Open Access Article
Open Access ArticleFunctional profiling of adenylation domains in nonribosomal peptide synthetases by competitive activity-based protein profiling†
Shota
Kasai‡
,
Sho
Konno‡
,
Fumihiro
Ishikawa
* and
Hideaki
Kakeya
*
Department of System Chemotherapy and Molecular Sciences, Division of Bioinformatics and Chemical Genomics, Graduate School of Pharmaceutical Sciences, Kyoto University, Sakyo, Kyoto 606-8501, Japan. E-mail: fishika@pharm.kyoto-u.ac.jp; scseigyo-hisyo@pharm.kyoto-u.ac.jp
First published on 4th September 2015
Abstract
We describe competitive activity-based protein profiling (ABPP) to accelerate the functional prediction and assessment of adenylation (A) domains in nonribosomal peptide synthetases (NRPSs) in proteomic environments. Using a library of sulfamoyloxy-linked aminoacyl-AMP analogs, the competitive ABPP technique offers a simple and rapid assay system for adenylating enzymes and provides insight into enzyme substrate candidates and enzyme active-site architecture.
Peptide-based natural products are structurally diverse and display biologically important activities, including a large number of clinical antimicrobial, anticancer and immunosuppressive drugs, virulence factors, and signaling molecules.1 Many of these small molecules are biosynthesized by large, highly versatile multifunctional enzymes known as nonribosomal peptide synthetases (NRPSs).2 Currently, a large number of biosynthetic gene clusters encoding NRPSs have been found in various organisms through recent genome sequencing projects.3 Functional analysis and engineering of these synthetases have focused on the heterologous expression of biosynthetic clusters in host organisms such as Streptomyces strains, yeast and Escherichia coli.4 Because of the multifunctionality of the NRPS family and method complications associated with predicting NRPS activities in proteomic samples, there has been significant interest in developing methods that assess NRPS activities directly in complex proteomes. Such approaches should accelerate both the functional characterization and manipulation of NRPSs. In addition, some of these megasynthetase enzymes pose particular difficulties in laboratory assessments. This is partly because these enzymes are large molecular proteins ranging in size between 300 and 800 kDa, and because of the general intractability of producer organisms to conventional genetic manipulation and heterologous expression.5 Chemical proteomic techniques of NRPS family members could provide highly complementary genetic approaches that facilitate the direct functional analysis of NRPSs in biological samples.
Adenylation (A) domains housed within all NRPS modules are essential catalytic motifs and function as gatekeepers to select amino acid building blocks used in the construction of peptide-based natural products. The A-domain selectively incorporates cognate amino acids into NRPs from a much larger monomer pool, including all 20 proteinogenic amino acids and a number of non-proteinogenic amino acids and aryl acids.6 The A-domain recognizes a cognate amino acid and converts it to the corresponding aminoacyl adenylate intermediate at the expense of ATP with the release of PPi (Fig. 1). The adenylated substrate subsequently undergoes nucleophilic attack by the terminal thiol group of the 4′-phosphopantetheine arm of a downstream thiolation (T) domain, leading to the formation of a thioester bound aminoacyl-S-T. The simple biochemical logic of the A-domains has made them attractive targets for metabolic engineering,7 mutasynthesis,8 combinatorial biosynthesis9 and directed evolution10 for the production of new metabolites. Therefore, A-domains should provide an ideal entry to assess NRPS-related biosynthetic pathways and engineered systems. The conventional methods to assay enzymatic activities of the A-domains in NRPSs rely on radioactive methods such as ATP-PPi exchange11 and the uptake of radiolabeled amino acids.12 In these methods, the assay poses several laborious, complicated and time-consuming handling steps involving radioactive materials and the labile thioester bond that holds radiolabeled amino acids to the synthetase. Other techniques using a continuous hydroxylamine release assay are limited to the analysis of purified proteins.13 Advanced techniques to not only rapidly verify expression, folding and activity parameters, but also accelerate the functional prediction and assessment of the A-domains of NRPSs in proteomes from native and heterologous systems are therefore desirable. Here, we describe the utility of competitive activity-based protein profiling (ABPP) to predict the substrate promiscuity of the A-domains in NRPS enzymes by coupling with a library of 5′-O-(N-aminoacyl)sulfamoyladenosine (aminoacyl-AMS) inhibitors (Fig. 2a).
 | ||
| Fig. 1 The adenylation reaction catalyzed by the A-domains of NRPSs. Modules are composed of thiolation (T), adenylation (A) and condensation (C) domains. | ||
 | ||
| Fig. 2 (a) Methods for proteomic analysis of the A-domains in NRPS enzymes through competitive ABPP. In a typical competitive ABPP experiment, proteomes treated with an inhibitor (or vehicle control) are incubated with an active site-directed probe for an A-domain. Samples are then photoactivated with UV light (365 nm), treated with a rhodamine (Rh)–azide reporter tag under click chemistry (CC) conditions, and monitored by SDS-PAGE coupled with in-gel fluorescence imaging. Modules are composed of thiolation (T), adenylation (A), condensation (C) and thioesterase (TE) domains. The rectangles represent nonspecific proteins. (b) The structures of the active site-directed proteomic probes.18,19 (c) The structures of cognate competitors of the probes in (b). | ||
An aminoacyl-AMS scaffold, a bioisosteric and chemically stable non-hydrolysable analog of the aminoacyl adenylate monophosphate intermediate has been used in the design of inhibitors that display tightly bound inhibitory characteristics for the A-domains in NRPS enzymes.14–17 We have recently developed active site-directed proteomic probes for the A-domains in NRPSs coupled to the aminoacyl-AMS scaffold with a photoreactive benzophenone and a clickable alkyne functionality at the 2′-OH group of the adenosine skeleton (Fig. 2b).18,19 The synthetic probes selectively target the cognate A-domains in NRPS enzymes in native proteomic environments. We envisaged that chemical proteomic strategies using a combination of a library of aminoacyl-AMP analogs should offer a direct readout of enzyme substrate specificity and the active-site architecture of A-domains in NRPSs. To this end, aminoacyl-AMP analogs 9, 10 and 13–25 were synthesized by the assembly of the AMS scaffold and protected NHS activated amino acids (Fig. 2c and Scheme S1, ESI†).
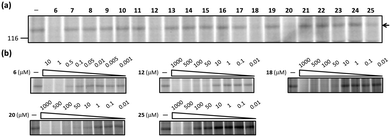
We first asked whether a competitive ABPP technique for the A-domains in NRPSs could provide details about enzyme substrate selectivity and enzyme active-site microenvironments in biological samples. This is because libraries of peptide-based ABPP probes have been applied to map active-site specificity in several classes of proteases.20,21 For the proof-of-principle experiments, we chose five A-domains in the gramicidin S synthetases, GrsA and GrsB in proteomic extracts from the gramicidin S producers, Aneurinibacillus migulanus ATCC 9999 and DSM 5759. Gramicidin S is a cyclic decapeptide biosynthesized by GrsA and GrsB peptide synthetases (Fig. S1, ESI†).22 The GrsA initiation module contains the domain structure A1 (L-Phe)-T-E and incorporates D-Phe into gramicidin S. GrsB consists of four NRPS modules, C-A2-T-C-A3-T-C-A4-T-C-A5-T-TE. The four A-domains (A2–A5) are housed within this protein and selectively incorporate their cognate amino acids, L-Pro, L-Val, L-Orn and L-Leu, respectively, in the NRPS assembly line. A library of aminoacyl-AMP analogs was tested towards endogenous GrsA in the context of a complex proteome using competitive proteomic profiling assays with probe 1. In the competitive ABPP assays, probe 1 was incubated with the A. migulanus ATCC 9999 proteomes in the absence or presence of aminoacyl-AMP analogs 6–25 for 10 min at room temperature and pH 8.0. These samples were then photoactivated with UV light (365 nm) for 30 min at 0 °C, treated with the rhodamine (Rh)–azide reporter tag under standard click chemistry (CC) conditions23 and monitored by SDS-PAGE coupled with in-gel fluorescence scanning. As expected, several compounds showed inhibitory activities towards the A-domain of GrsA (Fig. 3a and Fig. S2, ESI†). Five (6, 12, 18, 20 and 25) of the 20 compounds showed high inhibition characteristics of the A-domain of GrsA. The labeling of the A-domain of GrsA by probe 1 completely disappeared in the presence of compounds 6, 12, 18 and 20. In contrast, incubation of the A. migulanus ATCC 9999 proteome with 100 μM 25 exhibited moderate inhibition of the labeling of the A-domain of GrsA, with an estimated reduction of fluorescence band intensity by 56% when compared with the DMSO control. Compound 19 provided 100% inhibition of the A-domain of GrsA when tested at a concentration of 1 mM. Treatment of the A. migulanus ATCC 9999 proteome with the other aminoacyl-AMP analogs prior to the administration of 1 did not block labeling of the A-domain of GrsA. To estimate the strength of their interactions with the A-domain of GrsA in the proteome, dose–response competitive ABPP experiments with probe 1 were conducted for compounds 6, 12, 18, 20 and 25 at concentrations ranging from 1 nM to 1 mM (6: 1 nM–10 μM; 12, 18, 20 and 25: 10 nM–1 mM). In addition, we chose 17 and 23 that exhibited no binding characteristics towards the A-domain of GrsA for comparative activity studies. These data revealed clear differences in the inhibitor selectivity profiles of the A-domain of GrsA by 6, 12, 18, 20, and 25, providing IC50 values of 0.38 ± 0.14 μM, 23.0 ± 0.12 μM, 27.2 ± 0.22 μM, 9.9 ± 0.13 μM and 126 ± 0.18 μM, respectively (Fig. 3b and Fig. S3, ESI†). In contrast, 17 and 23 did not exhibit inhibitory activities with the A-domain of GrsA up to 1 mM (Fig. S4, ESI†). These results were comparable to a competitive ABPP study towards the A-domain of recombinant GrsA (Fig. S5 and S6, and Table S3, ESI†).
We next sought to demonstrate whether the inhibitor sensitivity profiles of the A-domain of GrsA could provide insights into enzyme substrate selectivity of adenylating enzymes. To assess the specific activities of the A-domain of recombinant holo-GrsA, the apparent steady-state kinetic parameters of the most active amino acid substrates (L-Phe, L-Leu, L-Met, L-Trp, and L-His) were examined using a coupled hydroxamate–MesG continuous spectrophotometric assay (Fig. S7, S8, S10–S12 and Table S1, ESI†).13 To investigate a correlation between the IC50 values and substrate tolerance, the measured Michaelis constants (Km) towards the selected substrates were compared. This is because aminoacyl-AMS molecules were originally designed not as transition-state analogs relating to adenylation reactions, but as analogs of the reaction intermediates. The Km value for L-Phe with the A-domain of recombinant GrsA was calculated to be 24.8 μM. The Km values for L-Leu, L-Met, L-Trp and L-His with the A-domain of GrsA were 2.85, 18.5, 1.49 and 17.5 mM, respectively. While a pronounced increase in Km values is observed among L-Leu, L-Met, L-Trp and L-His, the A-domain of GrsA was found to discriminate stringently between the selected amino acids. The Km values of L-Leu and L-Trp are approximately 6- and 12-fold lower than those of L-Met and L-His, respectively. In addition, L-Thr and L-Lys were tested because the corresponding compounds 17 and 23 did not provide inhibitor-sensitive profiles towards the A-domains of both recombinant and endogenous GrsA (Fig. S4 and S6, ESI†). Indeed, the Km value of the A-domain of recombinant GrsA with L-Thr was 246 mM, which is 15- to 160-fold higher than the Km values of the other substrates tested (Fig. S9 and Table S1, ESI†). Incubation of GrsA (2.8 μM) with L-Lys (100–500 mM) resulted in virtually no detectable enzymatic activity. Taken together, these data validate that the competitive ABPP technique using a combination of a library of aminoacyl-AMP analogs can be used to image enzyme substrate candidates and active-site architectures of the A-domains of NRPSs in proteomic samples.
As an ultimate application of the competitive ABPP, we evaluated the A-domains housed within the modular synthetase GrsB using probes 2, 3, 4 and 5. DSM 5759 expressed higher levels of active GrsB than ATCC 9999.19 Thus, the proteome from DSM 5759 was used to demonstrate our strategy. In the competitive ABPP experiments for the Pro-activating domain of GrsB, probe 2 was incubated with the A. migulanus DSM 5759 proteome in the absence or presence of aminoacyl-AMP analogs 6–25 (100 μM) (Fig. 4a and Fig. S14, ESI†). The labeling of the Pro-activating domain of GrsB was lost completely only by the addition of 7. A dose–response curve for proteomic inhibition of the Pro-activating domain of GrsB by 7 gave an IC50 value of 0.29 ± 0.09 μM (Fig. S15, ESI†). These inhibitor sensitivity profiles correspond identically to those of the recombinantly expressed homologous A-domain within the first module of tyrocidine synthetase TycB (TycB1) and correlate well with its substrate preferences (Fig. S13, S16 and S17, and Tables S2 and S4, ESI†).24 Indeed, a comparison of the predicted structure of the active site of A-domains from the Pro-activating domains of GrsB and TycB reveals that they are highly conserved,25,26 further demonstrating the utility of this ABPP technique.
In competitive ABPP assays for the Orn-activating domain of GrsB, probe 3 was used to visualize protein–ligand interactions. The competitive ABPP of aminoacyl-AMP analogs 6–25 revealed that 15, 23, 24 and the cognate competitor 8 conferred potent inhibition activities at a concentration of 100 μM with estimated reduction in fluorescence band intensities when compared to the DMSO control of 63, 84, 82 and 85%, respectively (Fig. 4b and Fig. S18, ESI†). Dose–response curves for proteomic inhibition of the Orn-activating domain of GrsB by 8, 15, 23 and 24 were constructed by competitive ABPP with probe 3, providing IC50 values of 18.5 ± 2.6 nM, 139 ± 0.33 μM, 8.2 ± 0.14 μM and 4.6 ± 0.15 μM, respectively (Fig. S19 and Table S5, ESI†). These results indicate that the Orn-activating domain of GrsB displays high substrate specificity towards the cognate amino acid substrate. Alignment analysis of the sequence from GrsB with the putative A-domains for L-Orn from tyrocidine synthetase TycC, fengycin synthetases Pps1 and FenC, and bacitracin synthetase BacB reveals a highly conserved active site structure.25,26 The conserved residues, Glu278 and Ser322, play a key role in substrate recognition through salt-bridge formation and hydrogen bonding with the side chain amino group of L-Orn. The binding characteristics of 8, 23 and 24 towards the Orn-activating domain of GrsB are attributed to the common electrostatic interactions between the carboxylate of Glu278 and the side chain functionalities of the compounds. Since the Orn-activating domain of HSAF synthetase can activate both L-Orn and L-Lys among the 16 amino acids tested,27 it is postulated that the Orn-activating domain of GrsB could accept L-Gln, L-Lys and L-Arg as alternative substrates.
To evaluate the Val- and Leu-activating domains of GrsB in competitive experiments, the DSM 5759 proteome was individually treated with 100 μM of each aminoacyl-AMP analog 6–25 before the addition of probes 4 and 5. Assessment of the inhibition potency revealed that five (12, 13, 18, 20 and 25) and three (10, 12 and 18) of the 20 compounds displayed high (≥90%) inhibition of the Val- and Leu-activating domains of GrsB, respectively (Fig. 4c and d, Fig. S20 and S22, ESI†). Inhibitors 12, 13, 18, 20 and 25 gave IC50 values of 2.8 ± 0.23 μM, 6.0 ± 0.10 μM, 0.42 ± 0.08 μM, 0.65 ± 0.04 μM and 2.2 ± 0.28 μM for the inhibition of the Val-activating domain of GrsB (Fig. S21 and Table S6, ESI†). A dose–response curve by the probe’s cognate competitor 11 afforded an IC50 value of 0.11 ± 0.04 μM (Fig. S21 and Table S6, ESI†). Dose–response curves for the inhibition of the Leu-activating domain of GrsB by 12 and 18 gave IC50 values of 1.7 ± 0.09 nM and 2.7 ± 0.22 μM, respectively (Fig. S23 and Table S7, ESI†). In contrast, 10 did not provide an inhibitor sensitive profile towards the Leu-activating domain of GrsB (Fig. S23, ESI†). These results emphasize not only the cognate amino acid specificity, but also the different inhibitor recognition patterns in the Val- and Leu-activating domains. The A-domains that activate amino acids with hydrophobic side chains show lower substrate selectivity than those that activate polar amino acids. Indeed, the Leu-activating domains of the surfactin synthetases SrfAA, SrfAB and SrfAC are known to activate L-Val and L-Ile with lower catalytic efficiency.28 The Val-activating domain of SrfAB activates L-Ile to a lesser extent.29 Using a combination of a large base of NRPS gene sequences,25,26 the competitive ABPP technique could provide functional characteristics and a molecular basis underpinning substrate recognition of the A-domains of NRPS enzymes.
In summary, we have demonstrated a simple, rapid and nonradioactive assay system to accelerate greatly the direct functional prediction and characterization of A-domains in NRPSs in proteomic environments. Using a library of aminoacyl-AMP analogs, the competitive ABPP technique offered insights into enzyme substrate candidates and enzyme active-site microenvironments. These techniques should serve as powerful diagnostic tools in metabolic engineering, mutasynthesis, combinatorial biosynthesis and directed evolution programs by quantifying expression, solubility, folding and the functional prediction of A-domains of NRPSs in native/heterologous systems. Using a range of non-proteinogenic amino acid sulfamoyl adenosines, these approaches can be further used to construct inhibitor sensitivity profiles for non-proteinogenic building blocks, which could allow the successful introduction of functionalized amino acid components into peptide-based natural products.
This work was partly supported by a Grant-in Aid for Young Scientists (B) 26750370 (F.I.) and research grants from the Japan Society of the Promotion of Science and the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT) (H.K.). We thank Prof. Mohamed Marahiel (Philipps-Universität Marburg, Germany) for providing the GrsA and TycB1 expression constructs.
Notes and references
- C. T. Walsh, Science, 2004, 303, 1805–1810 CrossRef CAS PubMed.
- M. A. Fischbach and C. T. Walsh, Chem. Rev., 2006, 106, 3468–3496 CrossRef CAS PubMed.
- H. B. Bode and R. Muller, Angew. Chem., Int. Ed., 2005, 44, 6828–6846 CrossRef CAS PubMed.
- H. Zhang, Y. Wang and B. A. Pfeifer, Mol. Pharmacol., 2008, 5, 212–225 CrossRef CAS PubMed.
- D. E. Cane and C. T. Walsh, Chem. Biol., 1999, 6, R319–R325 CrossRef CAS.
- G. H. Hur, C. R. Vickery and M. D. Burkart, Nat. Prod. Rep., 2012, 29, 1074–1098 RSC.
- J. Thirlway, R. Lewis, L. Nunns, M. A. Nakeeb, M. Styles, A.-W. Struck, C.-P. Smith and J. Micklefield, Angew. Chem., Int. Ed., 2012, 51, 7181–7184 CrossRef CAS PubMed.
- S. Weist, C. Kittel, D. Bischoff, B. Bister, V. Pfeifer, G. J. Nicholson, W. Wohlleben and R. D. Süssmuth, J. Am. Chem. Soc., 2004, 126, 5942–5943 CrossRef CAS PubMed.
- M. A. Fischbach, J. R. Lai, E. D. Roche, C. T. Walsh and D. R. Liu, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11951–11956 CrossRef CAS PubMed.
- K. Zhang, K. M. Nelson, K. Bhuripanyo, K. D. Grimes, B. Zhao, C. C. Aldrich and J. Yin, Chem. Biol., 2013, 20, 92–101 CrossRef CAS PubMed.
- L. G. Otten, M. L. Schaffer, B. R. M. Villiers, T. Stachelhaus and F. Hollfelder, Biotechnol. J., 2007, 2, 232–240 CrossRef CAS PubMed.
- H. Chen, B. Hubbard, S. E. O’Connor and C. T. Walsh, Chem. Biol., 2002, 9, 103–112 CrossRef CAS.
- D. J. Wilson and C. C. Aldrich, Anal. Biochem., 2010, 404, 56–63 CrossRef CAS PubMed.
- R. Finking, A. Neumüller, J. Solsbacher, D. Konz, G. Kretzschmar, M. Schweitzer, T. Krumm and M. A. Marahiel, ChemBioChem, 2003, 4, 903–906 CrossRef CAS PubMed.
- D. J. Wilson, C. Shi, A. M. Teitelbaum, A. M. Gulick and C. C. Aldrich, Biochemistry, 2013, 52, 926–937 CrossRef CAS PubMed.
- M. Miethke, P. Bisseret, C. L. Beckering, D. Vignard, J. Eustache and M. A. Marahiel, FEBS J., 2006, 273, 409–419 CrossRef CAS PubMed.
- J. A. Sundlov, C. Shi, D. J. Wilson, C. C. Aldrich and A. M. Gulick, Chem. Biol., 2012, 19, 188–198 CrossRef CAS PubMed.
- S. Konno, F. Ishikawa, T. Suzuki, N. Dohmae, M. D. Burkart and H. Kakeya, Chem. Commun., 2015, 51, 2262–2265 RSC.
- F. Ishikawa, S. Konno, T. Suzuki, N. Dohmae and H. Kakeya, ACS Chem. Biol., 2015 DOI:10.1021/acschembio.5b00097.
- D. Kato, K. M. Boatright, A. B. Berger, T. Nazif, G. Blum, C. Ryan, K. A. H. Chehade, G. S. Salvesen and M. Bogyo, Nat. Chem. Biol., 2005, 1, 33–38 CrossRef CAS PubMed.
- S. A. Sieber, S. Niessen, H. S. Hoover and B. F. Cravatt, Nat. Chem. Biol., 2006, 2, 274–281 CrossRef CAS PubMed.
- J. Kratzschmar, M. Krause and M. A. Marahiel, J. Bacteriol., 1989, 171, 5422–5429 CAS.
- A. E. Speers, G. C. Adam and B. F. Cravatt, J. Am. Chem. Soc., 2003, 125, 4686–4687 CrossRef CAS PubMed.
- H. D. Mootz and M. A. Marahiel, J. Bacteriol., 1997, 179, 6843–6850 CAS.
- T. Stachelhaus, H. D. Mootz and M. A. Marahiel, Chem. Biol., 1999, 6, 493–505 CrossRef CAS.
- G. L. Challis, J. Ravel and C. A. Townsend, Chem. Biol., 2000, 7, 211–224 CrossRef CAS.
- L. Lou, G. Qian, Y. Xie, J. Hang, H. Chen, K. Zaleta-Rivera, Y. Li, Y. Shen, P. H. Dussault, F. Liu and L. Du, J. Am. Chem. Soc., 2011, 133, 643–645 CrossRef CAS PubMed.
- G. Galli, F. Rodriguez, P. Cosmina, C. Pratesi, R. Nogarotto, F. de Ferra and G. Grandi, Biochim. Biophys. Acta, 1994, 1205, 19–28 CrossRef CAS.
- A. Elsner, H. Engert, W. Saenger, L. Hamoen, G. Venema and F. Bernhard, J. Biol. Chem., 1997, 272, 4814–4819 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supporting figures; procedures for the syntheses of aminoacyl-AMP analogs; complete gel images; full experimental details; and copies of 1H and 13C-NMR spectra. See DOI: 10.1039/c5cc04953a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |