruoslahti, M. J. Sailor and S. N. Bhatia, Nat. Mater., 2012, 10, 545–552 CrossRef PubMed.
- M. Helou, M. Reisbeck, S. F. Tedde, L. Richter, L. Bar, J. J. Bosch, R. H. Stauber, E. Quandt and O. Hayden, Lab Chip, 2013, 13, 1035–1038 RSC.
- K. Pakarinen, E. J. Petersen, L. Alvila, G. C. Waissi-Leinonen, J. Akkanen, M. T. Leppanen and J. V. Kukkonen, Environ. Toxicol. Chem., 2013, 32, 1224–1232 CrossRef CAS PubMed.
- V. K. Sharma, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2009, 44, 1485–1495 CrossRef CAS PubMed.
- K. Tiede, M. Hassellov, E. Breitbarth, Q. Chaudhry and A. B. Boxall, J. Chromatogr. A, 2009, 1216, 503–509 CrossRef CAS PubMed.
- K. Hund-Rinke and M. Simon, Environ. Sci. Pollut. Res. Int., 2006, 13, 225–232 CrossRef CAS.
- D. Y. Lyon, L. K. Adams, J. C. Falkner and P. J. Alvarezt, Environ. Sci. Technol., 2006, 40, 4360–4366 CrossRef CAS.
- T. Tong, C. T. Binh, J. J. Kelly, J. F. Gaillard and K. A. Gray, Water Res., 2013, 47, 2352–2362 CrossRef CAS PubMed.
- B. Nowack and T. D. Bucheli, Environ. Pollut., 2007, 150, 5–22 CrossRef CAS PubMed.
- J. A. Leenheer and J. P. Croue, Environ. Sci. Technol., 2003, 37, 18A–26A CrossRef CAS.
- K. J. Wilkinson, J. C. Negre and J. Buffle, J. Contam. Hydrol., 1997, 26, 229–243 CrossRef CAS.
- J. J. Alberts and M. Takács, Environ. Int., 1999, 25, 237–244 CrossRef CAS.
- J. McLaughlin and J. C. Bonzongo, Environ. Toxicol. Chem., 2012, 31, 168–175 CrossRef CAS PubMed.
- P. Verdugo, A. L. Alldredge, F. Azam, D. L. Kirchman, U. Passow and P. H. Santschi, Mar. Chem., 2004, 92, 67–85 CrossRef CAS PubMed.
- S. Sánchez-Cortés, O. Francioso, C. Ciavatta, J. V. García-Ramos and C. Gessa, J. Colloid Interface Sci., 1998, 198, 308–318 CrossRef.
- L. Chekli, S. Phuntsho, M. Roy, E. Lombi, E. Donner and H. K. Shon, Water Res., 2013, 47, 4585–4599 CrossRef CAS PubMed.
- A. Hitchman, G. H. Smith, Y. Ju-Nam, M. Sterling and J. R. Lead, Chemosphere, 2013, 90, 410–416 CrossRef CAS PubMed.
- L. Hou, D. Zhu, X. Wang, L. Wang, C. Zhang and W. Chen, Environ. Toxicol. Chem., 2013, 32, 493–500 CrossRef CAS PubMed.
- S. Lee, K. Kim, H. K. Shon, S. Kim and J. Cho, J. Nanopart. Res., 2011, 13, 3051–3061 CrossRef CAS.
- V. L. Pallem, H. A. Stretz and M. J. Wells, Environ. Sci. Technol., 2009, 43, 7531–7535 CrossRef CAS.
- D. P. Stankus, S. E. Lohse, J. E. Hutchison and J. A. Nason, Environ. Sci. Technol., 2011, 45, 3238–3244 CrossRef CAS PubMed.
- Z. Wang, J. Li, J. Zhao and B. Xing, Environ. Sci. Technol., 2011, 45, 6032–6040 CrossRef CAS PubMed.
- M. Kosmulski, J. Colloid Interface Sci., 2009, 337, 439–448 CrossRef CAS PubMed.
- G. Chen, X. Liu and C. Su, Environ. Sci. Technol., 2012, 46, 7142–7150 CrossRef CAS PubMed.
- S. Khan, A. Mukherjee and N. Chandrasekaran, J. Environ. Sci., 2011, 23, 346–352 CrossRef CAS.
- B. Xie, Z. Xu, W. Guo and Q. Li, Environ. Sci. Technol., 2008, 42, 2853–2859 CrossRef CAS.
- S. S. Khan, A. Mukherjee and N. Chandrasekaran, Water Res., 2011, 45, 5184–5190 CrossRef CAS PubMed.
- M. Erhayem and M. Sohn, Sci. Total Environ., 2014, 468–469, 249–257 CrossRef CAS PubMed.
- L. Liang, L. Luo and S. Zhang, Colloids Surf., A, 2011, 384, 126–130 CrossRef CAS PubMed.
- H. Hyung and J. H. Kim, Environ. Sci. Technol., 2008, 42, 4416–4421 CrossRef CAS.
- L. Chekli, S. Phuntsho, M. Roy and H. K. Shon, Sci. Total Environ., 2013, 461–462, 19–27 CrossRef CAS PubMed.
- M. Delay, T. Dolt, A. Woellhaf, R. Sembritzki and F. H. Frimmel, J. Chromatogr. A, 2011, 1218, 4206–4212 CrossRef CAS PubMed.
- D. Zhou and A. A. Keller, Water Res., 2010, 44, 2948–2956 CrossRef CAS PubMed.
- Y. Zhang, Y. Chen, P. Westerhoff and J. Crittenden, Water Res., 2009, 43, 4249–4257 CrossRef CAS PubMed.
- R. L. Johnson, G. O. Johnson, J. T. Nurmi and P. G. Tratnyek, Environ. Sci. Technol., 2009, 43, 5455–5460 CrossRef CAS.
- A. Quigg, W.-C. Chin, C.-S. Chen, S. Zhang, Y. Jiang, A.-J. Miao, K. A. Schwehr, C. Xu and P. H. Santschi, ACS Sustainable Chem. Eng., 2013, 1, 686–702 CAS.
- S. Ottofuelling, F. Von der Kammer and T. Hofmann, Environ. Sci. Technol., 2011, 45, 10045–10052 CrossRef CAS PubMed.
- T. Phenrat, T. C. Long, G. V. Lowry and B. Veronesi, Environ. Sci. Technol., 2009, 43, 195–200 CrossRef CAS.
- A. B. Giasuddin, S. R. Kanel and H. Choi, Environ. Sci. Technol., 2007, 41, 2022–2027 CrossRef CAS.
- J. Fabrega, S. R. Fawcett, J. C. Renshaw and J. R. Lead, Environ. Sci. Technol., 2009, 43, 7285–7290 CrossRef CAS.
- Z. Li, K. Greden, P. J. Alvarez, K. B. Gregory and G. V. Lowry, Environ. Sci. Technol., 2010, 44, 3462–3467 CrossRef CAS PubMed.
- J. Zhao, Z. Wang, Y. Dai and B. Xing, Water Res., 2013, 47, 4169–4178 CrossRef CAS PubMed.
- I. Blinova, A. Ivask, M. Heinlaan, M. Mortimer and A. Kahru, Environ. Pollut., 2010, 158, 41–47 CrossRef CAS PubMed.
- S. P. Yang, O. Bar-Ilan, R. E. Peterson, W. Heideman, R. J. Hamers and J. A. Pedersen, Environ. Sci. Technol., 2013, 47, 4718–4725 CrossRef CAS PubMed.
- M. A. Dobrovolskaia, J. D. Clogston, B. W. Neun, J. B. Hall, A. K. Patri and S. E. McNeil, Nano Lett., 2008, 8, 2180–2187 CrossRef CAS PubMed.
- M. A. Dobrovolskaia and S. E. McNeil, Nat. Nanotechnol., 2007, 2, 469–478 CrossRef CAS PubMed.
- A. Capriotti, G. Caracciolo, C. Cavaliere, V. Colapicchioni, S. Piovesana, D. Pozzi and A. Laganà, Chromatographia, 2014, 77, 755–769 CAS.
- G. J. Howlett, A. P. Minton and G. Rivas, Curr. Opin. Chem. Biol., 2006, 10, 430–436 CrossRef CAS PubMed.
- K. L. Planken and H. Colfen, Nanoscale, 2010, 2, 1849–1869 RSC.
- K. Nontapot, V. Rastogi, J. A. Fagan and V. Reipa, Nanotechnology, 2013, 24, 155701 CrossRef PubMed.
- K. E. Moore, M. Pfohl, F. Hennrich, V. S. Chakradhanula, C. Kuebel, M. M. Kappes, J. G. Shapter, R. Krupke and B. S. Flavel, ACS Nano, 2014, 8, 6756–6764 CrossRef CAS PubMed.
- U. Sakulkhu, M. Mahmoudi, L. Maurizi, J. Salaklang and H. Hofmann, Sci. Rep., 2014, 4, 5020 CAS.
- L. Bohmert, M. Girod, U. Hansen, R. Maul, P. Knappe, B. Niemann, S. M. Weidner, A. F. Thunemann and A. Lampen, Nanotoxicology, 2014, 8, 631–642 CrossRef PubMed.
- J. Ashby, S. Schachermeyer, S. Pan and W. Zhong, Anal. Chem., 2013, 85, 7494–7501 CrossRef CAS PubMed.
- C. Evans, J. Noirel, S. Y. Ow, M. Salim, A. G. Pereira-Medrano, N. Couto, J. Pandhal, D. Smith, T. K. Pham, E. Karunakaran, X. Zou, C. A. Biggs and P. C. Wright, Anal. Bioanal. Chem., 2012, 404, 1011–1027 CrossRef CAS PubMed.
- M. Maskos and R. Stauber, Comprehensive Biomaterials, 2011, vol. 3, pp. 329–339 Search PubMed.
- B. I. Ipe, A. Shukla, H. Lu, B. Zou, H. Rehage and C. M. Niemeyer, ChemPhysChem, 2006, 7, 1112–1118 CrossRef CAS PubMed.
- L. Treuel, S. Brandholt, P. Maffre, S. Wiegele, L. Shang and G. U. Nienhaus, ACS Nano, 2014, 8, 503–513 CrossRef CAS PubMed.
- G. U. Nienhaus, P. Maffre and K. Nienhaus, Methods Enzymol., 2013, 519, 115–137 CAS.
- P. Maffre, K. Nienhaus, F. Amin, W. J. Parak and G. U. Nienhaus, Beilstein J. Nanotechnol., 2011, 2, 374–383 CrossRef CAS PubMed.
- P. Maffre, S. Brandholt, K. Nienhaus, L. Shang, W. J. Parak and G. U. Nienhaus, Beilstein J. Nanotechnol., 2014, 5, 2036–2047 CrossRef CAS PubMed.
- D. Docter, C. Bantz, D. Westmeier, H. Galla, Q. Wang, C. J. Kirkpatrick, M. Maskos and R. H. Stauber, Beilstein J. Nanotechnol., 2014, 5, 1380–1392 CrossRef PubMed.
- G. Baier, C. Costa, A. Zeller, D. Baumann, C. Sayer, P. H. Araujo, V. Mailander, A. Musyanovych and K. Landfester, Macromol. Biosci., 2011, 11, 628–638 CrossRef CAS PubMed.
- R. Huang, R. P. Carney, K. Ikuma, F. Stellacci and B. L. Lau, ACS Nano, 2014, 8, 5402–5412 CrossRef CAS PubMed.
- Y. Cheng, M. Wang, G. Borghs and H. Chen, Langmuir, 2011, 27, 7884–7891 CrossRef CAS PubMed.
- S. S. Acimovic, M. A. Ortega, V. Sanz, J. Berthelot, J. L. Garcia-Cordero, J. Renger, S. J. Maerkl, M. P. Kreuzer and R. Quidant, Nano Lett., 2014, 14, 2636–2641 CrossRef CAS PubMed.
- J. N. Anker, W. P. Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. Van Duyne, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- S. Laera, G. Ceccone, F. Rossi, D. Gilliland, R. Hussain, G. Siligardi and L. Calzolai, Nano Lett., 2011, 11, 4480–4484 CrossRef CAS PubMed.
- A. Bhogale, N. Patel, J. Mariam, P. M. Dongre, A. Miotello and D. C. Kothari, Colloids Surf., B, 2014, 113, 276–284 CrossRef CAS PubMed.
- A. Astegno, E. Maresi, V. Marino, P. Dominici, M. Pedroni, F. Piccinelli and D. Dell'Orco, Nanoscale, 2014, 6, 15037–15047 RSC.
- R. Cukalevski, M. Lundqvist, C. Oslakovic, B. Dahlback, S. Linse and T. Cedervall, Langmuir, 2011, 27, 14360–14369 CrossRef CAS PubMed.
- V. Marino, A. Astegno, M. Pedroni, F. Piccinelli and D. Dell'Orco, Nanoscale, 2014, 6, 412–423 RSC.
- A. Ceccon, M. Lelli, M. D'Onofrio, H. Molinari and M. Assfalg, J. Am. Chem. Soc., 2014, 136, 13158–13161 CrossRef CAS PubMed.
- M. Calvaresi, F. Arnesano, S. Bonacchi, A. Bottoni, V. Calo, S. Conte, G. Falini, S. Fermani, M. Losacco, M. Montalti, G. Natile, L. Prodi, F. Sparla and F. Zerbetto, ACS Nano, 2014, 8, 1871–1877 CrossRef CAS PubMed.
- L. Calzolai, F. Franchini, D. Gilliland and F. Rossi, Nano Lett., 2010, 10, 3101–3105 CrossRef CAS PubMed.
- K. K. Chittur, Biomaterials, 1998, 19, 357–369 CrossRef CAS.
- J. Zhang and Y. B. Yan, Anal. Biochem., 2005, 340, 89–98 CrossRef CAS PubMed.
- B. Zhang, Q. Li, P. Yin, Y. Rui, Y. Qiu, Y. Wang and D. Shi, ACS Appl. Mater. Interfaces, 2012, 4, 6479–6486 CAS.
- G. J. Thomas, Jr., Annu. Rev. Biophys. Biomol. Struct., 1999, 28, 1–27 CrossRef PubMed.
- M. Lundqvist, I. Sethson and B. H. Jonsson, Langmuir, 2004, 20, 10639–10647 CrossRef CAS PubMed.
- J. Vidic, F. Haque, J. M. Guigner, A. Vidy, C. Chevalier and S. Stankic, Langmuir, 2014, 30, 11366–11374 CrossRef CAS PubMed.
- K. Mopper, A. Stubbins, J. D. Ritchie, H. M. Bialk and P. G. Hatcher, Chem. Rev., 2007, 107, 419–442 CrossRef CAS PubMed.
- M. B. Müller, D. Schmitt and F. H. Frimmel, Environ. Sci. Technol., 2000, 34, 4867–4872 CrossRef.
- A. J. Simpson, W. L. Kingery and P. G. Hatcher, Environ. Sci. Technol., 2003, 37, 337–342 CrossRef CAS.
- L. Berwick, P. F. Greenwood and R. J. Smernik, Water Res., 2010, 44, 3039–3054 CrossRef CAS PubMed.
- P. Schmitt-Kopplin and A. Kettrup, Electrophoresis, 2003, 24, 3057–3066 CrossRef CAS PubMed.
- T. Riedel and T. Dittmar, Anal. Chem., 2014, 86, 8376–8382 CrossRef CAS PubMed.
- F. D. Sahneh, C. M. Scoglio, N. A. Monteiro-Riviere and J. E. Riviere, Nanomedicine, 2015, 10, 25–33 CrossRef CAS PubMed.
- A. Hung, S. Mwenifumbo, M. Mager, J. J. Kuna, F. Stellacci, I. Yarovsky and M. M. Stevens, J. Am. Chem. Soc., 2011, 133, 1438–1450 CrossRef CAS PubMed.
- D. Dell'Orco, M. Lundqvist, S. Linse and T. Cedervall, Nanomedicine, 2014, 9, 851–858 CrossRef PubMed.
- H. M. Ding and Y. Q. Ma, Biomaterials, 2014, 35, 8703–8710 CrossRef CAS PubMed.
- Z. W. Ulissi, J. Zhang, V. Sresht, D. Blankschtein and M. S. Strano, Langmuir, 2015, 31, 628–636 CrossRef CAS PubMed.
- F. Darabi Sahneh, C. Scoglio and J. Riviere, PLoS One, 2013, 8, e64690 Search PubMed.
- M. Engelke, J. Koser, S. Hackmann, H. Zhang, L. Madler and J. Filser, Environ. Toxicol. Chem., 2014, 33, 1142–1147 CrossRef CAS PubMed.
- S. Lin, Y. Zhao and A. E. Nel, Small, 2013, 9, 1608–1618 CrossRef CAS PubMed.
- S. George, S. Lin, Z. Ji, C. R. Thomas, L. Li, M. Mecklenburg, H. Meng, X. Wang, H. Zhang, T. Xia, J. N. Hohman, J. I. Zink, P. S. Weiss and A. E. Nel, ACS Nano, 2012, 6, 3745–3759 CrossRef CAS PubMed.
- D. Cupi, N. B. Hartmann and A. Baun, Environ. Toxicol. Chem., 2015, 34, 497–506 CrossRef CAS PubMed.
- M. D. Fernandez, M. N. Alonso-Blazquez, C. Garcia-Gomez and M. Babin, Sci. Total Environ., 2014, 497–498, 688–696 CrossRef CAS PubMed.
- S. I. Gomes, A. M. Soares, J. J. Scott-Fordsmand and M. J. Amorim, J. Hazard. Mater., 2013, 254–255, 336–344 CrossRef CAS PubMed.
- A. Nel, T. Xia, H. Meng, X. Wang, S. Lin, Z. Ji and H. Zhang, Acc. Chem. Res., 2013, 3, 607–621 CrossRef PubMed.
- R. Damoiseaux, S. George, M. Li, S. Pokhrel, Z. Ji, B. France, T. Xia, E. Suarez, R. Rallo, L. Madler, Y. Cohen, E. M. Hoek and A. Nel, Nanoscale, 2012, 3, 1345–1360 RSC.
- P. Roach, D. Farrar and C. C. Perry, J. Am. Chem. Soc., 2006, 128, 3939–3945 CrossRef CAS PubMed.
- T. Patel, D. Telesca, R. Rallo, S. George, T. Xia and A. E. Nel, J. Agric. Biol. Environ. Stat., 2013, 18, 159–177 CrossRef PubMed.
- R. Liu, R. Rallo, R. Weissleder, C. Tassa, S. Shaw and Y. Cohen, Small, 2013, 9, 1842–1852 CrossRef CAS PubMed.
- Y. Cohen, R. Rallo, R. Liu and H. H. Liu, Acc. Chem. Res., 2013, 46, 802–812 CrossRef CAS PubMed.
- R. Liu, H. Y. Zhang, Z. X. Ji, R. Rallo, T. Xia, C. H. Chang, A. Nel and Y. Cohen, Nanoscale, 2013, 5, 5644–5653 RSC.
- J. Liu, S. Legros, F. von der Kammer and T. Hofmann, Environ. Sci. Technol., 2013, 47, 4113–4120 CrossRef CAS PubMed.
- M. A. Lancaster, M. Renner, C. A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Nature, 2013, 501, 373–379 CrossRef CAS PubMed.
- J. Kasper, M. I. Hermanns, C. Bantz, M. Maskos, R. Stauber, C. Pohl, R. E. Unger and J. C. Kirkpatrick, Part. Fibre Toxicol., 2011, 8, 6 CrossRef CAS PubMed.
- A. D. Lehmann, N. Daum, M. Bur, C. M. Lehr, P. Gehr and B. M. Rothen-Rutishauser, Eur. J. Pharm. Biopharm., 2011, 77, 398–406 CrossRef CAS PubMed.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef CAS PubMed.
- N. S. Bhise, J. Ribas, V. Manoharan, Y. S. Zhang, A. Polini, S. Massa, M. R. Dokmeci and A. Khademhosseini, J. Controlled Release, 2014, 190, 82–93 CrossRef CAS PubMed.
- T. Wagner, M. Dieckmann, S. Jaeger, S. Weggen and C. U. Pietrzik, Exp. Cell Res., 2013, 319, 1956–1972 CrossRef CAS PubMed.
- J. A. Kim, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 62–68 CrossRef CAS PubMed.
- Y. C. Toh, T. C. Lim, D. Tai, G. Xiao, D. van Noort and H. Yu, Lab Chip, 2009, 9, 2026–2035 RSC.
- V. N. Goral, Y. C. Hsieh, O. N. Petzold, J. S. Clark, P. K. Yuen and R. A. Faris, Lab Chip, 2010, 10, 3380–3386 RSC.
- T. A. Nguyen, T. I. Yin, D. Reyes and G. A. Urban, Anal. Chem., 2013, 85, 11068–11076 CrossRef CAS PubMed.
- G. Oberdorster, Environ. Health Perspect., 2012, 120, A13 CrossRef PubMed ; author reply.
- A. Nel, Y. Zhao and L. Madler, Acc. Chem. Res., 2013, 46, 605–606 CrossRef CAS PubMed.
- S. Lin, Y. Zhao, Z. Ji, J. Ear, C. H. Chang, H. Zhang, C. Low-Kam, K. Yamada, H. Meng, X. Wang, R. Liu, S. Pokhrel, L. Madler, R. Damoiseaux, T. Xia, H. A. Godwin, S. Lin and A. E. Nel, Small, 2013, 9, 1776–1785 CrossRef CAS PubMed.
- V. Sanchez-Lopez, J. M. Fernandez-Romero and A. Gomez-Hens, Anal. Chim. Acta, 2009, 645, 79–85 CrossRef CAS PubMed.
- T. Wang, J. Bai, X. Jiang and G. U. Nienhaus, ACS Nano, 2012, 6, 1251–1259 CrossRef CAS PubMed.
- J. Schaefer, C. Schulze, E. E. Marxer, U. F. Schaefer, W. Wohlleben, U. Bakowsky and C. M. Lehr, ACS Nano, 2012, 6, 4603–4614 CrossRef CAS PubMed.
- S. Linse, C. Cabaleiro-Lago, W. F. Xue, I. Lynch, S. Lindman, E. Thulin, S. E. Radford and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 8691–8696 CrossRef CAS PubMed.
- J. Brown, W. M. de Vos, P. S. DiStefano, J. Dore, C. Huttenhower, R. Knight, T. D. Lawley, J. Raes and P. Turnbaugh, Nat. Biotechnol., 2013, 31, 304–308 CrossRef CAS PubMed.
- J. K. Goodrich, J. L. Waters, A. C. Poole, J. L. Sutter, O. Koren, R. Blekhman, M. Beaumont, W. Van Treuren, R. Knight, J. T. Bell, T. D. Spector, A. G. Clark and R. E. Ley, Cell, 2014, 159, 789–799 CrossRef CAS PubMed.
- C. Human and Microbiome Project, Nature, 2012, 486, 215–221 CrossRef PubMed.
- D. L. Merrifield, B. J. Shaw, G. M. Harper, I. P. Saoud, S. J. Davies, R. D. Handy and T. B. Henry, Environ. Pollut., 2013, 174, 157–163 CrossRef CAS PubMed.
- K. Williams, J. Milner, M. D. Boudreau, K. Gokulan, C. E. Cerniglia and S. Khare, Nanotoxicology, 2014, 1–11 CAS.
- I. L. Bergin and F. A. Witzmann, Int. J. Biomed. Nanosci. Nanotechnol., 2013, 3 DOI:10.1504/IJBNN.2013.054515.
- C. F. Maurice, H. J. Haiser and P. J. Turnbaugh, Cell, 2013, 152, 39–50 CrossRef CAS PubMed.
- X. Sun, Z. Feng, T. Hou and Y. Li, ACS Appl. Mater. Interfaces, 2014, 6, 7153–7163 CAS.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- C. A. Falaschetti, T. Paunesku, J. Kurepa, D. Nanavati, S. S. Chou, M. De, M. Song, J. T. Jang, A. Wu, V. P. Dravid, J. Cheon, J. Smalle and G. E. Woloschak, ACS Nano, 2013, 7, 7759–7772 CrossRef CAS PubMed.
- R. H. Stauber, S. K. Knauer, N. Habtemichael, C. Bier, B. Unruhe, S. Weisheit, S. Spange, F. Nonnenmacher, V. Fetz, T. Ginter, S. Reichardt, C. Liebmann, G. Schneider and O. H. Kramer, Oncotarget, 2012, 3, 31–43 Search PubMed.
- O. Dassonville, A. Bozec, J. L. Fischel and G. Milano, Crit. Rev. Oncol. Hematol., 2007, 62, 53–61 CrossRef PubMed.
- L. Treuel, X. Jiang and G. U. Nienhaus, J. R. Soc., Interface, 2013, 10, 20120939 CrossRef PubMed.
- T. J. Maccormack, R. J. Clark, M. K. Dang, G. Ma, J. A. Kelly, J. G. Veinot and G. G. Goss, Nanotoxicology, 2012, 6, 514–525 CrossRef CAS PubMed.
Footnote |
| † Equal contributor note. |
| This journal is © The Royal Society of Chemistry 2015 |