Open Access Article
Open Access ArticleProbing the ligand recognition and discrimination environment of the globin-coupled oxygen sensor protein YddV by FTIR and time-resolved step-scan FTIR spectroscopy†
Andrea
Pavlou
a,
Markéta
Martínková
b,
Toru
Shimizu
b,
Kenichi
Kitanishi
b,
Martin
Stranava
b,
Andreas
Loullis
a and
Eftychia
Pinakoulaki
*a
aDepartment of Chemistry, University of Cyprus, PO Box 2037, 1678 Nicosia, Cyprus. E-mail: effiep@ucy.ac.cy
bDepartment of Biochemistry, Faculty of Science, Charles University in Prague, 128 43 Prague 2, Czech Republic
First published on 27th May 2015
Abstract
YddV is a newly discovered signal transducer heme protein that recognizes O2 and CO. Structural differences in the ligand-bound heme complex in YddV reflect variations in catalytic regulation by O2 and CO. Time-resolved step-scan (TRS2) FTIR studies of the wild type and of the important in oxygen recognition and stability of the heme Fe(II)–O2 complex L65M, L65T, Y43A, Y43F and Y43W mutants were performed to determine the site-specific protein dynamics following carbon monoxide (CO) photodissociation. These mutations were designed to perturb the electrostatic field near the iron-bound gaseous ligand (CO) and also to allow us to investigate the communication pathway between the distal residues of the protein and heme. TRS2-FTIR spectra of YddV–heme–CO show that the heme propionates are in protonated and deprotonated states. Moreover, the rate of decay of the vibrations of amide I is on a time scale that coincides with the rate of rebinding of CO, which suggests that there is coupling between ligation dynamics in the distal heme environment and (i) relaxation of the protein backbone and (ii) the environment sensed by the heme propionates. The fast recombination rates in L65M, L65T and Y43W imply a significant role of L65 and Y43 in controlling the ligand dynamics. The implications of these results with respect to the role of the heme propionates and the charged or proton-donating residues in the distal pocket, which are crucial for stabilizing bound gaseous ligands, are discussed.
Introduction
In the Escherichia coli genome, yddV and dos genes are organized as a bicistronic operon to regulate the synthesis and degradation of 3′,5′-cyclic diguanylic acid (c-di-GMP), respectively.1 Modulation of the overall conversion from GTP to pGpG via c-di-GMP is catalyzed by YddV (also termed EcDosC) and EcDOS (Escherichia coli direct oxygen sensor, also termed EcDosP). The product of the dos gene is the heme-based oxygen sensor protein EcDOS and the product of yddV from Escherichia coli (Ec) is a globin-coupled heme-based oxygen sensor protein, which displays diguanylate cyclase activity in response to the availability of oxygen.1–3 The structures and signal transduction mechanisms of the globin-coupled oxygen sensor protein are unique and differ from those of FixL and EcDOS, which contain the heme-bound PAS fold domain.2–7 The sensor domain of the globin-coupled oxygen sensor family has a globin fold at its N-terminus and contains a heme that acts as an O2-binding site, but with limited amino acid homology to myoglobin (Mb), hemoglobin (Hb), and the diguanylate cyclase (DGC) domain at its C-terminus. The globin fold lacks the D and half of the E helices of Mb and Hb. Seven heme-based oxygen sensor proteins with the globin fold, YddV, AfGcHK, HemAT-Bs, HemDGC, BpeGReg, AvGReg, and GsGCS, are known to date.2,3,7–13 Because of the importance of c-di-GMP as a messenger in bacteria and the increasing interest in understanding the connectivity between O2 and the metabolism of c-di-GMP,14 it is essential to elucidate the properties of newly discovered globin-coupled oxygen sensor proteins. In addition, little is known about the dynamics of signal transduction by globin-coupled oxygen sensors and the role of the distal environment in regulating the binding of O2 and CO. Determination of the structural dynamics of the protein moiety, which is associated with ligand binding/photodissociation from bound ligand(s) to heme Fe(II), is of particular importance for our interest in understanding the initial steps in the signalling mechanism.Of the known globin-coupled oxygen sensors, crystal structures have been reported for the sensor domain of HemAT from Bacillus subtilis (HemAT-Bs) and GsGCS.8,12 The sensor domain of HemAT-Bs is a homodimer with significant differences in the conformation of the distal residues (A91, T95, V89, Y70) that surround heme in each subunit.8 HemAT detects O2 by its sensor domain and transmits the signals via conformational changes to its functional domain, which exhibits activity associated with methyl-accepting chemotaxis. Resonance Raman (RR) studies revealed the formation of open and closed forms of six-coordinate O2-bound HemAT-Bs, which suggests the involvement of the distal residue T95 in regulating binding of O2via the observed formation of a H-bond with O2 coordinated to heme;15 the other distal residues are Y70, L92, R91 and V89. In addition, the formation of H-bonds between the ligand H86, which is proximal to heme iron, and the heme 6-propionate and between T95 and heme-bound O2 results in conformational changes in the E and B helices.16 On the other hand, based on the observation that mutation of Y70 made the dissociation constant of heme-bound O2 larger than that with the wild type, it was proposed that Y70 forms a H-bond with O2 bound to heme,17 in contrast to the conclusions based on RR experiments.15 In addition, UV Raman experiments indicated a change to a more hydrophilic environment around Y70 in ligand-bound forms, without the formation of a H-bond between Y70 and O2 or CO.18 Interestingly, it has also been proposed that the O2-bound form has a structural linkage with the signaling domain and that negative cooperative binding or heterogeneity play a key role in the signal transduction pathway.17 The different conclusions on the role of Y70 in the binding and recognition of the O2-signaling ligand, which were based on different experimental approaches, leave questions open. Recently, it was demonstrated that although the heme cavity recognizes ligands such as O2, NO and CO, the conformational changes that are induced in the protein distal site are not similar and only O2 alters the distal site in such a way that the conformational changes are transmitted to the effector domain.18
Based on catalytic activities of YddV in terms of the initial rate of product formation (c-di-GMP), similar turnover numbers (0.022 min−1) were measured for O2 and CO, which indicates that YddV recognizes both O2 and CO.3 In the absence of X-ray crystallographic data based on a comparative amino acid sequence study with well-known heme-based sensors, it has been proposed that H98 is proximal to the heme ligand. The isolated sensor domain YddV (YddV–heme) only forms a five-coordinate high-spin Fe(III) complex, which contains proximal H98 as the fifth ligand.3 At the distal site, it has been demonstrated that the conserved residues L65 and Y43 are important for the binding and stabilization of the heme Fe(II)–O2 complex.3,19,20 Extensive pH studies on L65 mutant proteins have revealed the presence of a H2O molecule as the sixth axial ligand, which is converted to OH− at alkaline pH, and the formation of a six-coordinate low-spin species.19 RR Soret excitation experiments on wild-type and Y43F mutant proteins have been reported.3 The RR frequencies of the wild-type protein oxygen complex with ν(Fe–O2) = 565 cm−1 and the CO complex with ν(Fe–CO) = 495 cm−1 are distinctly different from those reported for ν(Fe–O2) = 559 cm−1 and ν(Fe–CO) = 505 cm−1 for the Y43F mutant protein, which suggests that Y43 forms a H-bond with O2 and CO ligands bound to heme iron. Interestingly, the Y43A and Y43L mutant proteins exhibited very low affinities for O2 and therefore ν(Fe–O2) was not observed. In addition, ν(CO) for the wild-type and Y43F mutant proteins was reported at 1965 and 1959 cm−1, respectively, which indicates the formation of a single conformation.3
Fourier transform infrared (FTIR) spectroscopy has been extensively applied to monitor the environment distal to the heme of the heme-bound CO ligand and time-resolved step-scan FTIR (TRS2-FTIR) spectroscopy has proved to be a very powerful technique in studying transient changes at the level of individual amino acids during protein action and conformational changes to the protein backbone.21–27 YddV is a globin-coupled heme-based oxygen sensor protein, in which critical roles in the recognition of O2 and CO have been reported for Y43 and L65 residues, and therefore in this work we applied FTIR and TRS2-FTIR to probe the dynamics of wild-type YddV–heme as well as of different Y43 and L65 mutants of the protein. In the FTIR spectrum of CO-bound YddV–heme we detected two CO modes at 1962 and 1923 cm−1, which we attributed to neutral and strongly H-bonded conformers, respectively. In spectra of the L65M mutant a single vibration of C–O was observed at 1953 cm−1, which indicates a significant alteration in the distal structure upon the L65M mutation. On the other hand, although the CO adduct of the L65T mutant produced a low photoproduct yield (29%), the CO vibration was observed at the same frequency as in the wild type at 1962 cm−1, which suggests that the L65T mutation causes no significant alterations in the interactions of bound CO with the distal environment. Both Y43A and Y43W mutants produced photoproduct yields similar to that observed for the wild-type protein (>90%), but the C–O mode was observed at 1962 and 1953 cm−1, respectively. The Y43F mutant produced a low photoproduct yield and its C–O mode was at 1955 cm−1. TRS2-FTIR difference spectra of wild-type YddV–heme–CO subsequent to photodissociation of CO show that the heme propionates are in protonated and deprotonated forms and the observed constant for recombination of CO to heme iron is kWT = 528 s−1 at pD 8. Perturbation of amide I is observed upon photolysis of CO. The rate of decay of the vibrations of amide I and the heme propionates is on a time scale that coincides with the rate of rebinding of CO, which suggests that there is coupling between ligation dynamics in the distal heme environment and (i) protein relaxation and (ii) the environment sensed by the heme propionates. Based on the observed recombination constants for L65 and Y43 mutants, we conclude that rebinding is faster in L65 mutants than in Y43 mutants.
Materials and methods
Materials
All chemicals used were of the highest purity grade that was available from commercial sources and were used without further purification. Potassium phosphate was obtained from Sigma-Aldrich (St. Louis, MO, USA). Water, doubly distilled over quartz, was purified using a Milli-Q Plus system (EMD Millipore, Billerica, MA, USA). All glassware used for sample preparation was conditioned in advance by standing it for 24 h in 10% HCl Suprapur (Merck, Darmstadt, Germany).Overexpression and protein purification
Cloning, site-directed mutagenesis, overexpression in E. coli and purification of YddV–heme were performed as previously described.3,19,20 WT and Y43F, Y43L, Y43W, L65M, and L65T mutants of YddV–heme were prepared as described below. In brief, E. coli BL21 (DE3) (Stratagene, Novagen) was transformed with an appropriate plasmid (pET28a(+)/YddV–heme–Histag), plated on LB agar containing kanamycin (50 mg mL−1) and incubated at 37 °C overnight. On the following day, a single colony was inoculated in TB medium containing kanamycin and shaken overnight at 250 rpm and 37 °C. The culture medium was then added to fresh TB medium (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution) containing kanamycin and the mixture was again shaken at 250 rpm and 37 °C. Once the culture reached an OD600 of 1.2, the medium was cooled to 20 °C and protein expression was induced by the addition of 0.1 mM isopropyl β-D-thiogalactopyranoside and 0.45 mM δ-aminolevulinic acid, followed by further shaking for 24 h. E. coli cells were harvested by centrifugation for 30 min at 5000g and 4 °C, frozen in liquid nitrogen, and stored at −80 °C until protein extraction and purification. Frozen pellet cells were suspended in buffer A [50 mM Tris-HCl pH 8.0, 150 mM NaCl, 20 mM imidazole] containing 1 mM phenylmethanesulfonyl fluoride (PMSF) and lysed with 0.2 mg mL−1 lysozyme. The crude extract was sonicated and then centrifuged at 25
200 dilution) containing kanamycin and the mixture was again shaken at 250 rpm and 37 °C. Once the culture reached an OD600 of 1.2, the medium was cooled to 20 °C and protein expression was induced by the addition of 0.1 mM isopropyl β-D-thiogalactopyranoside and 0.45 mM δ-aminolevulinic acid, followed by further shaking for 24 h. E. coli cells were harvested by centrifugation for 30 min at 5000g and 4 °C, frozen in liquid nitrogen, and stored at −80 °C until protein extraction and purification. Frozen pellet cells were suspended in buffer A [50 mM Tris-HCl pH 8.0, 150 mM NaCl, 20 mM imidazole] containing 1 mM phenylmethanesulfonyl fluoride (PMSF) and lysed with 0.2 mg mL−1 lysozyme. The crude extract was sonicated and then centrifuged at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 1 h. The resulting supernatant was applied to a HisTrap HP column (GE Healthcare, Amersham, U.K.) that had been pre-equilibrated with buffer A containing PMSF. Recombinant proteins were retained due to their (His)6 tags and then eluted using a linear gradient from 20 to 300 mM imidazole in buffer A. Protein fractions were pooled and dialyzed against 50 mM pH 8.0 Tris-HCl buffer overnight. Finally, the purified proteins were quickly frozen in liquid nitrogen and stored at −80 °C.
000g for 1 h. The resulting supernatant was applied to a HisTrap HP column (GE Healthcare, Amersham, U.K.) that had been pre-equilibrated with buffer A containing PMSF. Recombinant proteins were retained due to their (His)6 tags and then eluted using a linear gradient from 20 to 300 mM imidazole in buffer A. Protein fractions were pooled and dialyzed against 50 mM pH 8.0 Tris-HCl buffer overnight. Finally, the purified proteins were quickly frozen in liquid nitrogen and stored at −80 °C.
UV-vis spectroscopy
Optical absorption spectra were recorded with a Shimadzu UV 1700 UV-visible spectrometer. A 2 μL sample of wild-type YddV–heme was diluted in 300 μL 50 mM Tris-HCl buffer pH 8 to achieve a final concentration of about 8 μM. Reduction of YddV was carried out under anaerobic conditions by adding a few grains of sodium dithionite to Tris buffer and then adding ∼3 μL of the dithionite solution to the YddV–heme sample. For the preparation of the carbon monoxide adduct, the dithionite-reduced sample was exposed to 1 atm CO (1 mM) in an anaerobic sealed quartz cuvette of path length 1 cm. Optical absorption spectra were also recorded before and after FTIR measurements to ensure the formation and stability of CO adducts.Sample preparation for FTIR measurements
The YddV–heme samples used for FTIR measurements had a concentration of ∼0.7–1.0 mM in 50 mM Tris for pH 8 and pD 8 after buffer exchange. The pD solutions prepared in D2O buffers were measured using a pH meter and assuming pD = pH (observed) + 0.4. Dithionite-reduced samples were exposed to 1 atm CO in an anaerobic cell to prepare the carbon monoxide adduct and transferred to a tightly sealed FTIR cell with two CaF2 windows under anaerobic conditions (path length (l) = 6 μm). CO gas was obtained from Linde.FTIR and TRS2-FTIR measurements
Static FTIR spectra were recorded with 4 cm−1 spectral resolution on a Bruker Vertex 80V spectrometer. For time-resolved step-scan FTIR measurements, 532 nm pulses from a Continuum Minilite II Nd:YAG laser (5 ns width, 10 Hz) were used as a pump light (4 mJ per pulse) to photolyze YddV–heme–CO adducts. A vacuum pump was used to evacuate the interferometer compartment to a final pressure of 3.2 mbar. The FTIR spectrometer was placed on a Newport VH optical vibration isolation table to ensure that vibrational background noise from environmental sources was avoided. For time-resolved experiments, a TTL (transistor–transistor logic) pulse provided by a digital delay pulse generator (Quantum Composers 9524) triggered in order flash lamps, a Q-switch, and the FTIR spectrometer. Pre-triggering the FTIR spectrometer to begin data collection before the laser fires allows fixed reference points to be collected at each mirror position. Changes in intensity were recorded with a photovoltaic MCT detector (Kolmar Technologies KV100-1B-7/190, response limit 850 cm−1) and digitized with a 180 kHz 24-bit analog-to-digital converter (ADC). A broadband interference optical filter (LP-4200, Spectrogon) with a short wavelength cut-off at 4.2 μm was used to limit the free spectral range from 4.2 to 11.8 μm. This led to a spectral range of 2633 cm−1, which was equal to an undersampling ratio of 6. Single-sided spectra were collected at 4 cm−1 spectral resolution, 6 μs time resolution, and 10 coadditions per data point. The total accumulation time for each measurement was 25 min and 15 measurements were collected and averaged. Blackman–Harris three-term apodization with 32 cm−1 phase resolution and the Mertz phase correction algorithm were used. The photoproduct yield at 6 μs was calculated by dividing the ΔA of the CO mode in the TRS2-FTIR spectrum at 6 μs by that in the FTIR spectrum recorded prior to the photodissociation experiment.Results and discussion
Fig. 1 shows a schematic representation of the heme site of YddV bound to CO. The optical absorption spectrum of Fe(III) as isolated YddV–heme, which is shown in Fig. 2 (trace A), displays a Soret maximum at 394 nm and a visible (Q-band) band at 506 nm, which is typical of five-coordinate, high-spin Fe(III) heme protein. The latter confirms the hitherto reported3 distinctive structural architecture of Fe(III) YddV as a 5-coordinate high-spin globin-coupled sensor compared to other GCS, which appear as 6-coordinate with low or high spin in the Fe(III) form, and provides evidence that there is no axial water or OH− coordinated to heme–Fe(III). In addition, a shoulder at 642 nm is observed, which is typical of a porphyrin-to-ferric iron charge transfer (CT) transition characteristic of ferric high-spin heme b (Fig. 2, trace A). The dithionite-reduced YddV–heme protein, the spectrum of which is shown in Fig. 2 trace B, displays a Soret maximum at 432 nm and a visible band at 560 nm, which indicates the formation of a 5-coordinate high-spin complex. Upon exposure of the reduced protein to 1 atm CO gas, the spectrum shown in Fig. 2 trace C with a Soret maximum at 421 nm and visible bands at 540 and 566 nm was obtained. The spectrum of the Fe(II)–CO complex indicates the formation of a six-coordinate low-spin species, as shown previously.3 | ||
| Fig. 1 Schematic representation of the proposed structure of the CO-bound form of YddV (residues are numbered for YddV), based on the crystal structure of the Fe(III)–CN adduct of HemAT-Bs (PDB 1OR4)8 and homology modeling. | ||
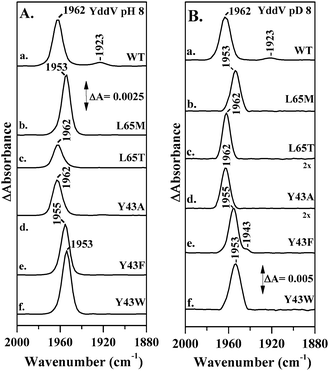
We detected two modes at 1962 and 1923 cm−1 in the FTIR spectrum of 12C16O-bound YddV–heme (Fig. 3A trace a); both are sensitive to isotopic substitution in CO and downshift to 1918 and 1878 cm−1, respectively, when the experiment is performed using isotopically labelled 13C16O (Fig. S1, ESI†). We assigned the νCO of YddV–heme based on the νCO values of Mb that were observed at 1910–1930 cm−1 (strong H-bonding interaction), 1940–1950 cm−1 (moderate H-bonding interaction) and 1960–1970 cm−1 (neutral).21 Therefore, we assigned the major band in the FTIR spectrum of CO-bound YddV–heme at 1962 cm−1 (neutral) and the minor band (strong H-bonding interaction) at 1923 cm−1 to C–O modes of the heme Fe(II)–CO complex. Detection of the major band has been reported in RR spectra (νCO = 1965 cm−1)3 but the strong H-bonding conformer is reported for the first time. Similar open (neutral) and closed (H-bonded) conformers have also been reported for CO-bound HemAT.28,29 In the spectrum of the L65M (Fig. 3A, trace b) mutant C–O modes are observed at 1953 cm−1, which is 9 cm−1 lower than that observed in the wild type, indicating significant alteration in the distal structure upon the L65M mutation. On the other hand, in the L65T mutant (Fig. 3A, trace c) the C–O mode was observed at the same frequency as in the wild type at 1962 cm−1, which suggests that the L65T mutation causes no significant alterations in the interactions of bound CO with the distal environment. In the Y43F (Fig. 3A, trace e) and Y43W mutants (Fig. 3A, trace f) the C–O mode is observed at 1955 and 1953 cm−1, respectively, but in the Y43A mutant (Fig. 3A, trace d) the C–O mode is observed at the same frequency as in the wild type. Notably, in none of the mutant proteins that were examined was the strong H-bonding conformer observed in the wild type at 1923 cm−1 detected.
We investigated the CO-bound YddV–heme complex at pD 8, aiming to explore the sensitivity to pH/pD of the distal site in the YddV–heme–CO complex (Fig. 3B). The observed sensitivity to pD includes decreased intensity of the strong H-bonding conformer at 1923 cm−1 (Fig. 3B, trace a) and broadening of the CO peak in the Y43W mutant (Fig. 3B, trace f), which indicates the involvement of multiple protein–ligand conformations that differ in the polarity of electrostatic interactions. In addition, a moderate hydrogen-bonded conformer was observed at 1943 cm−1 in the Y43F mutant (Fig. 3B, trace e). The rest of the data in D2O for all proteins that were examined are similar to those observed with pH. The absence of the strong H-bonding conformer in the L65 and Y43 mutant proteins demonstrates that both residues significantly inhibit its formation. The insensitivity of the CO mode of the major conformer in the L65T and Y43A mutant proteins indicates, in contrast to those in L65M, Y43W and Y43F, that these specific mutations are not directly involved in controlling the strength of the C–O bond.
Fig. 4A and C show TRS2-FTIR difference spectra in different spectral regions (td = 6 μs–7.4 ms, 4 cm−1 spectral resolution) of YddV–heme at pD 8 subsequent to photolysis of CO. The negative band at 1962 cm−1 in Fig. 4A originates from the photolyzed heme–CO complex (92% photoproduct yield at 6 μs), whereas the 1923 cm−1 conformer is not observed in the TRS2-FTIR spectra, which is presumably due to its much lower intensity. The continuous variability in the intensity of the CO mode associated with heme iron over a 6 μs–7.4 ms time scale is used to quantify rebinding of the ligand to heme. The final spectrum at 7.4 ms demonstrates that there is no irreversible light-induced effect on heme iron. The intensity of the Fe(II)–CO band at 1962 cm−1 was measured as a function of time to determine the rate of recombination of CO to heme iron (k = 528 s−1) at room temperature (Fig. 4B). The curve is a three-parameter fit to the experimental data according to pseudo-first-order kinetics. In FTIR difference spectra that were obtained upon photolysis of CO from heme iron, the appearance of signals in the amide I region (1620–1680 cm−1) can be attributed to changes in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes caused by perturbations in the polypeptide backbone with possible contributions from Gln/Asn side chains.30 Asymmetric COO− modes from deprotonated heme propionates and Glu and Asp side chains are expected in the 1530–1590 cm−1 region.27,30–32 The presence of the positive peak at 1670 cm−1 in time-resolved step-scan FTIR spectra of the YddV–heme–CO adduct subsequent to photodissociation of CO (Fig. 4C) can be tentatively assigned to the protonated form of the heme propionates, which indicates that the propionates are perturbed upon photodissociation of CO.31 We attributed the peaks/troughs at 1646/1656 cm−1 to the absorbance of amide I and the 1541/1562 cm−1 pair to the ν(COO−)asym of propionate(s).27,30–32 The intensities of the amide I mode at 1656 cm−1 (Fig. 4D) and of the heme propionate(s) mode at 1562 cm−1 (inset, Fig. 4D) were measured as a function of time and the corresponding rate constants (k1656 = 480 s−1 and k1562 = 474 s−1) were calculated as described previously for the CO mode. Therefore, the amide I and heme propionate(s) modes follow similar kinetics with rebinding of CO. Changes in intensity and/or shifts in frequency at 1636 cm−1 that could be attributed to Gln or Asn are present in the TRS2-FTIR spectra.30
O modes caused by perturbations in the polypeptide backbone with possible contributions from Gln/Asn side chains.30 Asymmetric COO− modes from deprotonated heme propionates and Glu and Asp side chains are expected in the 1530–1590 cm−1 region.27,30–32 The presence of the positive peak at 1670 cm−1 in time-resolved step-scan FTIR spectra of the YddV–heme–CO adduct subsequent to photodissociation of CO (Fig. 4C) can be tentatively assigned to the protonated form of the heme propionates, which indicates that the propionates are perturbed upon photodissociation of CO.31 We attributed the peaks/troughs at 1646/1656 cm−1 to the absorbance of amide I and the 1541/1562 cm−1 pair to the ν(COO−)asym of propionate(s).27,30–32 The intensities of the amide I mode at 1656 cm−1 (Fig. 4D) and of the heme propionate(s) mode at 1562 cm−1 (inset, Fig. 4D) were measured as a function of time and the corresponding rate constants (k1656 = 480 s−1 and k1562 = 474 s−1) were calculated as described previously for the CO mode. Therefore, the amide I and heme propionate(s) modes follow similar kinetics with rebinding of CO. Changes in intensity and/or shifts in frequency at 1636 cm−1 that could be attributed to Gln or Asn are present in the TRS2-FTIR spectra.30
In an effort to elucidate the environment that is sensed by bound CO, we investigated TRS2-FTIR spectra of the CO adducts of L65M (Fig. S2A, ESI†) and L65T (Fig. S2B, ESI†) YddV mutants and Y43A (Fig. S3A, ESI†), Y43F (Fig. S3B, ESI†) and Y43W (Fig. S3C, ESI†) mutant proteins subsequent to photolysis of CO. The intensity of Fe(II)–CO bands was measured as a function of time to determine the rate of recombination of CO to heme iron in each mutant (Fig. 5 and 6). The rates of recombination of CO to heme iron in wild-type YddV–heme and mutant proteins, as well as the yield of photoproduct formation at 6 μs, are summarized in Table 1. Close inspection of the data shows the large effect of the L65M and L65T mutants of YddV–heme protein on the recombination of CO. Both the rate constant of ligand association and the rate constant of ligand rebinding reflect the environment of access of the ligand to the binding site. In the case of the L65M mutant the yield of photoproduct is similar to that with the wild-type protein, but recombination of CO is faster (k = 1764 s−1). The L65T protein exhibits a low photoproduct yield at 6 μs (29%) and also fast recombination of CO (k = 2929 s−1). The fast recombination rates in both mutants imply a significant role of L65 in controlling the ligand dynamics. Data for the Y43 mutants demonstrate that the rate of recombination (k = 285 s−1–820 s−1) is dependent on the mutant and a higher barrier to recombination is formed in the case of the Y43A mutant. The Y43A mutant protein exhibited very low affinity for O2 and the Fe(II)–O2 complex was not detected in its RR spectrum3 but in the case of binding to CO, the frequency of CO, photodissociation yield, and rebinding constant are similar to those of the wild type. Furthermore, kinetic analysis of the amide I and heme propionate(s) modes in the Y43A mutant (Fig. S4, ESI†) reveals a similar behaviour to the wild-type protein; protein relaxation and perturbation of heme propionates correlate with recombination of CO. The photoproduct yield for the CO adduct of Y43F (34%) is significantly lower compared to that of the wild type. It should be noted that in the Y43F mutant, the frequencies of both Fe(II)–CO and Fe(II)–O2 were different compared to those in the wild type, which indicates that Y43 forms a H-bond with both O2 and CO ligands.3 On the other hand, the Y43W mutant exhibited a very high (>150 s−1) dissociation constant of O2 and the Fe(II)–O2 complex was not detected.3 In the case of the CO-bound Y43W mutant, the frequency of CO is 9 cm−1 lower than that of the wild type and a high photoproduct yield is observed, but the recombination rate is higher than that observed in the wild type.
| t 1 (μs) | k (s−1) | % photo-product at 6 μs | |
|---|---|---|---|
| WT | 1312 ± 73 | 528 | 92 |
| L65M | 393 ± 13 | 1764 | 91 |
| L65T | 237 ± 8 | 2929 | 29 |
| Y43A | 2424 ± 92 | 285 | 92 |
| Y43F | 1363 ± 32 | 508 | 34 |
| Y43W | 845 ± 22 | 820 | 92 |
| Amide I (WT) | 1443 ± 179 | 480 | |
| Heme prop. (WT) | 1461 ± 479 | 474 |
In YddV–heme the mutation of L65 to either Met (a residue of similar size and hydrophobicity) or Thr (a polar residue) results in significantly faster recombination rates of CO compared to that observed for the wild-type protein, even though ν(CO) is not affected in the L65T mutant. Therefore, our data suggest that L65 plays an important part in the kinetic barrier to ligand rebinding and no correlation between ν(CO) and the recombination rate is observed. Interestingly, the corresponding Leu residue in HemAT (L92) has been proposed as acting as part of the conformational gate for access of a ligand to the heme pocket, based on the low photoproduct yield that was observed for L92 mutants in continuous wave photolysis experiments; however, kinetic measurements were not performed.29 The mutation of Y43 to Ala, a non-polar residue with less steric hindrance than Tyr, creates a higher barrier to recombination of CO compared to the wild-type protein and Y43F and Y43W mutants. This appears to be a contradictory observation, as the expected increased available space in the heme pocket in the Y43A mutant compared to those in wild-type YddV–heme and Y43F and Y43W mutants would suggest an increase in the rate of recombination of CO.21 The diverse effects of Y43 mutations on the recombination rate of CO are challenging to interpret in the absence of a crystal structure of YddV that would allow the identification of possible interactions of Y43 with other distal site residues and/or internal water solvent molecule(s), which ultimately affect the rate of recombination of CO and result in its decrease in the Y43A mutant. Taken together, inspection of the recombination rates of CO and the absence of the minor H-bonded conformer in YddV–heme mutants suggest strong coupling of the L65 and Y43 residues to the properties of CO bound to heme iron. Comparison of the O2 and CO data demonstrates that L65 and Y43 are crucial for ligand recognition and discrimination and therefore for specific sensing of gases.3,19
The following discussion compares the behavior of the distal residues Y43 and L65, the role of the heme propionates and the relaxation of the protein backbone observed in YddV–heme–CO experiments to the corresponding features of the well-studied GCS protein HemAT. In comprehensive studies of HemAT, the unique specificity and tuning of the distal amino acid residues in creating the necessary conformational changes for ligand recognition were demonstrated. The behavior of T95 and Y70 upon binding of O2, CO, and NO to heme iron produced distinct ligand-bound conformations, which indicates that these residues are the major contributors to ligand recognition and discrimination, and L92 induced the necessary structural changes in T95 and Y70 to maintain the H-bonded conformers.15,28 Such specific recognition and discrimination of iron-bound gaseous ligands by the distal protein environment is crucial to intramolecular signal transduction by HemAT. For the signal transduction mechanism in HemAT, the G, H and B helices undergo significant displacement on ligand binding that has been suggested to trigger conformational changes from the sensor domain to the functional domain.8,18,33 In YddV, the crucial residues in the recognition of O2 and CO bound to heme iron are L65 and Y43, which facilitate the formation of H-bonded conformers; when those residues are mutated the H-bonded closed conformer of the heme–CO adduct is absent. The overall dynamics of the distal moiety of heme in YddV appears quite distinct from that in HemAT. In HemAT, the sensor domain displayed a signal for the amide I vibration in UV RR spectra in hundreds of nanoseconds subsequent to photolysis of CO and recovery in 50 μs, which indicates that the isolated sensor domain undergoes fast conformational changes to the protein backbone upon photolysis of CO.33 In YddV–heme relaxation of the amide vibration is completed with a rate constant of 480 s−1, which is similar to that of the rebinding of CO (k = 528 s−1), indicating a much slower process of conformational relaxation of the protein compared to HemAT. Such protein dynamics in HemAT has been proposed to play a crucial role in the signaling process. In HemAT it has been shown that upon binding of O2 to heme a H-bond forms between H86 and the heme propionate, thereby inducing a conformational change, which is communicated to the CE loop and the E helix.16,18 This conformational change is regarded as essential for signal transduction in HemAT. Hydrogen-bonding networks that include the heme propionates and are essential for transmitting CO–NO–O2 binding signals from heme to the protein have also been identified in myoglobin.34 We observed conformational changes in the protonated and deprotonated forms of the heme propionates in YddV–heme upon photodissociation of CO, which suggests that the heme propionates may be important in communicating changes from heme to the protein in YddV, similarly to HemAT.
Conclusions
In summary, our results demonstrate a significant role for L65 and Y43 in controlling the properties of the heme-bound ligand and ligand dynamics in YddV. Taken together, our results and those previously reported show that there are distinct differences in the interaction of YddV with CO and O2 compared to the well-studied sensor protein HemAT and that conformational changes in the protein upon ligand dissociation and rebinding are slower for YddV. Of particular interest is the role of the heme propionates in both YddV and HemAT. The heme propionates appear to behave in a similar way in both proteins upon ligand binding/dissociation. Experiments on the YddV–O2 adduct are under way to further explore the heme-protein communication pathway and the mechanism of intramolecular signal transduction.Acknowledgements
This work was funded by the European Regional Development Fund and the Republic of Cyprus through the Research Promotion Foundation (Grants ANAVATHMISI/PAGIO/0308/14 and PENEK/0609/41) and University of Cyprus internal grant to EP, and the Charles University in Prague (UNCE 204025/2012), Grant Agency of Charles University in Prague (756214) and the Grant Agency of the Czech Republic (grant 15-19883S) to MM.Notes and references
- M. M. Méndez-Ortiz, M. Hyodo, Y. Hayakawa and J. Membrillo-Hernádez, Genome-Wide Transcriptional Profile of Escherichia coli in Response to High Levels of the Second Messenger 3′,5′-Cyclic Diguanylic Acid, J. Biol. Chem., 2006, 281, 8090–8099 CrossRef PubMed.
- J. R. Tuckerman, G. Gonzalez, E. H. S. Sousa, X. Wan, J. A. Saito, M. Alam and M.-A. Gilles-Gonzalez, An Oxygen-Sensing Diguanylate Cyclase and Phosphodiesterase Couple for c-di-GMP Control, Biochemistry, 2009, 48, 9764–9774 CrossRef CAS PubMed.
- K. Kitanishi, K. Kobayashi, Y. Kawamura, I. Ishigami, T. Ogura, K. Nakajima, J. Igarashi, A. Tanaka and T. Shimizu, Important Roles of Tyr43 at the Putative Heme Distal Side in the Oxygen Recognition and Stability of the Fe(II)-O2 Complex of YddV, a Globin-Coupled Heme-Based Oxygen Sensor Diguanylate Cyclase, Biochemistry, 2010, 49, 10381–10393 CrossRef CAS PubMed.
- M. Martínková, K. Kitanishi and T. Shimizu, Heme-based Globin-Coupled Oxygen Sensors: Linking Oxygen to Functional Regulation of Diguanylate Cyclase, Histidine Kinase, and Methyl-Accepting Chemotaxis, J. Biol. Chem., 2013, 288, 27702–27711 CrossRef PubMed.
- Y. Sasakura, T. Yoshimura-Suzuki, H. Kurokawa and T. Shimizu, Structure-Function Relationships of EcDOS, a Heme-Regulated Phosphodiesterase from Escherichia coli, Acc. Chem. Res., 2006, 39, 37–43 CrossRef CAS PubMed.
- A. Moglich, R. A. Ayers and K. Moffat, Structure and Signalling Mechanism of Per-ARNT_Sim Domains, Structure, 2009, 17, 1282–1294 CrossRef CAS PubMed.
- S. Hou, R. W. Larsen, D. Boudko, C. W. Riley, E. Karatan, M. Zimmer, G. W. Ordal and M. Alam, Myoglobin-like Aerotaxis Transducers in Archaea and Bacteria, Nature, 2000, 403, 540–547 CrossRef CAS PubMed.
- W. Zhang and G. N. Phillips Jr., Structure of the Oxygen Sensor in Bacillus subtilis: Signal Transduction of Chemotaxis by Control of Symmetry, Structure, 2003, 11, 1097–1110 CrossRef CAS.
- X. Wan, J. R. Tuckerman, J. A. Saito, T. A. K. Freitas, J. S. Newhouse, J. R. Denery, M. Y. Galperin, G. Gonzalez, M. A. Gilles-Gonzalez and M. Alam, Globins Synthesize the Second Messenger Bis-(3–5)-Cyclic Diguanosine Monophosphate in Bacteria, J. Mol. Biol., 2009, 388, 262–270 CrossRef CAS PubMed.
- H. Sawai, S. Yoshioka, T. Uchida, M. Hyodo, Y. Hayakawa, K. Ishimori and S. Aono, Molecular Oxygen Regulates the Enzymatic Activity of a Heme-Containing Diguanylate Cyclase (HemDGC) for the Synthesis of Cyclic di-GMP, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 1804, 166–172 CrossRef CAS PubMed.
- L. Thijs, E. Vinck, A. Bolli, F. Trandafir, X. Wan, D. Hoogewijs, M. Coletta, A. Fago, R. E. Weber, S. Van Doorslaer, P. Ascenzi, M. Alam, L. Moens and S. Dewilde, Characterization of a Globin-Coupled Oxygen Sensor with a Gene-Regulating Function, J. Biol. Chem., 2007, 282, 37325–37340 CrossRef CAS PubMed.
- A. Pesce, L. Thijs, M. Nardini, F. Desmet, L. Sisinni, L. Gourlay, A. Bolli, M. Coletta, S. Van Doorslaer, X. Wan, M. Alam, P. Ascenzi, L. Moens, M. Bolognesi and S. Dewilde, HisE11 and HisF8 Provide Bis-histidyl Heme Hexa-Coordination in the Globin Domain of Geobacter sulfurreducens Globin-Coupled Sensor, J. Mol. Biol., 2009, 386, 246–260 CrossRef CAS PubMed.
- K. Kitanishi, K. Kobayashi, T. Uchida, K. Ishimori, J. Igarashi and T. Shimizu, Identification and Functional and Spectral Characterization of a Globin-coupled Histidine Kinase from Anaeromyxobacter sp. Fw109-5, J. Biol. Chem., 2011, 286, 35522–35534 CrossRef CAS PubMed.
- R. Hengge, Principles of c-di-GMP Signalling in Bacteria, Nat. Rev. Microbiol., 2009, 7, 263–273 CrossRef CAS PubMed.
- T. Ohta, H. Yoshimura, S. Yoshioka, S. Aono and T. Kitagawa, Oxygen Sensing Mechanism of HemAT from Bacillus subtilis: A Resonance Raman Spectroscopic Study, J. Am. Chem. Soc., 2004, 126, 15000–15001 CrossRef CAS PubMed.
- H. Yoshimura, S. Yoshioka, K. Kobayashi, T. Ohta, T. Uchida, M. Kubo, T. Kitagawa and S. Aono, Specific Hydrogen-Bonding Networks Responsible for Selective O2 Sensing of the Oxygen Sensor Protein HemAT from Bacillus subtilis, Biochemistry, 2006, 45, 8301–8307 CrossRef CAS PubMed.
- W. Zhang, J. S. Olson and G. N. Phillips Jr., Biophysical and Kinetic Characterization of HemAT, an Aerotaxis Receptor from Bacillus subtilis, Biophys. J., 2005, 88, 2801–2814 CrossRef CAS PubMed.
- S. F. El-Mashtoly, Y. Gu, H. Yoshimura, S. Yoshioka, S. Aono and T. Kitagawa, Protein Conformational Changes of HemAT-Bs upon Ligand binding Probed by Ultraviolet Resonance Raman Spectroscopy, J. Biol. Chem., 2008, 283, 6942–6949 CrossRef CAS PubMed.
- N. Nakajima, K. Kitanishi, K. Kobayashi, N. Kobayashi, J. Igarashi and T. Shimizu, Leu65 in the Heme Distal Side is Critical for the Stability of the Fe(II)-O2 Complex of YddV, a Globin-Coupled Oxygen Sensor Diguanylate Cyclase, J. Inorg. Biochem., 2012, 108, 163–170 CrossRef PubMed.
- M. Stranava, M. Martínková, M. Stiborová, P. Man, K. Kitanishi, L. Muchová, L. Vítek, V. Martínek and T. Shimizu, Introduction of Water into the Heme Distal Side by Leu65 Mutations of an Oxygen Sensor, YddV, Generates Verdoheme and Carbon Monoxide, Exerting the Heme Oxygenase Reaction, J. Inorg. Biochem., 2014, 140, 29–38 CrossRef CAS PubMed.
- T. Li, M. L. Quillin, G. N. Phillips Jr. and J. S. Olson, Structural Determinants of the Stretching Frequency of CO Bound to Myoglobin, Biochemistry, 1994, 33, 1433–1446 CrossRef CAS.
- K. Gerwert, Infrared and Raman Spectroscopy of Biological Materials, Marcel Dekker, NY, 2000, ch. 6, pp. 193–231 Search PubMed.
- D. Heibrink, H. Sigurdson, C. Bolwien, P. Brzezinski and J. Heberle, Transient Binding of CO to CuB in Cytochrome c Oxidase Is Dynamically Linked to Structural Changes around a Carboxyl Group: A Time-Resolved Step-Scan Fourier Transform Infrared Investigation, Biophys. J., 2002, 82, 1–10 CrossRef.
- K. Koutsoupakis, S. Stavrakis, E. Pinakoulaki, T. Soulimane and C. Varotsis, C. Direct Observation of the Equilibrium CuB-CO Complex and Functional Implications of the Transient Heme a3 Propionates in Cytochrome ba3-CO from Thermus thermophilus: FTIR and Time-Resolved Step-Scan FTIR Studies, J. Biol. Chem., 2002, 277, 32860–32866 CrossRef CAS PubMed.
- C. Koutsoupakis, E. Pinakoulaki, S. Stavrakis, V. Daskalakis and C. Varotsis, Time-resolved Step-Scan Fourier Transform Infrared Investigation of Heme-Copper Oxidases: Implications for O2 Input and H2O/H+ Output Channels, Biochim. Biophys. Acta, Bioenerg., 2004, 1655, 347–352 CrossRef CAS PubMed.
- E. Pinakoulaki and C. Varotsis, Time-Resolved Resonance Raman and Time-Resolved Step-Scan FTIR Studies of Nitric Oxide Reductase from Paracoccus denitrificans: Comparison of the Heme b3-FeB Site to that of the Heme-CuB in Oxidases, Biochemistry, 2003, 42, 14856–14861 CrossRef CAS PubMed.
- E. Pinakoulaki, C. Koutsoupakis, H. Sawai, A. Pavlou, Y. Kato, Y. Asano and S. Aono, Aldoxime Dehydratase: Probing the Heme Environment Involved in the Synthesis of the Carbon-Nitrogen Triple Bond, J. Phys. Chem. B, 2011, 115, 13012–13018 CrossRef CAS PubMed.
- E. Pinakoulaki, H. Yoshimura, S. Yoshioka, S. Aono and C. Varotsis, Recognition and Discrimination of Gases by the Oxygen-Sensing Signal Transducer Protein HemAT as Revealed by FTIR Spectroscopy, Biochemistry, 2006, 45, 7763–7766 CrossRef CAS PubMed.
- E. Pinakoulaki, H. Yoshimura, V. Daskalakis, S. Yoshioka, S. Aono and C. Varotsis, Two Ligand Binding Sites in the O2-Sensing Signal Transducer HemAT: Implications for Ligand Recognition/Discrimination and Signaling, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 14796–14801 CrossRef CAS PubMed.
- A. Barth, Infrared Spectroscopy of Proteins, Biochim. Biophys. Acta, 2007, 1767, 1073–1101 CrossRef CAS PubMed.
- J. Behr, P. Hellwig, W. Mantele and H. Michel, Redox Dependent Changes at the Heme Propionates in Cytochrome c Oxidase from Paracoccus denitrificans: Direct Evidence from FTIR Difference Spectroscopy in Combination with Heme Propionate 13C Labeling, Biochemistry, 1998, 37, 7400–7406 CrossRef CAS PubMed.
- C. Koutsoupakis, T. Soulimane and C. Varotsis, Probing the Q-Proton Pathway of ba3-Cytochrome c Oxidase by Time-Resolved Fourier Transform Infrared Spectroscopy, Biophys. J., 2004, 86, 2438–2444 CrossRef CAS.
- S. F. El-Mashtoly, M. Kubo, Y. Gu, H. Sawai, S. Nakashima, T. Ogura, S. Aono and T. Kitagawa, Site-Specific Protein Dynamics in Communication Pathway from Sensor to Signaling Domain of Oxygen Sensor Protein HemAT-Bs, J. Biol. Chem., 2012, 287, 19973–19984 CrossRef CAS PubMed.
- Y. Gao, S. F. El-Mashtoly, B. Pal, T. Hayashi, K. Harada and T. Kitagawa, Pathway of Information Transmission from Heme to Protein upon Ligand Binding/Dissociation in Myoglobin Revealed by UV Resonance Raman Spectroscopy, J. Biol. Chem., 2006, 281, 24637–24646 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S4 and Table S1. See DOI: 10.1039/c5cp01708d |
| This journal is © the Owner Societies 2015 |