Open Access Article
Open Access ArticleSupramolecular copolymers with stimuli-responsive sequence control†
Lorenzo
Albertazzi
,
Nick
van der Veeken
,
Matthew B.
Baker
,
Anja R. A.
Palmans
and
E. W.
Meijer
*
Institute for Complex Molecular Systems, Eindhoven University of Technology, 5612 AZ Eindhoven, The Netherlands. E-mail: e.w.meijer@tue.nl
First published on 15th September 2015
Abstract
We report a novel responsive supramolecular copolymer able to change its monomer sequence as a result of molecular stimuli. Nucleic acids and RNAse are used as molecular inputs, controlling the clustering of specific monomers along the polymer backbone. This opens new ways towards the molecular control of synthetic supramolecular networks.
Stimuli-responsive molecular materials, i.e. materials able to change their structure or their functionality in response to specific molecular cues, are a promising class of molecules for several applications.1,2 Nature is a source of inspiration for these synthetic architectures; indeed most of the cellular structures are tightly controlled by a combination of signalling molecules and enzymatic activities.3 The design of synthetic structures that undergo controlled structural changes in response to multiple inputs is an important step towards mimicking the complex pathways of signal transduction.4 Supramolecular systems play a pivotal role to achieve stimuli-responsiveness because the weak non-covalent interactions that are the basis of supramolecular architectures can be easily tuned by a variety of inputs such as light, temperature, molecular interactions and enzymatic activity.1 In this framework, enzymes are particularly interesting to apply as a trigger because of their easy availability and their high chemical selectivity. Several examples of micelles,5,6 nanoparticles7 and hydrogels8,9 able to assemble or disassemble in presence of a specific enzymatic activity have been reported in the literature. However, only few examples of enzyme responsive one-dimensional (1D) supramolecular aggregates, e.g. supramolecular polymers, have been reported.10,11 Interestingly in most of the supramolecular systems the output is a complete assembly/disassembly process while few examples of structures able to undergo a subtle structural change such as a change in shape or chemical composition, have been reported. A beautiful example of the versatility of one-dimensional aggregates to undergo complex structural changes in response of the chemical environment was reported by George and coworkers.11 They showed a change of the helicity in a supramolecular polymers in response to ATPase activity.
We have recently investigated the ability of pegylated benzene-1,3,5-tricarboxamide (BTA) supramolecular polymers to self-assemble in water into μm-long 1D fibers harnessing a combination of hydrogen-bonding and hydrophobic interactions.12,13 Detailed studies of BTA properties in water13,14 and the potential of this material for biomedical purposes such as siRNA delivery15 have been recently reported. In particular we showed that, due to the reversibility of the supramolecular interactions, BTA polymers display a dynamic behaviour, i.e. they continuously assemble and disassemble by exchanging monomers between aggregates. This is a typical feature of biological systems and it is at the basis of their responsive nature.3 We demonstrated how this dynamic behaviour can be exploited to trigger a change in the monomer distributions along the polymer upon binding of a multivalent nucleic acid such as single stranded DNA (ssDNA): the ssDNA acted as a recruiter, binding specific monomers along the polymer and clusters them, which resulted in spatiotemporal control of the monomer distribution.16 The control of polymer sequence is a major challenge towards the design of synthetic materials able of repair and adaptation.17,18 Moreover, we demonstrated that BTA polymers behaved super-selectively, i.e. a non-linear dependence on recruiter length was observed. This allows a sharper change to stimuli: whereas ssDNA longer than a critical threshold possessed strong clustering capabilities, the shorter strands resulted in no changes of the monomer sequence.
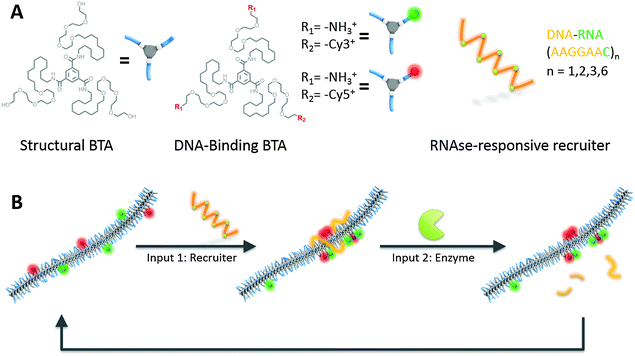
Here, we report a BTA supramolecular polymer that changes its monomer sequence in response to two different molecular inputs with: (i) a multivalent anionic nucleic acid able to cluster specific monomers into a block copolymer-like conformation and (ii) an RNAse enzyme able to degrade the recruiter and restore a random copolymer distribution. This process is shown schematically in Fig. 1. We prepared a supramolecular polymer by the co-assembly of two different types of monomers: (i) neutral non-functional BTA monomers and (ii) amine-functionalized cationic monomers able to bind nucleic acid via electrostatic interactions. The DNA-binding monomers are labelled with fluorescent dyes, either Cy3 or Cy5. These dyes form a good Förster resonance energy transfer (FRET) pair with a Förster radius of 5 nm. As FRET is strongly dependent on the average dye distance, we can use it to distinguish between random distribution and blocky structures. In the latter case, the dye-labeled monomers are in closer proximity, resulting in a higher FRET efficiency. After co-assembly and equilibration, which results in a random distribution of the dye-labelled BTAs along the 1D polymer chain, a negatively charged nucleic acid recruiter is added. In our design the recruiter consist of a hybrid DNA–RNA strand containing 7 DNA bases (AAAGGAA) and one RNA base (c). Strands of different length (8 to 48) have been obtained by repeating the AAAGGAAc sequence. The presence of the RNA base makes the recruiter sensitive to the enzyme RNAse,19 capable to selectively cleave the DNA–RNA connections. Upon the addition of RNAse, all the sequences will be cleaved to a 8mer strand. Therefore as shown in Fig. 1B our supramolecular system is designed to respond to two different molecular inputs with a reversible change and, in addition, can potentially undergo multiple cycles.
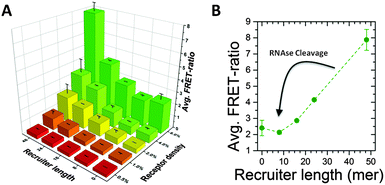
First, we demonstrated the ability of the cleavable recruiter to induce clustering of dye-labelled monomers along the polymer backbone. Fig. 2A shows the FRET ratio, e.g. the ratio of the fluorescence of the Cy5-donor and the Cy3-acceptor, respectively, after 3 days of incubating the supramolecular polymer with the cleavable recruiters. The 3D plot shows the FRET ratio for different DNA–RNA length (8 to 48mer) and different densities of recruiter-binding monomers (0.5% to 8%) and these results are compared with the FRET ratio in absence of the recruiter (see Fig. S2 and S3, ESI†).
FRET efficiencies rise after the addition of the recruiter indicating monomer clustering and the maximum FRET ratio is obtained with the combination of the longest recruiter with the highest density of cationic monomers (see also Fig. S4, ESI†). Interestingly, the trend of the FRET ratio with the recruiter length is strongly non-linear. Below a threshold of 16-mer no clustering is observed while a strong increase in FRET ratio is observed for longer strands. This observation can be explained as a consequence of the multivalency of the system. These results are in good agreement to our previous report and demonstrate a super-selective behaviour of the multivalent supramolecular polymers.16,20 This non-linearity is of great importance for the responsiveness, as the DNA–RNA strands obtained after incubation with RNAse falls below the threshold (see Fig. 2B) and cannot induce clustering. Therefore the enzyme will turn an effective recruiter into a non-effective one leading to the loss of clustering along the structure.
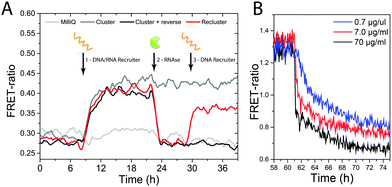
Fig. 3 shows the results of the reversible clustering experiments. Fig. 3A shows the time-dependence of the FRET ratio upon addition of the cleavable recruiter (time point 1) and RNAse (time point 2). Clearly apparent, the DNA–RNA recruiter induces an increase in FRET ratio that reaches a plateau after about 12 h while the signal is constant in the control when only MilliQ water is added (light-grey curve). After 24 h, RNAse is added to the sample, which results in a sudden drop of the FRET efficiency that goes back to the initial level. This indicates that the clustered dye-labelled BTAs become randomly distributed along the polymer chain as a result of the ability of RNAse to cleave the recruiter, reconstituting the starting situation. When only MilliQ water is added at time point 2 (dark-grey curve), the level of FRET ratio is unaffected, indicating that the dye-labelled BTAs remain clustered. A more detailed plot with the average of 4 individual measurements is reported in Fig. S6 (ESI†). These results show how the two different molecular inputs (a nucleic acid strand and an enzyme) can control the fine structure of a supramolecular polymer in water. The rate of the disassembly is dependent of the enzyme concentration as shown in Fig. 3B; therefore the kinetics of the release of clustering of the dye-labelled monomers can be tuned, opening up the way to kinetic control over supramolecular pathways.
Finally, we show the possibility to restore the clustering by adding a DNA-only recruiter (24mer) to the supramolecular polymer. In this case, the DNA recruiter is of identical size but not susceptible to degradation by the RNAse that is still present in the solution. The red curve in Fig. 3A shows an increase of the FRET ratio upon addition of the fresh recruiter (time point 3). This increase in FRET signal indicates that the system is undergoing another event of monomer clustering, highlighting the dynamic nature of this supramolecular polymer system.
In conclusion, we presented a supramolecular polymer in which the monomer sequence can be controlled by external stimuli. In this example multivalent binders and enzymes allow the reversible switching from a supramolecular block copolymer to a random copolymer in a controlled fashion. The presented work paves the way to more complex systems where the supramolecular polymers respond to multiple enzymes and recruiters in a reaction network involving positive and negative feedback loops. Although the molecular system proposed here is far from the complexity of cell signalling, it represents an important step towards the design of synthetic supramolecular systems mimicking the pathway complexity of biological systems.
The research leading to this manuscript has received funding from the Netherlands Institute for Regenerative Medicine (NIRM). LA thanks financial support of NWO (VENI program 722.014.010). ARA and EWM acknowledge financial support from the Dutch Ministry of Education, Culture and Science (Gravity program 024.001.035) and the European Research Council (FP7/2007–2013, ERC Grant Agreement 246829). The ICMS animation studio is acknowledged for providing the artwork.
Notes and references
- A. P. Blum, J. K. Kammeyer, A. M. Rush, C. E. Callmann, M. E. Hahn and N. C. Gianneschi, J. Am. Chem. Soc., 2014, 137, 2140–2154 CrossRef PubMed.
- M. Zelzer, S. J. Todd, A. R. Hirst, T. O. McDonald and R. V. Ulijn, Biomater. Sci., 2012, 1, 11–39 RSC.
- D. A. Fletcher and R. D. Mullins, Nature, 2010, 463, 485–492 CrossRef CAS PubMed.
- H. Wu, Cell, 2013, 153, 287–292 CrossRef CAS PubMed.
- A. J. Harnoy, I. Rosenbaum, E. Tirosh, Y. Ebenstein, R. Shaharabani, R. Beck and R. J. Amir, J. Am. Chem. Soc., 2014, 136, 7531–7534 CrossRef CAS PubMed.
- J. Guo, J. Zhuang, F. Wang, K. R. Raghupathi and S. Thayumanavan, J. Am. Chem. Soc., 2014, 136, 2220–2223 CrossRef CAS PubMed.
- M.-P. Chien, A. S. Carlini, D. Hu, C. V. Barback, A. M. Rush, D. J. Hall, G. Orr and N. C. Gianneschi, J. Am. Chem. Soc., 2013, 135, 18710–18713 CrossRef CAS PubMed.
- J. Hu, G. Zhang and S. Liu, Chem. Soc. Rev., 2012, 41, 5933–5949 RSC.
- R. J. Williams, A. M. Smith, R. Collins, N. Hodson, A. K. Das and R. V. Ulijn, Nat. Nanotechnol., 2009, 4, 19–24 CrossRef CAS PubMed.
- D. S. Guo, T.-X. Zhang, Y.-X. Wang and Y. Liu, Chem. Commun., 2013, 49, 6779–6781 RSC.
- M. Kumar, P. Brocorens, C. Tonnelé, D. Beljonne, M. Surin and S. J. George, Nat. Commun., 2014, 5, 5793 CrossRef CAS PubMed.
- C. M. A. Leenders, L. Albertazzi, T. Mes, M. M. E. Koenigs, A. R. A. Palmans and E. W. Meijer, Chem. Commun., 2013, 49, 1963–1965 RSC.
- M. B. Baker, L. Albertazzi, I. K. Voets, C. M. A. Leenders, A. R. A. Palmans, G. M. Pavan and E. W. Meijer, Nat. Commun., 2015, 6, 6234 CrossRef CAS PubMed.
- L. Albertazzi, D. van der Zwaag, C. M. A. Leenders, R. Fitzner, R. W. van der Hofstad and E. W. Meijer, Science, 2014, 344, 491–495 CrossRef CAS PubMed.
- M. Bakker, C. Lee, E. W. Meijer, P. Y. W. Dankers and L. Albertazzi, submitted.
- L. Albertazzi, F. J. Martinez-Veracoechea, C. M. A. Leenders, I. K. Voets, D. Frenkel and E. W. Meijer, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12203–12208 CrossRef CAS PubMed.
- Y. Ura, J. M. Beierle, L. J. Leman, L. E. Orgel and M. R. Ghadiri, Science, 2009, 325, 73–77 CrossRef CAS PubMed.
- J. Niu, R. Hili and D. R. Liu, Nat. Chem., 2013, 5, 282–292 CrossRef PubMed.
- B. R. Kelemen, T. A. Klink, M. A. Behlke, S. R. Eubanks, P. A. Leland and R. T. Raines, Nucleic Acids Res., 1999, 27, 3696–3701 CrossRef CAS.
- F. J. Martinez-Veracoechea and D. Frenkel, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10963–10968 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5cc06951c |
| This journal is © The Royal Society of Chemistry 2015 |