Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ultrafast non-radiative decay of gas-phase nucleosides
Simone De
Camillis
a,
Jordan
Miles
a,
Grace
Alexander
a,
Omair
Ghafur
b,
Ian D.
Williams
a,
Dave
Townsend
bc and
Jason B.
Greenwood
*a
aCentre for Plasma Physics, School of Mathematics and Physics, Queen's University Belfast, Belfast BT7 1NN, UK. E-mail: j.greenwood@qub.ac.uk
bInstitute of Photonics and Quantum Sciences, Heriot-Watt University, Edinburgh EH14 4AS, UK
cInstitute of Chemical Sciences, Heriot-Watt University, Edinburgh EH14 4AS, UK
First published on 13th August 2015
Abstract
The ultrafast photo-physical properties of DNA are crucial in providing a stable basis for life. Although the DNA bases efficiently absorb ultraviolet (UV) radiation, this energy can be dissipated to the surrounding environment by the rapid conversion of electronic energy to vibrational energy within about a picosecond. The intrinsic nature of this internal conversion process has previously been demonstrated through gas phase experiments on the bases, supported by theoretical calculations. De-excitation rates appear to be accelerated when individual bases are hydrogen bonded to solvent molecules or their complementary Watson–Crick pair. In this paper, the first gas-phase measurements of electronic relaxation in DNA nucleosides following UV excitation are reported. Using a pump–probe ionization scheme, the lifetimes for internal conversion to the ground state following excitation at 267 nm are found to be reduced by around a factor of two for adenosine, cytidine and thymidine compared with the isolated bases. These results are discussed in terms of a recent proposition that a charge transfer state provides an additional internal conversion pathway mediated by proton transfer through a sugar to base hydrogen bond.
Introduction
DNA bases are strong ultraviolet (UV) chromophores which absorb at wavelengths in the range of 240–290 nm.1 These molecules are the basis for life despite the fact that the pre-biotic Earth was bathed in ultraviolet radiation, suggesting that they are able to effectively quench excess absorbed energy very rapidly without generating photolesions. Similarly these ultrafast energy quenching mechanisms are relevant, for example, to the extent to which ultraviolet exposure leads to skin cancer and the efficacy of sunscreens.2 As such, the energy relaxation and dissipation mechanisms operating in these systems have been the subject of hundreds of experimental and theoretical studies over the last 20 years.Measurements of ultrafast UV quenching in DNA were first made possible in solution through the development of the transient absorption and fluorescence up-conversion techniques,3,4 which have been used to observe ultrafast dynamics in DNA components from individual bases through to double helix strands. Theoretical techniques which can accurately describe the transient molecular dynamics observed have developed in parallel and have been essential in interpreting the experimental results.5 However, the complexity of the DNA structures means that further theoretical and experimental advances are required for a complete understanding of the ultrafast de-excitation dynamics. Complementary gas-phase experiments have been crucial in understanding the intrinsic properties of the isolated base molecules and validating the theoretical methodology. The first such measurements, undertaken by Kang et al.,6 studied the dynamics of low-lying ππ* states following initial excitation with 267 nm laser pulses and subsequent tracking of the evolution in multiphoton ionization probabilities by a delayed 800 nm pulse. These pump–probe results showed that the non-radiative de-excitation in isolated RNA and DNA nucleobases occurred within a few picoseconds or less. A subsequent study using a 400 nm probe and much better time resolution of around 80 fs7 revealed a bi-exponential decay mechanism. A fast component (<200 fs) was interpreted as wavepacket motion to the minimum of the ππ* potential energy surface and population transfer via a conical intersection to a state of nπ* character. Subsequent non-radiative transitions to the ground state were then assigned to a longer, picosecond process. A key reaction coordinate for this process corresponds to out-of-plane deformations of the aromatic ring system, which sends the nπ* and ππ* excited states towards conical intersections with the ground state.8–12
These formative experiments were followed up by more detailed studies which have mainly focussed on adenine since it is relatively easy to produce in the gas phase and has only one dominant tautomer. Of particular interest was the extent to which additional electronic states formed via Rydberg/valance mixing were contributing to the de-excitation dynamics. Such states exhibit predominantly 3s character in the Franck–Condon region, but evolve increasingly towards πσ* character at extended N–H distances.13 Initial studies of methylated and deuterated adenine showed no change in the lifetimes.7,14 However, time resolved photoelectron spectra (TRPES) obtained by Satzger et al.15 suggested that de-excitation from the ππ* state does proceed via a πσ* state (in competition with the nπ* internal conversion pathway). This contradicted theoretical calculations that predicted the πσ* state lies too high in energy to be accessed by a 267 nm pump.10,16 Ashfold et al.17 and, Evans and Ullrich18 agreed with this theoretical interpretation through measurements of H atom production and TRPES respectively, indicating πσ* states were only involved for wavelengths less than 238 nm. Similarly, for the other nucleobases a number of studies suggest that πσ* states can only be accessed at shorter wavelengths, but interpretation of the data is complicated by the presence of many more structural conformers.19–23
There have many more studies of other gas phase nucleobase derivatives (for example3,4,14,24–28) which demonstrate that, except for simple methylation, de-excitation rates following UV excitation are generally slower for chemically modified bases and even for some isomers. These observations can, in some cases, be explained by the constraint put on out-of-plane deformations, but more generally modified nucleobases have energy barriers which restrict access to conical intersections. Indeed the canonical bases exploited for genetic encoding appear to have been chosen by natural selection as they are photochemically more stable than other variants.29
For the nucleo-bases/sides/tides in solution, the lifetimes are found to be shorter than for the isolated bases in the gas-phase.30,31 The fact that Watson–Crick pairs in solution have even faster rates of decay32,33 suggests that H-bonding to the base opens up additional quenching mechanisms involving proton transfer. For guanine–cytosine base pairs, Sobolewski and Domcke34,35 have proposed that population of an inter-base charge transfer state moves an electron from guanine to cytosine followed in the same direction by a proton involved in one of the base-to-base H-bonds.
Gas phase studies of ultrafast dynamics in DNA components larger than the individual bases have yet to be undertaken. However, resonant two-photon ionization spectroscopic studies using nanosecond laser pulses have provided some indirect insight. For example, the observation of spectra which are broader for base pairs than for individual bases suggests that rapid decay via charge transfer states is also playing a role in the gas phase.36 For some nucleosides, it has also proved difficult to produce significant amounts of parent ions in the mass spectrum which suggests that ionization is being suppressed due to a faster excited state decay rate than for the bases.37,38 This observation recently motivated Tuna et al.39 to show that a charge transfer state with a conical intersection to the ground state exists in adenosine which can be reached as the distance between a proton on the ribose sugar and the N3 atom of the base reduces. This state was found to be present in both the syn and anti conformers and the conical intersections were significantly lower in energy than those intrinsic to the base. The only other theoretical work investigating electronic relaxation for a nucleoside was performed by Zgierski and Alavi40 who concluded for cytidine that the radiationless decay mechanisms were qualitatively similar to cytosine.
For DNA strands, the character of the excited state and hence its decay mechanisms depends strongly on the conformation of the helix. For tightly wound strands (A-type helix) where the adjacent bases are stacked on top of each other, the photo-excitation is de-localised between the two neighbouring bases which results in slower decay. If the bases are not well stacked (B-type helix), then the de-excitation is shorter and the decay mechanism is proposed to be similar to that of gas phase monomers.41 Therefore gas phase studies of DNA building blocks are important for understanding photo-protection mechanisms in long strands as well as providing benchmarks for the theoretical calculations which can be more easily compared with isolated molecules.
Given that studies of isolated nucleosides are a natural next step in bottom-up understanding of DNA photophysics, it is perhaps surprising that little theoretical work and no ultrafast pump–probe experiments have been undertaken to date. In the present study we have sought to remedy this situation by obtaining the first reported gas-phase measurements of ππ* excited state decay in all of the DNA nucleosides following UV excitation. The very low vapour pressures and strong propensity for thermal decomposition exhibited by these species at elevated temperatures has, up to now, made such an undertaking experimentally challenging. We have overcome these issues, however, through the use of a “soft” laser desorption method based on back-irradiation of thin foil targets coated with the molecular samples of interest.42,43 Similar approaches have previously been successfully applied to the study of dissociative electron attachment in thymidine.44
Experiment
To produce gas-phase targets, nucleosides were desorbed directly from solid samples deposited on a 10 μm stainless steel foil which was integrated into the repeller electrode of a time-of-flight mass spectrometer.42,43 The samples (99% purity) were obtained from Sigma-Aldrich and used without further purification. The foil was irradiated with a 455 nm CW laser on the opposite side to that on which the samples were deposited to increase the temperature to ∼150–250 °C. Unlike conventional molecular beams produced by ovens, evaporation using the present arrangement has the advantage that once desorbed the molecules do not undergo several cycles of condensation and re-evaporation, and they reach the interaction region quickly (in 10s of microseconds) due to the short travel distance from the foil (4–5 mm). This reduces the likelihood of thermal decomposition of the labile nucleoside species.Femtosecond laser pulses used to excite and then subsequently ionize the nucleosides were obtained from a Coherent Libra system which produced 800 nm, 1 mJ pulses at a repetition rate of 1 kHz. These were converted into the third harmonic (267 nm) using thin β-barium borate crystals as the non-linear medium. Residual fundamental pulses were delayed using a computer controlled translation stage and re-combined with the 267 nm pulses to facilitate 267 nm + 800 nm pump–probe measurements. Both beams were focussed into the interaction region where they crossed the molecular plume at a distance of 10–15 mm from the focal position, with pulse energies of 8 μJ and 30 μJ and peak intensities of 2–5 × 1011 W cm−2 and 1–2 × 1013 W cm−2, respectively. Ionization potentials (IPs) of the nucleosides under consideration here have not, to the best of our knowledge, been previously reported. However, IPs for all four species have been estimated from photoelectron studies performed on a series of methyl-substituted derivatives.45 On the basis of this work it seems reasonable to assume that a minimum of three 800 nm photons are required to facilitate ionization from all of the excited bases. The pulse length for the 800 nm probe was measured by autocorrelation to be 130 fs, while ionization of Xe gas yielded a cross correlation of 190 fs which was consistent with the 267 nm probe also having a pulse length of 130 fs. For the pump–probe measurements, time of flight spectra were acquired for about half a minute at each time delay which was incremented in 27 fs steps between −500 fs and +2500 fs. A Xe cross correlation was performed before each acquisition to obtain an accurate zero delay position.
Results and discussion
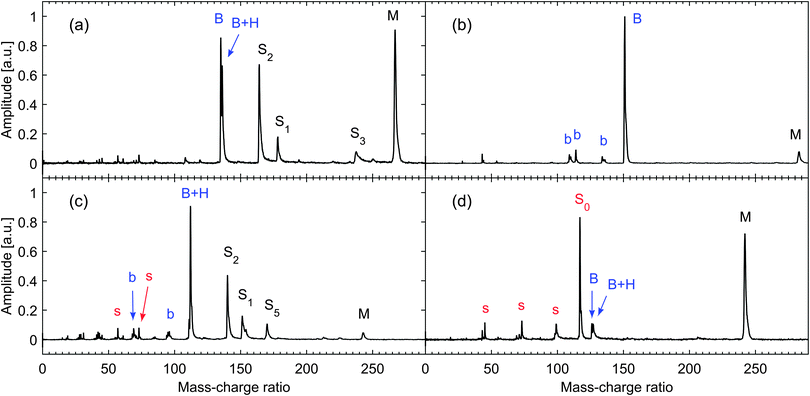
We have previously reported mass spectra for each of the nucleosides using one-colour (1+1) resonance-enhanced multiphoton ionization (REMPI) at 267 nm with low interaction intensity (see ESI in ref. 43). These show that parent molecular ions make up more than 90% of the ion yield for thymidine and adenosine. This can be regarded as a lower limit on the proportion of intact molecules in the sample plume desorbed from the foil surface since at the pump intensities employed (5 × 1011 W cm−2) there was a reasonable probability of the intact cation absorbing additional photons leading to dissociation. For guanosine and cytidine, while a strong parent peak was still observed in the spectrum, the largest peak corresponds to base ions. For cytidine this peak is nearly all due to protonated base ions which cannot originate from prior decomposition of the nucleoside into the nucleobase. For guanosine, about 40% of the ions in the mass spectrum were due to the parent, so we cannot rule out some decomposition prior to the laser interaction. However, this reduced parent yield may be a consequence of greater vibrational energy being deposited in the cation due to its lower ionization potential (8.0 eV45). Therefore we are confident that the molecular plumes generated in our experiments are dominated by the intact, isolated nucleosides.Mass spectra for the nucleosides obtained with the UV pump and IR probe pulses spatially and temporally overlapped are shown in Fig. 1 and have considerably more fragment ions than UV only REMPI spectra. We have attributed all significant peaks in the mass spectra to nucleoside fragments which agree with assignments found in other studies (for example NIST electron impact mass spectra46). It is interesting to note that while the resonant excitation selectively ionizes an electron from the base, the presence of the intact sugar group ions (or fragments thereof) indicates that charge has been transferred from base to sugar prior to dissociation. This is particularly noticeable for thymidine which is the only DNA nucleoside where the binding energy of the highest occupied molecular orbital in the base is greater than in the sugar,47 making the charge transfer energetically favourable. This subject has been discussed more fully by Poully et al.43
The variation in the thymidine parent ion yield and separately the sum of all the fragments is shown in Fig. 2 as a function of pump–probe delay time. It is evident that the excited state dynamics can be clearly observed through enhancement in the fragment ion yield for delays slightly greater than zero. As our analysis showed no difference in the dynamics between different fragments within the uncertainties, the yield from all fragments was summed to improve the statistics. Only a small proportion of this increase in fragment ions originates from loss in the parent ion, which is fairly insensitive to the pump–probe delay. Similar behaviour was found for the other nucleosides. The major contribution to the parent ions in the mass spectra arises from resonantly enhanced one-colour 1+1 ionisation by the pump pulse only. The lack of dynamics in the parent ion yield can be attributed to the high probability of dissociative ionisation out of the excited state by the moderately intense probe pulse so that signal enhancement is only observed for fragment ions. As it was not possible to guarantee a constant target density during the acquisition due to slow depletion of the sample on the foil, we have analysed the excited state dynamics by normalising the yield of all fragments to the parent ion yield. In this way we have accounted for the variations in target density without influencing the underlying dynamics. These results are presented for all the nucleosides in Fig. 3.
As noted by Kang et al.,6 following excitation by the pump, the probe pulses can resonantly populate higher lying states when the two pulses are temporally overlapped – introducing a zero delay enhancement to the signal. Therefore, they fitted their results for gas-phase bases with the sum of a Gaussian function centred at zero pump–probe delay and a single exponential convolved by the cross-correlation of the laser pulses. For our data we use a similar approach but include a second exponential function as our better time resolution allows us to observe the fast decay component previously identified by Canuel et al.7 To fully optimise the fitting process, we also introduced a third very long-lived component which arises due to parent ions generated directly by the pump being subsequently broken into fragment ions by absorption of probe photons.
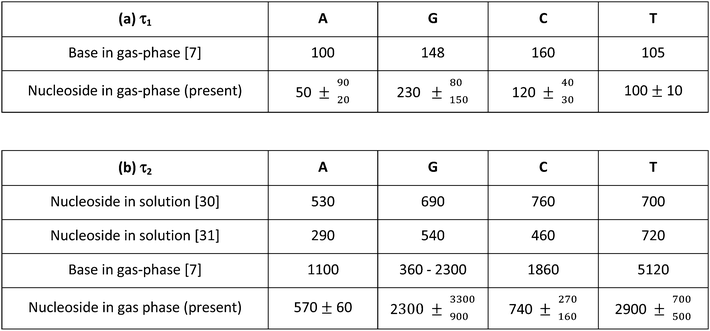
The two lifetimes obtained for each nucleoside are presented in Table 1 along with the results of previous measurements performed at similar pump wavelengths on both the isolated bases (in the gas phase) and the nucleosides (in solution). The short lifetime components (described by a time-constant τ1) are consistent with the values obtained for the isolated bases by Canuel et al.7 within the present uncertainties. We therefore attribute this extremely rapid decay (τ1 = 50–230 fs) to wavepacket motion away from the Franck–Condon region of the initially prepared ππ* potential energy surface following excitation, which may also include internal conversion to an nπ* state. The similarity of the base and nucleoside τ1 lifetimes suggests that there is little difference in their excited state potential energy surfaces near the Franck–Condon region.
In contrast, the second, slightly longer-lived component (described by a time-constant τ2) is around a factor of two shorter for adenosine, thymidine and cytidine than in the corresponding free base. This decay is a consequence of relaxation back to the ground state. In the case of guanosine, the situation appears somewhat different as we obtain a τ2 lifetime which is longer than for guanine. We note, however, that the lifetime previously reported for guanine (360 fs) is rather uncertain as later analysis including all fragment ions suggested that this was actually much longer (2300 fs).48 Theoretical calculations and simulated trajectories for guanine show that fast decay to the ground state (220 fs) is indeed present in the biologically relevant tautomer and proceeds directly from the initially populated ππ* state without the involvement of an nπ* state.12 The longer measured lifetime may be due to tautomers expected to be present in the experimental target. While the number of base tautomers present is restricted since the sugar is bound to the N9 atom, there are still at least three possible tautomers of the nucleoside.49 Without a means of pre-selecting different conformers and the fact that the present guanosine measurements have relatively poor statistics, it is not possible to determine if different tautomers are influencing the measured time constants (230, 2300 fs).
So and Alavi have previously reported excited state energies for adenosine, guanosine, cytidine and uridine (structurally very similar to thymidine) using density functional theory (B3LYP) in conjunction with the 311++G(d,p) and aug-cc-pVDZ basis sets.50 For comparative purposes, equivalent calculations were also reported for the isolated bases. In all cases, the addition of the sugar group appeared to have only a small influence on the energies of the lowest-lying singlet ππ* and nπ* states (the former of which is optically bright and is the principle state directly excited at 267 nm) – both in relative and absolute terms. Critically within the context of this current work, no clear systematic shifts in energy between these states were seen across the various nucleoside systems. This is perhaps unsurprising as the lowest-lying ππ* and nπ* states are predominantly localized on the aromatic pyrimidine/purine ring systems in all cases. So and Alavi50 also revealed states of πσ* character present in the low energy excited state manifold in the isolated bases are no longer seen in this energy region in the corresponding nucleosides (with the exception of guanosine). Such states have previously been implicated as playing a role in the dynamical relaxation of the bases following UV excitation. As discussed previously in the Introduction, the importance of this pathway following excitation in the 267 nm region is not fully resolved, although it is generally thought to be relatively minor.
For adenosine, thymidine and cytidine, the calculations outlined above clearly suggest that the significantly shorter overall excited state lifetimes observed (compared to the isolated bases) cannot be rationalized simply in terms of changes in the relative excited state energy orderings and/or spacings. This would therefore imply that there is an additional decay mechanism operating in the nucleosides which is not present in the bases. To date there have been limited theoretical investigations of the decay mechanisms for isolated nucleosides. Calculations of the decay pathway in cytidine suggest that transition to the ground state occurs via a biradical state which is qualitatively similar to the cytosine base.40 In contrast, Tuna et al.39 calculate the presence of a charge transfer state in two separate conformers of adenosine which intersect with the ground state as a proton is transferred from the sugar to base via an intramolecular H-bond (5′-OH⋯N3 in the syn conformer and 2′-OH⋯N3 in the anti conformer). As these conical intersections are about 0.6 eV lower than the conical intersections corresponding to ring puckering in the base, this could lead to faster de-excitation. It is interesting to note that we do not observe a shorter lifetime in guanosine despite the similarity of its structure to adenosine. However, our measured lifetimes are consistent with previous qualitative observations that guanosine and adenine parent ions can be produced in resonant two photon spectroscopy experiments but not adenosine.37,38
There is currently no theory supporting the existence of charge transfer states in cytidine and thymidine, but the influence of intra-molecular H-bonding between the base and sugar can be expected – as revealed in the DFT optimized ground state geometries previously reported by So and Alavi, where all nucleoside systems exhibit similar base-sugar interactions.50 It therefore seems reasonable to assume (as a starting point for future discussion) that this charge transfer state may potentially also be present in all cases and actively participate in the relaxation dynamics. Finally, it is interesting to note that the lifetimes for bases and nucleosides in solution are considerably shorter than for gas phase bases but in good agreement with our values for adenosine and cytidine. Similarly very short lifetimes can be found in H-bonded base pairs. These observations indicate that population of charge transfer states which stimulate proton migration to the base from the sugar, solvent or another base accelerates the de-excitation process.
Conclusion
The use of “soft” laser desorption methods have permitted the first reported dynamical study of excited state excess energy redistribution in gas-phase DNA nucleosides following UV irradiation. Time-resolved ion-yield measurements reveal that the lifetimes for internal conversion to the ground state following excitation are reduced by around a factor of two for adenosine, cytidine and thymidine when compared with the corresponding isolated bases. This is not the case for guanosine, which may be a consequence of several conformers/tautomers being present in the desorbed sample. Results are discussed in terms of a recent proposition that a charge transfer state in adenosine provides an additional internal conversion pathway mediated by proton transfer through a sugar to base hydrogen bond. Previously reported ground state geometry calculations for other DNA nucleosides appear to also suggest that this mechanism may operate more generally. However, more comprehensive theory looking at an expanded range of excited state properties in these systems would be highly desirable. This is in addition to more detailed gas-phase experimental studies using an expanded range of UV excitation wavelengths and/or exploiting more differential measurements (for example, time-resolved photoelectron imaging). We hope and anticipate that this communication will serve as a stimulus for such undertakings in the near future.Acknowledgements
This work was supported by the STFC Laser Loan Scheme, the Leverhulme Trust (RPG-2012-735), and the UK's Engineering and Physical Sciences Research Council (EP/M001644/1). GA is supported by EPSRC's Doctoral Training Grant scheme and JM by the Northern Ireland Department of Employment and Learning.References
- M. Barbatti, A. J. A. Aquino and H. Lischka, The UV absorption of nucleobases: semi-classical ab initio spectra simulations, Phys. Chem. Chem. Phys., 2010, 12, 4959–4967 RSC.
- E. M. M. Tan, M. Hilbers and W. J. Buma, Excited-State Dynamics of Isolated and Microsolvated Cinnamate-Based UV-B Sunscreens, J. Phys. Chem. Lett., 2014, 5, 2464–2468 CrossRef CAS PubMed.
- C. T. Middleton, K. de La Harpe, C. Su, Y. K. Law, C. E. Crespo-Hernández and B. Kohler, DNA Excited-State Dynamics: From Single Bases to the Double Helix, Annu. Rev. Phys. Chem., 2009, 60, 217–239 CrossRef CAS PubMed.
- K. Kleinermanns, D. Nachtigallová and M. S. de Vries, Excited state dynamics of DNA bases, Int. Rev. Phys. Chem., 2013, 32, 308–342 CrossRef CAS PubMed.
- L. González, D. Escudero and L. Serrano-Andrés, Progress and Challenges in the Calculation of Electronic Excited States, ChemPhysChem, 2012, 13(1), 28–51 CrossRef PubMed.
- H. Kang, K. T. Lee, B. Jung, Y. J. Ko and S. K. Kim, Intrinsic Lifetimes of the Excited State of DNA and RNA Bases, J. Am. Chem. Soc., 2002, 124, 12958–12959 CrossRef CAS PubMed.
- C. Canuel, M. Mons, F. Piuzzi, B. Tardivel, I. Dimicoli and M. Elhanine, Excited states dynamics of DNA and RNA bases: Characterization of a stepwise deactivation pathway in the gas phase, J. Chem. Phys., 2005, 122, 074316 CrossRef PubMed.
- S. Perun, A. L. Sobolewski and W. Domcke, Ab Initio Studies on the Radiationless Decay Mechanisms of the Lowest Excited Singlet States of 9H-Adenine, J. Am. Chem. Soc., 2005, 127, 6257–6265 CrossRef CAS PubMed.
- S. Perun, A. L. Sobolewski and W. Domcke, Conical intersections in Thymine, J. Phys. Chem. A, 2006, 110, 13238–13244 CrossRef CAS PubMed.
- S. Perun, A. L. Sobolewski and W. Domcke, Photostability of 9H-adenine: mechanisms of the radiationless deactivation of the lowest excited singlet states, Chem. Phys., 2005, 313, 107–112 CrossRef CAS PubMed.
- M. Z. Zgierski, S. Patchkovskii, T. Fujiwara and E. C. Lim, On the Origin of the Ultrafast Internal Conversion of Electronically Excited Pyrimidine Bases, J. Phys. Chem. A, 2005, 109, 9384–9387 CrossRef CAS PubMed.
- M. Barbatti, J. J. Szymczak, A. J. A. Aquino, D. Nachtigallová and H. Lischka, The decay mechanism of photoexcited guanine – A nonadiabatic dynamics study, J. Chem. Phys., 2011, 134, 014304 CrossRef PubMed.
- H. Reisler and A. I. Krylov, Interacting Rydberg and valence states in radicals and molecules: experimental and theoretical studies, Int. Rev. Phys. Chem., 2009, 28, 267–308 CrossRef CAS PubMed.
- H. Kang, B. Jung and S. K. Kim, Mechanism for ultrafast internal conversion of adenine, J. Chem. Phys., 2003, 118, 6717–6719 CrossRef CAS PubMed.
- H. Satzger, D. Townsend, M. Z. Zgierski, S. Patchkovskii, S. Ullrich and A. Stolow, Primary processes underlying the photostability of isolated DNA bases: Adenine, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10196–10201 CrossRef CAS PubMed.
- W. C. Chung, Z. G. Lan, Y. Ohtsuki, N. Shimakura, W. Domcke and Y. Fujimura, Conical intersections involving the dissociative 1πσ* state in 9H-adenine: a quantum chemical ab initio study, Phys. Chem. Chem. Phys., 2007, 9, 2075–2084 RSC.
- M. N. R. Ashfold, B. Cronin, A. L. Devine, R. N. Dixon and M. G. D. Nix, The Role of πσ* Excited States in the Photodissociation of Heteroaromatic Molecules, Science, 2006, 312, 1637–1640 CrossRef CAS PubMed.
- N. L. Evans and S. Ullrich, Wavelength Dependence of Electronic Relaxation in Isolated Adenine Using UV Femtosecond Time-Resolved Photoelectron Spectroscopy, J. Phys. Chem. A, 2010, 114, 11225–11230 CrossRef CAS PubMed.
- E. Nir, C. Plützer, K. Kleinermanns and M. de Vries, Properties of isolated DNA bases, base pairs and nucleosides examined by laser spectroscopy, Eur. Phys. J. D, 2002, 20, 317–329 CrossRef CAS PubMed.
- J. González-Vázquez, L. González, E. Samoylova and T. Schultz, Thymine relaxation after UV irradiation: the role of tautomerization and πσ* states, Phys. Chem. Chem. Phys., 2009, 11, 3927–3934 RSC.
- K. Kosma, C. Schröter, E. Samoylova, I. V. Hertel and T. Schultz, Excited-State Dynamics of Cytosine Tautomers, J. Am. Chem. Soc., 2009, 131, 16939–16943 CrossRef CAS PubMed.
- M. Schneider, R. Maksimenka, F. J. Buback, T. Kitsopoulos, L. R. Lago and I. Fischer, Photodissociation of thymine, Phys. Chem. Chem. Phys., 2006, 8, 3017–3021 RSC.
- M. Schneider, C. Schon, I. Fischer, L. Rubio-Lago and T. Kitsopoulos, Photodissociation of uracil, Phys. Chem. Chem. Phys., 2007, 9, 6021–6026 RSC.
- J.-W. Ho, H.-C. Yen, W.-K. Chou, C.-N. Weng, L.-H. Cheng, H.-Q. Shi, S.-H. Lai and P.-Y. Cheng, Disentangling Intrinsic Ultrafast Excited-State Dynamics of Cytosine Tautomers, J. Phys. Chem. A, 2011, 115, 8406–8418 CrossRef CAS PubMed.
- H. Kang, J. Chang, S. H. Lee, T. K. Ahn, N. J. Kim and S. K. Kim, Excited-state lifetime of adenine near the first electronic band origin, J. Chem. Phys., 2010, 133, 154311 CrossRef PubMed.
- M. Kotur, T. C. Weinacht, C. Zhou, K. A. Kistler and S. Matsika, Distinguishing between relaxation pathways by combining dissociative ionization pump probe spectroscopy and ab initio calculations: A case study of cytosine, J. Chem. Phys., 2011, 134, 184309 CrossRef PubMed.
- A. S. Chatterley, C. W. West, G. M. Roberts, V. G. Stavros and J. R. R. Verlet, Mapping the Ultrafast Dynamics of Adenine onto Its Nucleotide and Oligonucleotides by Time-Resolved Photoelectron Imaging, J. Phys. Chem. Lett., 2014, 5, 843–848 CrossRef CAS PubMed.
- C. Z. Bisgaard, H. Satzger, S. Ullrich and A. Stolow, Excited-State Dynamics of Isolated DNA Bases: A Case Study of Adenine, ChemPhysChem, 2009, 10, 101–110 CrossRef CAS PubMed.
- L. Serrano-Andrés and M. Merchán, Are the five natural DNA/RNA base monomers a good choice from natural selection? A photochemical perspective, J. Photochem. Photobiol., C, 2009, 10, 21–32 CrossRef PubMed.
- J. Peon and A. H. Zewail, DNA/RNA nucleotides and nucleosides: direct measurement of excited-state lifetimes by femtosecond fluorescence up-conversion, Chem. Phys. Lett., 2001, 348, 255–262 CrossRef CAS.
- J.-M. L. Peacourt, J. Peon and B. Kohler, Ultrafast Internal Conversion of Electronically Excited RNA and DNA Nucleosides in Water, J. Am. Chem. Soc., 2000, 122, 9348–9349 CrossRef.
- N. K. Schwalb and F. Temps, Ultrafast Electronic Relaxation in Guanosine is Promoted by Hydrogen Bonding with Cytidine, J. Am. Chem. Soc., 2007, 129, 9272–9273 CrossRef CAS PubMed.
- G. Groenhof, L. V. Schäfer, M. Boggio-Pasqua, M. Goette, H. Grubmüller and M. A. Robb, Ultrafast Deactivation of an Excited Cytosine-Guanine Base Pair in DNA, J. Am. Chem. Soc., 2007, 129, 6812–6819 CrossRef CAS PubMed.
- A. L. Sobolewski and W. Domcke, Ab initio studies on the photophysics of the guanine–cytosine base pair, Phys. Chem. Chem. Phys., 2004, 6, 2763–2771 RSC.
- A. L. Sobolewski, W. Domcke and C. Hättig, Tautomeric selectivity of the excited-state lifetime of guanine/cytosine base pairs: the role of electron-driven proton-transfer processes, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17903–17906 CrossRef CAS PubMed.
- A. Abo-Riziq, A. Grace, E. Nir, M. Kabelac, P. Hobza and M. S. de Vries, Photochemical selectivity in guanine–cytosine base-pair structures, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 20–23 CrossRef CAS PubMed.
- E. Nir and M. S. de Vries, Fragmentation of laser-desorbed 9-substituted adenines, Int. J. Mass Spectrom., 2002, 219, 133–138 CrossRef CAS.
- H. Asami, K. Yagi, M. Ohba, S. Urashima and H. Saigusa, Stacked Base-Pair Structures of Adenine Nucleosides Stabilized by the Formation of Hydrogen-Bonding Network Involving the Two Sugar Groups, Chem. Phys., 2013, 419, 84–89 CrossRef CAS PubMed.
- D. Tuna, A. L. Sobolewski and W. Domcke, Mechanisms of Ultrafast Excited-State Deactivation in Adenosine, J. Phys. Chem. A, 2014, 118(1), 122–127 CrossRef CAS PubMed.
- M. Z. Zgierski and S. Alavi, Quantum chemical study of biradical decay channels in cytidine nucleosides, Chem. Phys. Lett., 2006, 426, 398–404 CrossRef CAS PubMed.
- C. E. Crespo-Hernández and B. Kohler, Influence of secondary structure on electronic energy relaxation in adenine homopolymers, J. Phys. Chem. B, 2004, 108, 11182–11188 CrossRef.
- C. R. Calvert, L. Belshaw, M. J. Duffy, O. Kelly, R. B. King, A. G. Smyth, T. J. Kelly, J. T. Costello, D. J. Timson, W. A. Bryan, T. Kierspel, P. Rice, I. C. E. Turcu, C. M. Cacho, E. Springate, I. D. Williams and J. B. Greenwood, LIAD-fs scheme for studies of ultrafast laser interactions with gas phase biomolecules, Phys. Chem. Chem. Phys., 2012, 14, 6289–6297 RSC.
- J.-C. Poully, J. Miles, S. De Camillis, A. Cassimi and J. B. Greenwood, Proton irradiation of DNA nucleosides in the gas phase, Phys. Chem. Chem. Phys., 2015, 17, 7172–7180 RSC.
- I. Bald, I. Dabkowska and E. Illenberger, Probing Biomolecules by Laser-Induced Acoustic Desorption: Electrons at Near Zero Electron Volts Trigger Sugar–Phosphate Cleavage, Angew. Chem., Int. Ed., 2008, 47, 8518–8520 CrossRef CAS PubMed.
- C. Yu, T. J. O'Donnell and P. R. LeBreton, Ultraviolet Photoelectron Studies of Volatile Nucleoside Models. Vertical Ionization Potential Measurements of Methylated Uridine, Thymidine, Cytidine, and Adenosine, J. Phys. Chem., 1981, 85, 3851–3855 CrossRef CAS.
- P. J. Lindstrom and W. G. Mallard, NIST Standard Reference Database, National Institute of Standards and Technology, Gaithersburg MD 20899, 2005 Search PubMed.
- D. Ghosh, A. Golan, L. K. Takahashi, A. I. Krylov and M. Ahmed, A VUV Photoionization and Ab Initio Determination of the Ionization Energy of a Gas-Phase Sugar (Deoxyribose), J. Phys. Chem. Lett., 2012, 3, 97–101 CrossRef CAS.
- M. Mons, I. Dimicoli and F. Piuzzi, Radiation induced molecular phenomena in nucleic acids: a comprehensive theoretical and experimental analysis, Isolated Guanine: Tautomerism, Spectroscopy and Excited State Dynamics, Springer, Dordrecht, London, 2008, pp. 343–367 Search PubMed.
- K. B. Bravaya, O. Kostko, S. Dlogikh, A. Landau, M. Ahmed and A. I. Krylov, Electronic Structure and Spectroscopy of Nucleic Acid Bases: Ionization Energies, Ionization-Induced Structural Changes, and Photoelectron Spectra, J. Phys. Chem. A, 2010, 114, 12305–12317 CrossRef CAS PubMed.
- R. So and S. Alavi, Vertical Excitation Energies for Ribose and Deoxyribose Nucleosides, J. Comput. Chem., 2007, 28, 1776–1782 CrossRef CAS PubMed.
| This journal is © the Owner Societies 2015 |