Open Access Article
Open Access ArticleInnovative use of the taxol binding peptide overcomes key challenges of stable and high drug loading in polymeric nanomicelles†
Jennifer
Logie
ab,
Christopher K.
McLaughlin
ab,
Roger Y.
Tam
ab and
Molly S.
Shoichet
*abc
aDepartment of Chemical Engineering and Applied Chemistry, University of Toronto, 160 College St, Toronto, ON M5S3E1, Canada. E-mail: molly.shoichet@utoronto.ca
bInstitute for Biomaterials and Biomedical Engineering, University of Toronto, 160 College St, Toronto, ON M5S3E1, Canada
cDepartment of Chemistry, University of Toronto, 160 College St, Toronto, ON M5S3E1, Canada
First published on 23rd June 2015
Abstract
Despite widespread clinical use, delivery of taxane chemotherapeutics remains a challenge due to poor solubility and lack of selectively. Polymeric nanomicelle strategies have been pursued to overcome these issues; however current formulations are often limited by low drug loading and poor serum stability. To achieve a drug delivery system that addresses these issues, poly(D,L-lactide-co-2-methyl-2-carboxytrimethylene carbonate)-g-poly(ethylene glycol) was covalently modified with the taxol binding peptide – a peptide from the β-tubulin-taxane binding site. This modification resulted in drug loadings five times higher than unmodified polymers, which is significantly higher than typical hydrophobic modifications, including with benzyl and docetaxel functionalization. Unlike many formulations with high drug loading, these nanomicelles were stable in serum for up to 24 h and maintained docetaxel cytotoxicity. By incorporating the taxane binding peptide into the polymer chemistry, a new twist was applied to an old problem, which is broadly applicable to other polymeric micelle systems and drug-peptide combinations in general.
Taxanes, such as docetaxel (DTX) and paclitaxel, are used clinically against a wide range of cancers including breast, lung and pancreatic, and are one of the most prescribed cancer chemotherapeutics. Docetaxel is arguably the biggest oncology product ever developed, with a global market value of over $3 billion in 2010,1 and sales of Taxotere (Sanofi) at $500 million in 2013 even after being off-patent for three years.2 Despite their widespread use, delivery remains a challenge. Current formulations are poorly soluble and thus require the use of excipients, such as Polysorbate 80 or Cremophor EL, that cause a number of side effects such as hypersensitivity and hemolysis.3,4 Furthermore, these potent drugs lack specificity, and cause a variety of dose-limiting side effects such as neutropenia and thrombocytopenia.5
Polymeric nanoparticle micelles, comprised of a hydrophobic core and hydrophilic corona, have been widely investigated to improve the pharmacokinetics of taxanes; however, low drug encapsulation and limited serum stability have hindered their clinical translation.6–11 To address these issues, both the formulation and the affinity of the drug in the micelle have been pursued. On the one hand, while techniques such as nanoprecipitation have been shown to produce high drug loadings, encapsulation is transient and often comes at the cost of micelle stability.12 Modifications to the polymer core, on the other hand, have shown great promise at increasing drug loading without jeopardizing the stability of the delivery vehicle itself.13,14
Several strategies have been pursued to improve loading in polymeric nanomicelles. Traditionally, increasing the ratio of hydrophobic to hydrophilic polymer block lengths has improved loading efficiency;15 however, this reduces the amount of shielding and often results in rapid micelle dissociation upon dilution.16,17 Moreover, high drug loading often comes at the expense of stability in serum conditions, resulting in nanomicelles that release drug prematurely.18,19 More specific chemical modifications have shown increased loading without hindering the stability of the vehicles. For example, Hennink et al. increased taxane loading by covalently bonding aromatic groups to the core that facilitated pi-pi stacking between drug and polymer.9 Yang et al. exploited hydrophobic interactions by incorporating cholesterol onto a polycarbonate backbone to increase paclitaxel loading.11 Others have explored direct drug conjugation to the hydrophobic block of an amphiphilic polymer, which increased loading of free drug by inducing crystallinity within the core.8,20,21 While these systems resulted in loadings greater than those previously achieved, even higher loadings are required to achieve optimal dosing and overcome excessive use of excipients required for solubility and stability in serum. To this end, we designed a core modification in our polymeric nanomicelles that is specific to taxanes, thereby using a known structural motif in a new way to solve a problem that has been plaguing the field for decades.
Taxanes achieve their potency by interacting specifically with β-tubulin and arresting cell division. The native binding site of the taxanes has been identified as a specific taxol binding peptide (TBP) sequence, PGFAPLTSRGSQQYAA, on the M-loop of β-tubulin.22,23 We hypothesized that incorporation of this taxol binding peptide (TBP) into the hydrophobic backbone of our polymeric nanomicelles would enhance docetaxel loading without compromising serum stability. To test this hypothesis, we covalently modified the carboxylic acid functional groups of poly(D,L-lactide-co-2-methyl-2-carboxy-trimethylene carbonate)-graft-poly(ethylene glycol), P(LA-co-TMCC)-g-PEG, with TBP and compared its drug loading to more common hydrophobic modification strategies, including the use of benzyl and docetaxel groups (Fig. 1). Polymer-TBP (PTBP) nanomicelles were further investigated for serum stability and in vitro cytotoxicity.
To modify P(LA-co-TMCC) carboxylic acids, a series of strategies were exploited. To achieve the benzylated polymer backbone, the palladium catalyzed hydrogenolysis conditions for TMCC benzyl deprotection were controlled (Scheme S1, ESI†). The highest degree of substitution achieved was six benzyl groups per backbone, representing 50% of the TMCC monomers or 5% of all monomers (see 1H-NMR in Fig. S1, ESI†). To synthesize the docetaxel modified polymer backbone, Steglich esterification conditions were used whereby the 2′ hydroxyl groups of DTX were coupled to the P(LA-co-TMCC) carboxylic acids by carbodiimide chemistry (Scheme S2, ESI†).21 An average of 2.5 DTX per backbone was achieved, as calculated by 1H-NMR (Fig. S2, ESI†). P(LA-co-TMCC), with either benzyl (PBn) or docetaxel (PDTX) modifications, was subsequently modified by grafting terminally functionalized MeO-PEG-NH2 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) to the P(LA-co-TMCC) carboxylic acids backbone using N,N′-diisopropylcarbodiimide (DIC) and hydroxybenzotriazole (HOBt) coupling, followed by purification through a Sepharose CL4B column to remove unreacted PEG. Three PEG chains per backbone were grafted in all cases to achieve high stability.24
000 g mol−1) to the P(LA-co-TMCC) carboxylic acids backbone using N,N′-diisopropylcarbodiimide (DIC) and hydroxybenzotriazole (HOBt) coupling, followed by purification through a Sepharose CL4B column to remove unreacted PEG. Three PEG chains per backbone were grafted in all cases to achieve high stability.24
An alternate synthetic route was required to covalently modify the P(LA-co-TMCC) carboxylic acids with TBP due to potential cross-reactivity of the amino acid side chains. Here, the backbone carboxylic acid groups were first modified with 3,3′-dithiobis(propionic dihydrazide) (DTP)25,26 using DIC and HOBt as coupling agents, thereby introducing protected thiol functional groups, followed by MeO-PEG-NH2 grafting, as described above. After purification, reduction of the disulfides using dithiolthreitol (DTT) produced a thiolated polymer, which was then reacted with maleimide-functionalized TBP by a Michael addition reaction to produce PTBP with an average of 0.8 peptides per backbone (Scheme S3 and Fig. S3, ESI†). Unreacted thiols were quenched with N-(2-hydroxyethyl)-maleimide. All modified polymers, represented in Fig. 1, self-assembled by dialysis to form uniform polymeric micelles with diameters <200 nm and polydispersity indexes <0.2, as determined by dynamic light scattering (DLS, Table S1, ESI†). Additionally, PTBP micelles were further characterized by transmission electron microscopy (TEM), exhibiting a spherical shape and size under dehydrated conditions of 36 ± 11 nm, and a zeta potential of −2.41 ± 0.06 mV (Fig. 2 and Fig. S4, ESI†).
 | ||
| Fig. 2 Characterization of PTBP micelles show: (a) narrow distribution of 0.15 and diameter of 108 nm by DLS and (b) representative fields of view obtained by TEM (scale bar is 50 nm). | ||
To investigate differences in drug loading between modified polymers, micelles were formulated by dialysis with docetaxel, and the encapsulated drug was quantified using high performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS). To eliminate differences due to changes in molar mass, and thus the hydrophilic-hydrophobic ratio after chemical modification, all loadings were normalized to the mass of the hydrophobic backbone.
As shown in Fig. 3, all of the polymer modifications formed nanomicelles with significantly higher docetaxel loadings compared to the unmodified polymer (p < 0.05, Fig. 3). PBn and PDTX showed loadings consistent with those observed with other polymer systems, including polycaprolactone-PEG and polyhydroxypropyl methacrylamide-PEG.9,21 Impressively, PTBP formed micelles with loadings significantly higher than all other hydrophobically-modified formulations (p < 0.05) and five times higher than the control (49 ± 14%, p < 0.001), suggesting a high affinity of the drug with the peptide-polymer core.
To further explore whether the increased docetaxel loading in the PTBP formulation was due to a specific multivalent interaction or simply due to increased hydrophobicity, we synthesized a scrambled peptide sequence that had the same overall hydrophobicity and isoelectric point (of 9.34) as the TBP, but with the taxane-specific PLTSR amino acid sequence scrambled. In addition, the phenylalanine residue, which is known to interact with the 3′ phenyl ring of docetaxel, was scrambled within the peptide sequence. Notably, cell lines with point mutations in the microtubule at either the phenylalanine or within PLTSR are DTX resistant.27,28 Interestingly, the scrambled peptide–polymer conjugate (PSCR) showed significantly less encapsulated docetaxel than PTBP and only a modest increase in drug loading relative to unmodified polymer controls (two times higher, Fig. 3), which is similar to that of PBn and PDTX. Given that hydrophobic interactions in general can increase loading and we specifically maintained a similar hydrophobicity in the scrambled vs. normal taxol binding peptide, we attribute this modest increase in loading in PSCR nanomicelles to general hydrophobic interactions with the drug, similar to those observed with PBn and PDTX. Importantly, these data suggest that a specific interaction between TBP and docetaxel accounts for the significantly greater docetaxel loading in PTBP.
The high loading of the PTBP suggests multivalent interactions within the core, in which binding of the peptide to the drug facilitates drug-drug stacking. Characterizing the specific interaction between the taxol binding peptide and the drug is an analytical challenge due to the poor solubility of both the docetaxel and the peptide. Classic techniques for measuring affinity, such as isothermal titration calorimetry, require aqueous experimental conditions that are not achievable with these molecules. Organic solvents required to solubilize the binding partners in techniques such as 1H NMR neither reflects the native environment within the polymeric nanomicelle nor the cell, and have been reported to cause a change in the conformation of both short peptide sequences and docetaxel itself.29,30
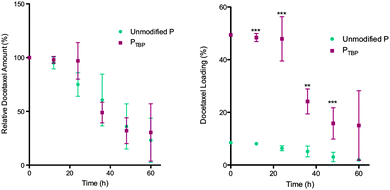
As higher drug loadings can result in reduced micelle stability in the presence of serum proteins,6 we were particularly interested in characterizing the stability of these high docetaxel loaded nanomicelles. The reduction in stability is typically associated with protein adsorption onto the nanoparticle surface which can cause premature drug release due to a partitioning effect between the hydrophobic core and hydrophobic pockets of the proteins.31 This lack of stability limits in vivo efficacy and is evident with in vitro studies. To determine whether the high docetaxel loading observed with PTBP alters micelle stability, we investigated their in vitro serum stability relative to unmodified polymeric nanomicelles. Nanomicelles were incubated with fetal bovine serum (FBS) in phosphate buffered saline (PBS, 1X) at 37 °C.1,24 At 0, 24 and 48 h, an aliquot of the nanomicelle solution was taken and separated from serum proteins using fast protein liquid chromatography (FPLC). The docetaxel in the nanomicelle fraction was quantified using HPLC-MS/MS. The compiled data in Fig. 4 is presented: (a) as docetaxel amount that is encapsulated, relative to time zero, as a function of time; and (b) as absolute drug loading over time, assuming no polymer is lost to protein adsorption. Drug loaded PTBP nanomicelles showed no significant differences (p = 0.89) in docetaxel release compared to the unmodified polymer (Fig. 4a), demonstrating that the higher docetaxel loaded PTBP nanomicelles were as stable in serum as the unmodified formulations. This is noteworthy because nanomicelles loaded with more chemotherapeutic are often less stable in serum.19,32,33 Moreover, the absolute drug loading was higher at all time points up to 48 h in PTBP nanomicelles (Fig. 4b) and remained relatively unchanged up to 24 h. Importantly, stable drug loadings in serum solutions for 24 h is considered long, and provides sufficient time for tumour accumulation in vivo.1 After 24 h, a decrease in docetaxel is observed (to ∼35% at 48 h), suggesting release of payload due to micelle dissociation.24 Importantly, these data show that the increased drug loading observed with PTBP micelles does not come at the expense of kinetic nanomicelle stability, which is key to ultimate use in vivo.
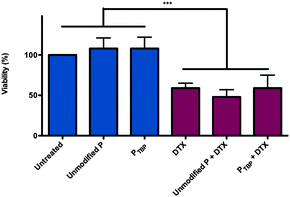
Polymeric cytocompatibility and docetaxel-loaded nanomicelle cytotoxicity are both critical for future in vivo applications. To demonstrate the cytocompatibility of the PTBP (without encapsulated docetaxel) and the cytotoxicity of docetaxel loaded PTBP micelles, polymeric nanomicelles were incubated with the human epithelial breast cancer line, SKBR-3 cells. Cell viability was assessed after 48 h and normalized to untreated cells (Fig. 5).
Importantly, both unmodified polymeric nanomicelles and PTBP nanomicelles, without encapsulated docetaxel, showed no cytotoxicity relative to untreated cells, demonstrating cytocompatibility of both polymers. Since free DTX is highly toxic with an IC50 of 10 ng mL−1 (Fig. S5, ESI†), it was used as a positive control to test the cytotoxicity of DTX when loaded in both unmodified and PTBP nanomicelles. Using dose-matched controls, we observed no significant difference between DTX and the encapsulated DTX, suggesting that encapsulation did not impede the mechanism of action of the drug within the cell. Importantly, all DTX formulations were cytotoxic compared to controls.
Achieving high drug loading while maintaining both serum stability and cytocompatibility are critical for ultimate use as clinically relevant polymeric nanomicelle formulations. Here we show, for the first time, that by incorporating the natural binding site of the drug with the β-tubulin peptide into our polymeric nanomicelle design, we achieve enhanced loading without jeopardizing either kinetic serum stability or drug toxicity. This strategy has broad applicability to other polymeric systems. The affinity demonstrated with P(LA-co-TMCC)-g-PEG is not specific to this polymer, but rather to the taxol binding peptide and taxanes. By incorporating this peptide onto other polymer scaffolds, the delivery of taxanes chemotherapeutics can be enhanced. Furthermore, a rational design approach using known peptide mimetics could form the basis for the encapsulation of other drugs,34 thereby overcoming key limitations of high drug loading (without the usual loss of serum stability) of polymeric nanomicelles.
We thank Professor Mitchell Winnik and Joey Hang Zhou (University of Toronto) for use of their GPC, Mr Steven Doyle and Mr Battista Calvieri (University of Toronto Microscopy Imaging Laboratory) for assistance with TEM imaging, and Pearson Wu who helped with benzyl deprotection experiments. We wish to acknowledge the Canadian Foundation for Innovation and the Ontario Research Fund for funding of the Centre for Spectroscopic Investigation of Complex Organic Molecules and Polymers, and Dr Darcy Burns (CSICOMP) who helped us with NMR. We are grateful for partial funding from the Natural Sciences and Engineering Research Council of Canada (MSS, PGSD to JL and PDF to CKM) and the Canadian Institutes of Health Research (MSS).
Notes and references
- X. Zhao, Z. Poon, A. C. Engler, D. K. Bonner and P. T. Hammond, Biomacromolecules, 2012, 13, 1315–1322 CrossRef CAS PubMed.
- Sanofi, 2014, http://en.sanofi.com/Images/38294_20150205_Results_en.pdf.
- A. K. Singla, A. Garg and D. Aggarwal, Int. J. Pharm., 2002, 235, 179–192 CrossRef CAS.
- R. B. González, J. Huwyler and F. Boess, Biopharm. Drug Dispos., 2004, 25, 37–49 CrossRef PubMed.
- R. Herbst and F. Khuri, Cancer Treat. Rev., 2003, 29, 407–415 CrossRef CAS.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, Mol. Pharmaceutics, 2008, 5, 505–515 CrossRef CAS PubMed.
- K. Park, ACS Nano, 2013, 7, 7442–7447 CrossRef CAS PubMed.
- A. S. Mikhail and C. Allen, Biomacromolecules, 2010, 11, 1273–1280 CrossRef CAS PubMed.
- Y. Shi, M. J. van Steenbergen, E. A. Teunissen, L. Novo, S. Gradmann, M. Baldus, C. F. van Nostrum and W. E. Hennink, Biomacromolecules, 2013, 14, 1826–1837 CrossRef CAS PubMed.
- R. R. Sawant and V. P. Torchilin, Int. J. Pharm., 2009, 374, 114–118 CrossRef CAS PubMed.
- A. L. Z. Lee, S. Venkataraman, S. B. M. Sirat, S. Gao, J. L. Hedrick and Y. Y. Yang, Biomaterials, 2012, 33, 1921–1928 CrossRef CAS PubMed.
- S. M. D'Addio and R. K. Prud'homme, Adv. Drug Delivery Rev., 2011, 63, 417–426 CrossRef PubMed.
- C. K. McLaughlin, J. Logie and M. S. Shoichet, Isr. J. Chem., 2013, 53, 670–679 CAS.
- X. Ke, V. W. L. Ng, R. J. Ono, J. M. W. Chan, S. Krishnamurthy, Y. Wang, J. L. Hedrick and Y. Y. Yang, J. Controlled Release, 2014, 193, 9–26 CrossRef CAS PubMed.
- M. Elsabahy, M. Dufresne and J. Leroux, Handbook of Materials for Nanomedicine, 2011 Search PubMed.
- M. G. Carstens, C. Rijcken, C. F. Nostrum and W. E. Hennink, in Multifunctional Pharmaceutical Nanocarriers, ed. V. Torchilin, Springer, New York, 2008, ch. 9, vol. 4, pp. 263–308 Search PubMed.
- D. Sutton, S. Wang, N. Nasongkla, J. Gao and E. E. Dormidontova, Exp. Biol. Med., 2007, 232, 1090–1099 CrossRef CAS PubMed.
- J. Liu, P. Zahedi, F. Zeng and C. Allen, J. Pharm. Sci., 2008, 97, 3274–3290 CrossRef CAS PubMed.
- P. Couvreur, Adv. Drug Delivery Rev., 2013, 65, 21–23 CrossRef CAS PubMed.
- T. Nakanishi, S. Fukushima, K. Okamoto, M. Suzuki, Y. Matsumura, M. Yokoyama, T. Okano, Y. Sakurai and K. Kataoka, J. Controlled Release, 2001, 74, 295–302 CrossRef CAS.
- M. Shahin and A. Lavasanifar, Int. J. Pharm., 2010, 389, 213–222 CrossRef CAS PubMed.
- S. Rao, L. He, S. Chakravarty, I. Ojima, G. A. Orr and S. B. Horwitz, J. Biol. Chem., 1999, 274, 37990–37994 CrossRef CAS PubMed.
- J. P. Snyder, J. H. Nettles, B. Cornett, K. H. Downing and E. Nogales, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5312–5316 CrossRef CAS PubMed.
- J. Logie, S. C. Owen, C. K. McLaughlin and M. S. Shoichet, Chem. Mater., 2014, 26, 2847–2855 CrossRef CAS.
- X. Z. Shu, Y. Liu, Y. Luo, M. C. Roberts and G. D. Prestwich, Biomacromolecules, 2002, 3, 1304–1311 CrossRef CAS PubMed.
- K. Vulic and M. S. Shoichet, J. Am. Chem. Soc., 2012, 134, 882–885 CrossRef CAS PubMed.
- J. J. Field, J. F. Díaz and J. H. Miller, Chem. Biol., 2013, 20, 301–315 CrossRef CAS PubMed.
- T. Hara, K. Ushio, M. Nishiwaki, J. Kouno, H. Araki, Y. Hikichi, M. Hattori, Y. Imai and M. Yamaoka, Cell Biol. Int., 2010, 34, 177–184 CrossRef CAS PubMed.
- H. J. Williams, A. I. Scott, R. A. Dieden, C. S. Swindell, L. E. Chirlian, M. M. Francl, J. M. Heerding and N. E. Krauss, Can. J. Chem., 1994, 72, 252 CrossRef CAS.
- P. E. Wright, H. J. Dyson and R. A. Lerner, Biochemistry, 1988, 27, 7167–7175 CrossRef CAS.
- J. Liu, F. Zeng and C. Allen, J. Controlled Release, 2005, 103, 481–497 CrossRef CAS PubMed.
- K. M. Huh, S. C. Lee, Y. W. Cho, J. Lee, J. H. Jeong and K. Park, J. Controlled Release, 2005, 101, 59–68 CrossRef CAS PubMed.
- H. Burt, X. Zhang, P. Toleikis, L. Embree and W. L. Hunter, Colloids Surf., B, 1999, 16, 161–171 CrossRef CAS.
- S. Wieczorek, E. Krause, S. Hackbarth, B. Röder, A. K. H. Hirsch and H. G. Börner, J. Am. Chem. Soc., 2013, 135, 1711–1714 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5cc04282h |
| This journal is © The Royal Society of Chemistry 2015 |