Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorophore ATCUN complexes: combining agent and probe for oxidative DNA cleavage†‡
C.
Wende
and
N.
Kulak
*
Institut für Chemie und Biochemie, Freie Universität Berlin, Fabeckstr. 34/36, 14195 Berlin, Germany. E-mail: nora.kulak@fu-berlin.de; Fax: +49 30 838 454697; Tel: +49 30 838 54697
First published on 22nd June 2015
Abstract
DNA can be oxidatively cleaved by copper complexes of the ATCUN peptide (amino terminal Cu(II)- and Ni(II)-binding motif). In order to investigate the fate of the metal ion throughout this process, we have exploited quenching/dequenching effects of conjugated fluorophores.
The ATCUN motif has been studied for more than 50 years.1 During this time the understanding of this binding motif has evolved from a small metal binding site in natural proteins like albumin – therein used for the transport of ions in blood plasma – to effective DNA cleavers with antitumoral activity.2 The latter ones soon found their way into bioinorganic chemistry due to their simple synthesis and modification and high affinity towards Cu(II) and Ni(II) ions.3,4
The simplest peptide mimicking the ATCUN motif is the tripeptide Gly–Gly–His. Several characteristics of it have been studied: binding properties,3 RNA interaction,4,5 DNA cleavage activity,6–8in vitro behaviour.9 Variations of the amino acid sequence allow an increase in binding affinity towards Cu(II) ions as well as DNA and RNA as targets.7,10–12 Imperiali et al. were able to improve the selectivity of the peptide for Cu(II) ions by exchanging both glycines (Gly) by 2,3-diaminopropionic acid (Dap) and β-alanine (β-Ala), respectively. Thereby, the introduction of an amino group at the N-terminus rendered the coupling with functional molecules like fluorophores possible, and lead to the development of a selective Cu(II) fluorescent chemosensor.13
Based on this work we have demonstrated that the quenching of different fluorophores by Cu(II) ions in fluorophore ATCUN complex conjugates can be used for monitoring the fate of the metal ion during oxidative DNA cleavage. This new approach allows us to follow the cleavage process not only by conventional agarose gel electrophoresis but also by fluorescence, having function (nuclease) and reporter (fluorophore) combined in one system.
There is only one example in the literature where a Cu(II) based DNA cleaving agent was equipped with a fluorophore. In that case a ligand exhibiting fluorescence was used for monitoring cell uptake.14 As to the best of our knowledge, no system has been described so far where an artificial nuclease was linked to a reporter molecule for investigating the status of the nucleolytic metal ion. This could be of interest for gaining better understanding of such a metal initiated cleavage reaction. A reporter molecule being part of the agent itself is supposed to be more reliable than an external reporter due to spatial proximity.
The peptides 1a–3a (Fig. 1) with the sequence fluorophore R-Dap-β-Ala-His-Ser-Ser-CONH2 were synthesized manually on solid support using standard Fmoc strategy (ESI,‡ S-1).15 The yield was determined by UV/vis spectroscopy exploiting the extinction of the coupled fluorophores (rhodamine B in 1a, dansyl chloride in 2a, and fluorescein isothiocyanate (FITC) for 3a, ESI,‡ S-2).§
The sequence of peptide a was developed by Imperiali et al. with the aim of improving metal binding properties when compared to Gly–Gly–His.13a The exchange of N-terminal glycine by Dap was performed in order to allow coupling of a fluorophore. Imperiali et al. have shown that the fluorescence quenching of Cu(II) ions becomes more efficient when the linker length of the peptide backbone to the fluorophore decreases. Whereas, the introduction of two serine residues at the C-terminus serves the increase of water solubility of the peptide.13a
The corresponding Cu(II) complexes 1b–3b were synthesized according to a literature procedure by incubation of the peptides 1a–3a with copper(II) chloride in aqueous solution.6 Afterwards their ability to cleave DNA was tested.
It is known that Cu(II) complexes of peptides with the ATCUN motif are able to cut plasmid DNA in the presence of reducing agents like ascorbic acid (Asc).6 Starting from concentration-dependent gel electrophoresis experiments, we could show that the complexes 1b–3b cleave plasmid DNA pBR322 efficiently (Fig. 2 and ESI,‡ S-3).
As expected all complexes cleave DNA only in the presence of ascorbic acid (lane 2 in Fig. 2 and lanes 2, 4 and 6 in Fig. S3.1–S3.4 in ESI‡).6,7,12 The degree of cleavage increased with increasing complex concentration. A concentration as low as 25 μM was sufficient to cleave supercoiled plasmid DNA (form I) to at least 30% into the open circular form II. Complex 3b even lead to a double strand break of DNA at this concentration (form III). At a concentration of 75 μM all complexes cleaved plasmid DNA almost completely into form III, and even smaller fragments were observed. At a concentration as low as 1 μM 1b ∼10% of the DNA was cleaved (Fig. S3.5, ESI‡).
The differences in cleavage efficiency can be explained by the different bulk of the dyes and their linkers, respectively. Due to the length and characteristics of the different linkers (1b: amide, 2b: sulfonic amide, 3b: thiourea) a closer (1b) or more distant (3b) approach of the dye to the copper centre and thus a change in DNA interaction is expected. Also the orientation of the fluorophores might play a role. In comparison to the unfunctionalised Cu(II) peptide complex b (R = H) and Cu(II)–Gly–Gly–His cleavage activity of the fluorophore carrying peptides 1b–3b is somewhat lower (Fig. S3.6, ESI,‡ for Cu(II)–Gly–Gly–His see also the literature6).
In order to investigate whether an incubation time longer than one hour would result in a change in cleavage efficiency, DNA was incubated with complex 1b also for two and four hours (Fig. S3.7, ESI‡). An increase in incubation time lead to an increase in cleavage yield. When the incubation time was doubled, the amount of form II DNA decreased due to formation of form III and fragments (black line in Fig. S3.7, ESI‡), indicating a somewhat linear relationship.
Assuming the fluorescence properties of the dyes can be used for the indirect detection of DNA cleavage by sensing the catalytic Cu species, it is important to consider that the fluorescence of rhodamine B in TRIS-HCl buffer at pH 7.4 is quenched completely. In order to be able to exploit fluorescence properties in this case, one has to work at lower pH values.2 Thus, the general stability of the Cu(II) complexes at different pH values was studied with peptide 2a (Fig. S4.1, ESI‡). The absorption spectra of peptide 2a in the presence of 1 equivalent Cu(II) at pH 2–4 shows only a broad band at 800 nm for Cu(II) ions in water.17a The intensity of d–d transition of the Cu(II)-peptide complex at 525–550 nm increases with increasing pH value while the band for Cu(II) decreases. These data suggest that the ATCUN complexes are stable over a wide pH range.§
Based on previously published studies6 and based on the fact that DNA cleavage with the complexes 1b–3b happens only in the presence of ascorbic acid as a reductant, the cleavage mechanism of DNA is most likely oxidative. In order to investigate the reaction mechanism and identify potential ROS that are involved in the cleavage process 1b–3b were incubated with plasmid DNA and several literature-known ROS scavengers (Table 1). The quenching effects were the same for all complexes including Cu(II)–Gly–Gly–His suggesting that the mechanism of cleavage is the same for these compounds. The results are exemplarily shown for 1b in Fig. 3.
Our results show that DNA cleavage with the ATCUN peptide is most likely carried out by hydroxyl radicals (lanes 3 and 4) as well as peroxo species (lane 6) since cleavage is inhibited in the presence of tert-butanol, DMSO and catalase. This is in accordance with results obtained by Cowan et al.6 Additionally, sodium azide exhibits a strong quenching effect (lane 5), which indicates the presence of singlet oxygen. In order to exclude that this effect is due to a contamination in the azide solution, a control experiment was carried out. Therein, the complexes were incubated with DNA as before, however, this time in deuterated water (Fig. S3.8 for 1b, S3.9 for 2b and 3b, ESI‡). Singlet oxygen has a lifetime in H2O of approximately 2 μs.22 In D2O the lifetime is increased by factor 10. Therefore reactions depending on singlet oxygen are more distinctive in D2O.25 This is also true in this case, since DNA cleavage was 2–3 times stronger regarding the formation of linear DNA for the compounds tested in D2O, indicating the presence of singlet oxygen.
It is known from the literature that the fluorescence of several dyes can be quenched by paramagnetic Cu(II) ions.23 Imperiali et al. showed that the binding of these ions by the ATCUN motif leads to quenching of the fluorescence of a dansyl labelled peptide.13a The effect of Cu(II) complexation on the fluorescence of peptides 1a–3a was thus studied using fluorescence spectroscopy. A decrease of fluorescence was observed with increasing Cu(II) concentration with a rest fluorescence of about 14% at an 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio ligand
1 ratio ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu(II) (Fig. S5.1, ESI,‡ exemplarily 1a).
Cu(II) (Fig. S5.1, ESI,‡ exemplarily 1a).
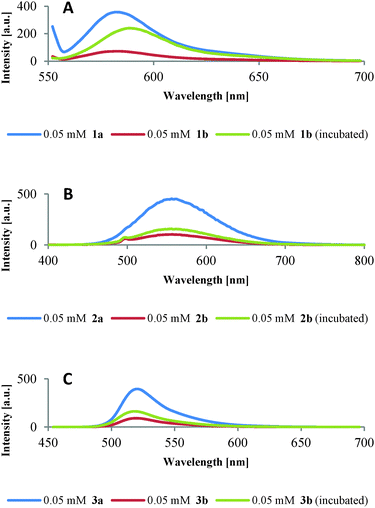
When complexes 1b–3b (Fig. 4, red lines) were incubated with plasmid DNA under gel electrophoresis conditions in the presence of ascorbate (Fig. 4, green lines), a reconstitution of fluorescence was observed indicating the disappearance of Cu(II) species. For comparison peptides 1a–3a are also shown (Fig. 4, blue lines).
During the course of these experiments, it is probable that Cu(I) ions and Cu-“oxo” type species like Cu-OOH and Cu-OH are produced. Since neither Cu(I) ions (d10 ion) nor Cu-“oxo” species lead to fluorescence quenching by energy transfer between donor and acceptor,23c,26 fluorescence is reconstituted.
Cleavage of DNA is supposed to happen when those Cu(I) ions are reoxidized by molecular oxygen leading to the generation of ROS (reactive oxygen species) (Scheme 1).6,11,24 Also the direct involvement of Cu-“oxo” species is feasible.
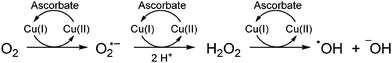 | ||
| Scheme 1 Mechanism of ROS production by Cu(II) ions in the presence of ascorbate.24 | ||
For Cu(II)–ATCUN complexes it was recently shown by cyclovoltammetric experiments that a Cu(III)/Cu(II) redox cycle might be involved in DNA cleavage instead.7,25 For investigation of the redox cycle in the present case the Cu(II)-peptide 2b was studied by NMR spectroscopy under an inert atmosphere in D2O-based TRIS-HCl buffer. The spectrum reveals typical Cu(II) paramagnetic species. After addition of ascorbic acid the NMR spectrum showed sharp, diamagnetic peaks (Fig. S6-1, ESI‡). It is thus very likely that Cu(I) is produced by reduction with ascorbic acid under the conditions applied. Furthermore, electrophoresis experiments in the presence of the Cu(I) specific chelator neocuproine showed an inhibition of cleavage activity by ATCUN complexes (Fig. S3.10, ESI‡). Both experiments are indications for the essential role of Cu(I) in DNA cleavage.
The incomplete reconstitution of fluorescence (Fig. 4) might be due to damage of the fluorescence dyes by ROS, as e.g. known for rhodamine B and hydroxyl radicals.20 Also, an interaction of the dyes with ascorbate is conceivable: incubation of 1b–3b with an excess of ascorbate as used in the samples for gel electrophoresis revealed a small quenching effect (Fig. S5.2, ESI‡). An on/off switching of the fluorescence of complexes 1b and 2b was observed by alternately titrating the reducing agent ascorbic acid and the oxidizing agent hydrogen peroxide with only a small loss of intensity in the second cycle (Fig. S5.3, ESI‡). Whereas for the dansyl labelled peptide 2b the switching is very distinct and might be considered a proof of concept, for the rhodamine B labelled peptide 1b the switching (at pH 5) is less pronounced. The reversibility of the process suggests the importance of Cu(II) quenching as against dye damaging by ascorbate, albeit the small loss in intensity throughout the experiment could be attributed to such a damage. The fluorophores can thus be used as indicators for the fate of Cu(II) ions responsible for DNA cleavage.
The three dyes applied in this study exhibit some advantages but also disadvantages concerning synthesis, fluorescence properties and stability (S-7, ESI‡). For example, hydroxyl radicals generated during the redox reactions assumed (Scheme 1), could damage FITC and rhodamine B after incubation times longer than 1 h (not shown).27 The experiments should thus be carried out within less than 1 h. Since short experimental times are already feasible, this does not pose any disadvantage.
In conclusion, investigation of DNA cleavage by agarose gel electrophoresis showed that fluorophore-carrying peptides 1a–3a coordinated to Cu(II) (1b–3b) are efficient DNA cleaving agents in the presence of a reducing agent. Plasmid DNA is cleaved even at very low concentrations. Due to the linked sterically bulky dyes and thus the hindered approach of the complexes to DNA, our system is, however, less efficient under very similar conditions than the Cu(II) complexes of Gly–Gly–His, Lys–Gly–His–Lys and the unfunctionalised peptide a (in the case of Lys–Gly–His–Lys the positive charge of the lysine side chains supports interaction with DNA6). Still, the cleavage activity of 1b–3b, which is in the low micromolar range (cf. Fig. S3.5, ESI‡), is close to the one of Cu(phen)2, the first reported and one of the most efficient oxidative chemical nucleases.28 Experiments regarding the mechanism of DNA cleavage lead to the conclusion that DNA is cleaved oxidatively by hydroxyl radicals, peroxo species and singlet oxygen.
By spectroscopic means (fluorescence, NMR) we demonstrated that Cu(II) is reduced to Cu(I) which is required to reduce O2 to superoxide and subsequently produces H2O2. Hydrogen peroxide can then interact with Cu(I) to form metal-oxo species and hydroxyl radicals that induce DNA cleavage (Scheme 1).24 Quenched fluorescence of the Cu(II)-coordinated peptides 1b–3b is regained when the Cu(II) ions react during this process. These new complexes not only initiate cleavage of DNA, but at the same time display changes of the oxidation state of the involved Cu(II) ions. Thus, our system simultaneously comprises a DNA cleaving agent and a redox-sensitive probe. After this proof of concept application of the herein described Cu(II) fluorescent peptides in cell experiments is conceivable (except for 1b which requires working at pH 5).
The authors would like to thank Biprajit Sarkar for helpful discussions. Simone Friedle, Maximilian Wende and the Kulak Group are acknowledged for proofreading the manuscript.
Notes and references
- T. Peters Jr., Biochim. Biophys. Acta, 1960, 39, 546–547 CrossRef CAS.
- E. Kimoto, H. Tanaka, J. Gyotoku, F. Morishige and L. Pauling, Cancer Res., 1983, 43, 824–828 Search PubMed; Z. Yu, M. Han and J. A. Cowan, Angew. Chem., Int. Ed., 2015, 54, 1901–1905 CrossRef CAS PubMed.
- S.-J. Lau, T. P. A. Kruck and B. Sarkar, J. Biol. Chem., 1974, 249, 5878–5884 CAS.
- E. C. Long, Acc. Chem. Res., 1999, 32, 827–836 CrossRef CAS.
- Y. Jin and J. A. Cowan, J. Biol. Inorg. Chem., 2007, 12, 637–644 CrossRef CAS PubMed.
- Y. Jin and J. A. Cowan, J. Am. Chem. Soc., 2005, 127, 8408–8415 CrossRef CAS PubMed.
- Y. Jin, M. A. Lewis, N. H. Gokhale, E. C. Long and J. A. Cowan, J. Am. Chem. Soc., 2007, 129, 8353–8361 CrossRef CAS PubMed.
- J. C. Joyner, J. Reichfield and J. A. Cowan, J. Am. Chem. Soc., 2011, 133, 15613–15626 CrossRef CAS PubMed.
- A. C. Matias, N. Villa dos Santos, R. Chelegão, C. S. Nomura, P. A. Fiorito and G. Cerchiaro, J. Inorg. Biochem., 2012, 116, 172–179 CrossRef CAS PubMed.
- J. C. Joyner, W. F. Hodnick, A. S. Cowan, D. Tamuly, R. Boyd and J. A. Cowan, Chem. Commun., 2013, 49, 2118–2120 RSC; S. Bradford, Y. Kawarasaki and J. A. Cowan, J. Inorg. Biochem., 2009, 103, 871–875 CrossRef CAS PubMed.
- C. Harford and B. Sarkar, Acc. Chem. Res., 1997, 30, 123–130 CrossRef CAS.
- D. F. Shullenberger, P. D. Eason and E. C. Long, J. Am. Chem. Soc., 1993, 115, 11038–11039 CrossRef CAS.
- A. Torrado, G. K. Walkup and B. Imperiali, J. Am. Chem. Soc., 1998, 120, 609–610 CrossRef CAS; T. R. Young, C. J. K. Wijekoon, B. Spyrou, P. S. Donnelly, A. G. Wedd and Z. Xiao, Metallomics, 2015, 7, 567–578 RSC.
- S. S. Bhat, A. A. Kumbhar, H. Heptullah, A. A. Khan, V. V. Gobre, S. P. Gejji and V. G. Puranik, Inorg. Chem., 2011, 50, 545–558 CrossRef CAS PubMed.
- S. I. Kirin, F. Noor, N. Metzler-Nolte and W. Mier, J. Chem. Educ., 2007, 84, 108–111 CrossRef CAS.
- L. Fülöp, B. Penke and M. Zarándi, J. Pept. Sci., 2001, 7, 397–401 CrossRef PubMed.
- H. Korpi, P. J. Figiel, E. Lankinen, P. Ryan, M. Leskelä and T. Repo, Eur. J. Inorg. Chem., 2007, 2465–2471 CrossRef CAS PubMed; K. P. Neupane, A. R. Aldous and J. A. Kritzer, Inorg. Chem., 2013, 52, 2729–2735 CrossRef PubMed.
- M. von Piechowski, M.-A. Thelen, J. Hoigné and R. E. Bühler, Ber. Bunsen-Ges. Phys. Chem., 1992, 96, 1448–1454 CrossRef CAS PubMed.
- E. L. Hegg and J. N. Burstyn, Inorg. Chem., 1996, 35, 7474–7481 CrossRef CAS.
- A. Sreedhara, J. D. Freed and J. A. Cowan, J. Am. Chem. Soc., 2000, 122, 8814–8824 CrossRef CAS.
- S.-H. Chiou, J. Biochem., 1983, 94, 1259–1267 CAS.
- J. G. Parker and W. D. Stanbro, J. Photochem., 1984, 25, 545–547 CrossRef.
- G. K. Rollefson and R. W. Stoughton, J. Am. Chem. Soc., 1941, 63, 1517–1520 CrossRef CAS; W.-Y. Lin and H. E. van Wart, J. Inorg. Biochem., 1988, 32, 21–38 CrossRef; J. Brunner and R. Krämer, J. Am. Chem. Soc., 2004, 126, 13626–13627 CrossRef PubMed.
- L. Guilloreau, S. Combalbert, A. Sournia-Saquet, H. Mazarguil and P. Faller, ChemBioChem, 2007, 8, 1317–1325 CrossRef CAS PubMed.
- C. Hureau, H. Eury, R. Guillot, C. Bijani, S. Sayen, P.-L. Solari, E. Guillon, P. Faller and P. Dorlet, Chem. – Eur. J., 2011, 17, 10151–10160 CrossRef CAS PubMed.
- L. Fabbrizzi, M. Licchelli, P. Pallavicini, A. Perotti, A. Taglietti and D. Sacchi, Chem. – Eur. J., 1996, 2, 75–82 CrossRef CAS PubMed; G. De Santis, L. Fabbrizzi, M. Licchelli, C. Mangano and D. Sacchi, Inorg. Chem., 1995, 34, 3581–3582 CrossRef.
- S. Feng, X. Chen, J. Fan, G. Zhang, J. Jiang and X. Wei, Anal. Lett., 1998, 31, 463–474 CrossRef CAS PubMed; J. Fan, H.-Q. Guo and S.-L. Feng, J. Fluoresc., 2007, 17, 257–264 CrossRef PubMed.
- J. M. Veal, K. Merchant and R. L. Rill, Nucleic Acids Res., 1991, 19, 3383–3388 CrossRef CAS PubMed.
Footnotes |
| † Dedicated to Stephen J. Lippard on the occasion of his 75th birthday. |
| ‡ Electronic supplementary information (ESI) available: Experimental section, determination of peptide yields, DNA cleavage, UV/vis, fluorescence and NMR experiments, properties of fluorophores. See DOI: 10.1039/c5cc04508h |
| § There is indication from the literature that the thiourea bond (as in peptide 3a) is not stable during cleavage from the solid support with TFA (trifluoroacetic acid).16 Under the conditions chosen, however, the coupling of FITC on solid support was possible, though the yield was low. Whereas the basic ATCUN sequence does not bind Cu(II) at pH values lower than 6,11,17 at pH 5 the rhodamine B labelled peptide 2a shows Cu(II) binding as well as fluorescence emission. This allows application of the presented concept also at non-physiological pH values. |
| This journal is © The Royal Society of Chemistry 2015 |