 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Small molecule binding to a G-hairpin and a G-triplex: a new insight into anticancer drug design targeting G-rich regions†
Arivazhagan
Rajendran‡
a,
Masayuki
Endo
*bc,
Kumi
Hidaka
a,
Marie-Paule
Teulade-Fichou
d,
Jean-Louis
Mergny
e and
Hiroshi
Sugiyama
*abc
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: hs@kuchem.kyoto-u.ac.jp
bInstitute for Integrated Cell-Material Sciences (WPI-iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: endo@kuchem.kyoto-u.ac.jp
cCREST, JST, Sanbancho, Chiyoda-ku, Tokyo 102-0075, Japan
dInstitut Curie, UMR 176 CNRS, Campus Universitaire Paris-Sud, 91405 Orsay, France
eUniv. Bordeaux, INSERM, U869, ARNA Laboratory, 2 rue Robert Escarpit, Pessac, F-33607, France
First published on 13th April 2015
Abstract
To gain new insights into G-quadruplex–drug interactions, we captured the solution-state structures of the complexes between a drug-like small molecule and a G-hairpin/G-triplex. Our results indicated that the ligand initially binds to the intermediates and induces stepwise folding into a quadruplex.
G-quadruplex-forming sequences are attractive therapeutic targets because of the high abundance of G-rich DNA regions in the human genome. Among the G-rich regions, telomeres have attracted particular interest as anticancer therapy targets because these regions are responsible for the stability of the human chromosome. Telomere is a repetitive nucleotide sequence of TTAGGG that has a duplex region followed by single-stranded 3′-overhang of up to a few hundred nucleobases at each end of a chromosome.1 It is a noncoding region that protects the genetic data, making it possible for cells to divide, and it plays a vital role in both aging and cancer. It is composed of a nucleoprotein complex called shelterin, which protects the telomere ends from the DNA-damage-response machinery.2 G-quadruplex-based anticancer therapy depends on the development and application of drugs that can replace the shelterin complex competitively by inducing the quadruplex structure and thereby lead to telomere dysfunction, which culminates in cell death.3 Several quadruplex binders of natural origin4–6 and synthetic molecules7–10 or metal complexes11 have been recognized. Their selectivity towards the quadruplex over B-form DNA was studied under bulk conditions. However, to the best of our knowledge, the molecular mechanism of quadruplex–drug interaction has never been investigated either experimentally or by theoretical methods. Thus, the interaction between the quadruplex-binders/chaperones and the quadruplex intermediates such as G-hairpins and G-triplexes is still unknown at the molecular level. Indeed, even theoretical discussions on this aspect of drug binding to these in-pathway intermediates are notably absent from the literature. This is mainly because the involvement of such intermediates in the folding process of a quadruplex is still not well understood. There have been an increasing number of experimental12–20 and theoretical21–25 studies on these intermediates in quadruplex folding; however, these studies only assumed their involvement. We have recently demonstrated the direct and single-molecule visualization of the solution-state structures of these intermediates (induced by salt20) using DNA origami26–30 and atomic force microscopy (AFM).31–36 Here, we present new insights into the binding of drug–quadruplex intermediates and propose the molecular mechanisms involved in the drug-induced quadruplex folding. By using our single-molecule method, we present the structures of drug-intermediate complexes with nanometer resolution. Other methods failed to provide concrete evidence on the presence of these intermediates and could not even hypothesize such drug-intermediate interactions. The ligand that was used for our studies was bisquinolinium pyridine dicarboxamide bearing a linker terminated by biotin (PDC-biotin, Fig. 1a).37
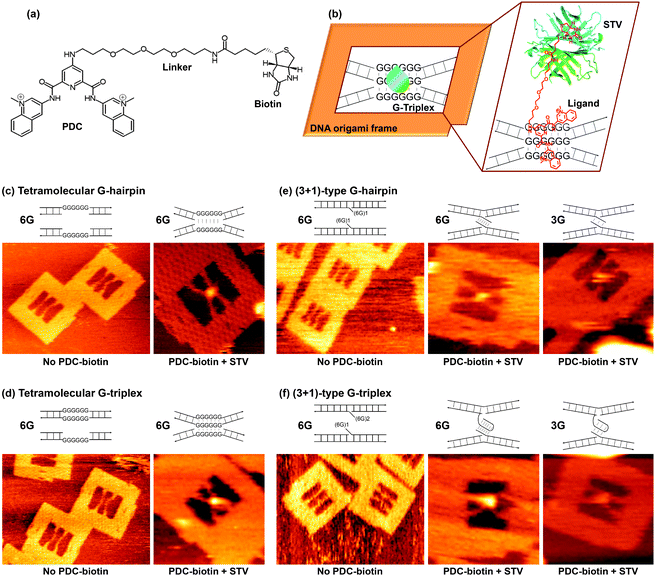 | ||
| Fig. 1 (a) Line drawing of the structure of the PDC-biotin ligand. (b) Schematic explanation of the DNA origami method for the analysis of ligand-induced formation of intermediate structures. The binding of STV to the biotin present in the ligand is also shown. Representative zoom-in AFM images of the DNA origami frame with incorporated duplexes for the tetramolecular antiparallel (c and d) and (3 + 1)-type (e and f) structures. The bright spot at the middle of X-shape indicates the STV-biotin present in the ligand binding. For comparison, the images recorded in the absence of a ligand are taken from our parallel study, ref. 20. [M13mp18] = 10 nM; [Staples] = 40 nM; [Tris-HCl] = 20 mM, pH 7.6; [MgCl2] = 5 mM (no ligand) or 10 mM (with ligand); [EDTA] = 1 mM; [KCl] = 0 mM; [PDC-biotin] = 1 μM; [STV] = 0.2 μM. Image size: 300 × 225 nm (no ligand) and 125 × 125 nm (with ligand). | ||
A DNA origami frame (Fig. 1b)28,29 was used to structurally and stoichiometrically control the DNA sequences,38 and the drug-intermediate binding for tetramolecular antiparallel and (3 + 1)-type structures (in which three G-tracts are present in one strand and one G-tract in another strand) were investigated.20 For the formation of the tetramolecular G-hairpin, we adopted two unique Watson–Crick duplexes each containing six contiguous Gs opposite to a nick in the complementary strand (see, Fig. 1c and Fig. S1, ESI†). Thus, only two G-repeat strands were used out of the four participating strands, which could lead to the formation of a G-hairpin structure specifically. In the case of the tetramolecular G-triplex, the top duplex contained six G–G mismatches in the middle, whereas the bottom duplex contained six contiguous Gs opposite to a nick in the complementary strand (Fig. 1d). This leaves only three strands with G-repeat sequences in the middle, which in turn can produce a G-triplex structure specifically. To bring the duplexes closer and to promote the formation of the desired intermediates, we incorporated structural flexibility into the strands by increasing their length. As a result, the lengths of the top and bottom duplexes used were 67 and 77 bp respectively, whereas the length between the two connecting sites in the origami was 64 bp.
The (3 + 1)-type G-hairpin was formed by adopting two duplexes, each containing a single-stranded overhang with a GGGGGGTTN (where N = A or T) sequence (Fig. 1e and Fig. S2, ESI†). In the case of the (3 + 1)-type G-triplex, the top duplex contained two repeats of the GGGGGGTTN sequence, whereas the bottom duplex contained one such sequence (Fig. 1f). We also investigated the (3 + 1)-type structures that contained three contiguous Gs, as they are naturally abundant (Fig. S3, ESI†). All the duplex DNAs used here contained single-stranded regions, each 16 bases in length at both termini, which are needed for their attachment inside the origami frame through complementary base pairing. Small-molecule-induced conformational switching was observed by using AFM by monitoring the topological changes of the incorporated duplexes from parallel (no structure) to X-shaped (formation of a notable intermediate structure). The presence of the ligand in the intermediate structure was confirmed by the localization of streptavidin (STV) using STV-biotin (biotin present in the ligand) binding as a pixel-enhancing marker in the AFM image. Note, (3 + 1) represents the type of G-quadruplex, though the hairpin and triplex may better be denoted (1 + 1) and (2 + 1), respectively.
Regarding the general experimental procedure, we have prepared the DNA origami and attached the duplexes of interest in each case. The origami assembly was then purified by passing through a Sephacryl S-300 filtration column to remove the excess staples and unattached duplexes. The PDC-biotin ligand was then added to the purified origami assembly, which was then immobilized on a mica surface. After gently washing the surface to remove the unbound origami assembly, STV was added on the surface and the system was incubated for 5 min. Imaging was carried out in PDC-biotin-free buffer after washing the mica surface to remove excess STV. For the detailed experimental procedure, see the ESI.†
Initially, we investigated the tetramolecular hairpin structure. The G-repeat sequences are not able to form the hairpin structure in the absence of quadruplex inducers such as PDC-biotin and salts (K+ and Mg2+), although a minor but significant amount (27%) of the hairpin structure was formed even in the presence of 5 mM Mg2+. Thus, a majority of the incorporated strands adopted a parallel form inside the origami frame (Fig. 1c). Interestingly, addition of PDC-biotin induced topological changes of the duplexes from parallel to X-shape by bringing the duplexes closer at the G-repeat region. Furthermore, the yield of the X-shape in this case was doubled when compared with that obtained under PDC-biotin-free conditions. A representative zoom-in AFM image is given in Fig. 1c, and the zoom-out image in Fig. S4, ESI.† The calculated yields in each case are summarized in Table 1. This topological change indicated that the ligand induces the formation of a tetramolecular antiparallel hairpin structure. The localization of STV in the center of the X-shape, as indicated by the bright spot in the AFM image, further confirmed that the ligand is present in the G-rich core, and consequently provided evidence for the ligand-hairpin binding. Among the four participating strands, only two strands contained the G-repeats and thus the incorporated strands can only form the hairpin structure, ignoring the possibility of other types of structure such as the G-triplex and the G-quadruplex. A similar trend was also observed for the tetramolecular antiparallel triplex structure (Fig. 1d and Fig. S4, ESI†). In this case, 24% of strands formed X-shape in the absence of a ligand, and addition of a ligand induced the formation of X-shape in as many instances as 44%.
| Sequences | G-hairpin | G-triplex | G-quadruplex | |||
|---|---|---|---|---|---|---|
| No liganda | With ligand | No liganda | With ligand | No ligand | With ligand | |
| a For comparison, these data are taken from our parallel studies in ref. 20. Numbers in the parentheses indicate the yield of X-shapes with bound STV. [Tris-HCl] = 20 mM, pH 7.6; [MgCl2] = 5 mM (no ligand) or 10 mM (with ligand); [EDTA] = 1 mM; [KCl] = 0 mM; [PDC-biotin] = 1 μM; [STV] = 0.2 μM. b For comparison, these data are taken from our parallel study in ref. 38. Numbers in parentheses and concentrations are as listed for ref. 20. c For comparison, these data are taken from our parallel study in ref. 39. Numbers in parentheses and concentrations are as listed for ref. 20. | ||||||
| Tetramolecular antiparallel structures | ||||||
| 6 G-repeat (%) | 27 | 54 (15) | 24 | 44 (34) | 18b | 69 (21)c |
| Counted origami | 416 | 174 | 153 | 149 | 331 | 329 |
| (3 + 1)-type structures | ||||||
| 6 G-repeat (%) | 24 | 43 (12) | 29 | 55 (30) | 26a | 64 (33) |
| Counted origami | 532 | 112 | 346 | 159 | 406 | 137 |
| 3 G-repeat (%) | 6 | 23 (10) | 11 | 29 (10) | 16a | 40 (16) |
| Counted origami | 345 | 154 | 432 | 205 | 331 | 138 |
In a similar way, we have tested the (3 + 1)-type hairpin structures (see Fig. 1e and Fig. S5 in the ESI†). For structures with six G-repeats, about 24% of the duplexes adopted parallel form in the absence of PDC-biotin, whereas this amount increased to 43% when PDC-biotin was added. Sequences containing three Gs were also tested, and the obtained X-shapes in the absence and presence of the ligand were 6 and 23%, respectively. These observations indicated that the ligand induces the (3 + 1)-type hairpin structure with a reasonable yield. It is worth mentioning here that three contiguous Gs avoid the potential caveat that may be possible with the relatively long six G-repeats. In the latter case, if a zero nucleotide chain reversal loop is feasible, the quadruplex may still be formed with two G3 runs from the same strand. However, it is not possible with the shorter three contiguous Gs, and thus evidencing the ligand-induced formation of the G-hairpin.
The (3 + 1)-type triplexes were then investigated (Fig. 1f and Fig. S5, ESI†). As can be seen from Table 1, 29% of structures adopted X-shape for sequences with six G-repeats in the absence of a PDC-biotin ligand and this amount was increased to 55% when the origami assembly was incubated with the ligand. Similar changes were also noticed with sequences containing three G-repeats. In this case, 11 and 29% of the structures adopted X-shape in the absence and in the presence of a ligand, respectively (Fig. 1f and Fig. S6, ESI†). In all cases, the localization of STV in the center of X-shape indicated clearly that the ligand is present in the G-rich core, and consequently evidenced ligand-intermediate binding. For comparison, the yields of G-quadruplex structures in each case are listed in Table 1. In two of the three systems, the yields of the ligand-induced structures follow the order: hairpin < triplex < quadruplex, reflecting the energetics of the structures, with lowest energy conformation for the quadruplex and the highest energy structure for the hairpin.
The binding of STV-biotin in the ligand was also characterized by analyzing the height profile. A representative graph of the height analysis of the protein particle inside an origami frame is given in Fig. 2. The estimated height of the origami frame and STV are 2.07 and 5.06 nm, respectively. These values are in good agreement with the theoretically expected values of 2 and 5 nm, respectively, for the origami and STV.39,40
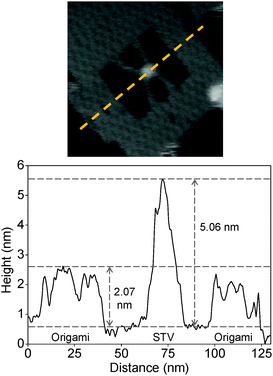 | ||
| Fig. 2 The graph shows a representative height profile estimated from the image shown above. The yellow broken line in the image indicates the location where the height was measured. | ||
Sequences with the G to T mutation were also tested in selected cases to check whether the formation of X-shape was due to the formation of a characteristic structure or whether the strands simply make physical contact for a short time. The mutated sequences failed to adopt X-shape and exhibited 100% parallel form, which indicated that the X-shape formed in the case of G-repeat sequences is due to the formation of a notable intermediate structure (Fig. S7, ESI†). Moreover, we have previously reported that STV alone can neither bind nor induce the formation of X-shape in G-rich sequences, which demonstrates further that the formation of X-shape is due to the ligand-induced formation of the intermediate structure.39 Note, the direct observation of the G-hairpin and G-triplex intermediates as they form in the G-quadruplex folding is still difficult by our (perhaps any other) method. However, we have frozen each intermediate inside the origami frame that cannot fold further into the final quadruplex structure. The hypothesis here is that if the ligand can induce such a structure, then it should be formed during the quadruplex folding even inside the cell.
In concluding remarks, by using the DNA origami as a novel scaffold to control the strand polarity, stoichiometry, and the number of G-repeats or G-repeat containing strands, we have investigated the formation of G-quadruplex intermediates such as G-hairpin and G-triplex structures induced by a G-quadruplex-binding PDC-biotin ligand. This ligand successfully bound to the G-repeat regions and induced the formation of intermediate structures for both tetramolecular and (3 + 1)-type systems. Our studies strongly support the assumptions that the G-hairpin and the G-triplex are formed as in-pathway intermediates of G-quadruplex folding (irrespective of whether it is induced by salt20 or a ligand). Furthermore, we could capture the solution-state structures of the intermediate-ligand complex with nanometer precision, while no parallel report exists in the literature. Moreover, we would like to point out that not even a theoretical discussion exists in the literature on drug binding to these intermediates. Thus, this communication constitutes the first report of research into an unprecedented area of intermediate-ligand binding. Based on these results, we could make a general hypothesis that the G-quadruplex-binding ligands are not merely quadruplex binders, but they would rather initially bind to the in-pathway intermediates and induce stepwise folding into a quadruplex structure. These mechanistic investigations shed light on drug-intermediate binding and could help in the development of novel anticancer drugs targeting G-rich regions. Regarding the demerits of this study, it is difficult to apply these results directly to the human genome inside cells because such conditions are completely different. Furthermore, the spatial resolution of the analysis of the intermediate-ligand complex should be improved to a few Angstroms.
We sincerely thank for the CREST grant from the Japan Science and Technology Corporation (JST), grants from the WPI program (WPI-iCeMS, Kyoto University) and JSPS KAKENHI (grant numbers 24310097, 24225005, 24104002 and 26620133). Financial support from The Mitsubishi Foundation and The Sekisui Chemical Grant Program to M.E. is also acknowledged.
Notes and references
- V. L. Makarov, Y. Hirose and J. P. Langmore, Cell, 1997, 88, 657–666 CrossRef CAS.
- T. d. Lange, Genes Dev., 2005, 19, 2100–2110 CrossRef PubMed.
- S. Muller, D. A. Sanders, M. D. Antonio, S. Matsis, J.-F. Riou, R. Rodriguez and S. Balasubramanian, Org. Biomol. Chem., 2012, 10, 6537–6546 Search PubMed.
- M. Franceschin, L. Rossetti, A. D'Ambrosio, S. Schirripa, A. Bianco, G. Ortaggi, M. Savino, C. Schultes and S. Neidle, Bioorg. Med. Chem. Lett., 2006, 16, 1707–1711 CrossRef CAS PubMed.
- L. Guittat, P. Alberti, F. Rosu, S. V. Miert, E. Thetiot, L. Pieters, V. Gabelica, E. D. Pauw, A. Ottaviani, J. F. Riou and J.-L. Mergny, Biochimie, 2003, 85, 535–547 CrossRef CAS.
- M. Y. Kim, H. Vankayalapati, S. Kazuo, K. Wierzba and L. H. Hurley, J. Am. Chem. Soc., 2002, 124, 2098–2099 CrossRef CAS PubMed.
- D. Y. Sun, B. Thompson, B. E. Cathers, M. Salazar, S. M. Kerwin, J. O. Trent, T. C. Jenkins, S. Neidle and L. H. Hurley, J. Med. Chem., 1997, 40, 2113–2116 CrossRef CAS PubMed.
- F. X. G. Han, R. T. Wheelhouse and L. H. Hurley, J. Am. Chem. Soc., 1999, 121, 3561–3570 CrossRef CAS.
- S. M. Gowan, R. Heald, M. F. G. Stevens and L. R. Kelland, Mol. Pharmacol., 2001, 60, 981–988 CAS.
- J. T. Kern and S. M. Kerwin, Bioorg. Med. Chem. Lett., 2002, 12, 3395–3398 CrossRef CAS.
- S. N. Georgiades, N. H. A. Karim, K. Suntharalingam and R. Vilar, Angew. Chem., Int. Ed., 2010, 49, 4020–4034 CrossRef CAS PubMed.
- D. Koirala, T. Mashimo, Y. Sannohe, Z. Yu, H. Mao and H. Sugiyama, Chem. Commun., 2012, 48, 2006–2008 RSC.
- M. Boncina, J. Lah, I. Prislan and G. Vesnaver, J. Am. Chem. Soc., 2012, 134, 9657–9663 CrossRef CAS PubMed.
- R. D. Gray, R. Buscaglia and J. B. Chaires, J. Am. Chem. Soc., 2012, 134, 16834–16844 CrossRef CAS PubMed.
- A. Y. Q. Zhang and S. Balasubramanian, J. Am. Chem. Soc., 2012, 134, 19297–19308 CrossRef CAS PubMed.
- W. Li, X.-M. Hou, P.-Y. Wang, X.-G. Xi and M. Li, J. Am. Chem. Soc., 2013, 135, 6423–6426 CrossRef CAS PubMed.
- V. Limongelli, S. D. Tito, L. Cerofolini, M. Fragai, B. Pagano, R. Trotta, S. Cosconati, L. Marinelli, E. Novellino, I. Bertini, A. Randazzo, C. Luchinat and M. Parrinello, Angew. Chem., Int. Ed., 2013, 52, 2269–2273 CrossRef CAS PubMed.
- N. An, A. M. Fleming and C. J. Burrows, J. Am. Chem. Soc., 2013, 135, 8562–8570 CrossRef CAS PubMed.
- P. M. Yangyuoru, A. Y. Q. Zhang, Z. Shi, D. Koirala, S. Balasubramanian and H. Mao, ChemBioChem, 2013, 14, 1931–1935 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka and H. Sugiyama, Angew. Chem., Int. Ed., 2014, 53, 4107–4112 CrossRef CAS PubMed.
- R. Stefl, T. E. Cheatham III, N. Spackova, E. Fadrna, I. Berger, J. Koca and J. Sponer, Biophys. J., 2003, 85, 1787–1804 CrossRef CAS.
- T. Mashimo, H. Yagi, Y. Sannohe, A. Rajendran and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 14910–14918 CrossRef CAS PubMed.
- P. Stadlbauer, M. Krepl, T. E. Cheatham III, J. Koca and J. Sponer, Nucleic Acids Res., 2013, 41, 7128–7143 CrossRef CAS PubMed.
- V. Gabelica, Biochimie, 2014, 105, 1–3 CrossRef CAS PubMed.
- P. Stadlbauer, L. Trantirek, T. E. Cheatham III, J. Koca and J. Sponer, Biochimie, 2014, 105, 22–35 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Angew. Chem., Int. Ed., 2012, 51, 874–890 CrossRef CAS PubMed.
- M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 1592–1597 CrossRef CAS PubMed.
- M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, Angew. Chem., Int. Ed., 2010, 49, 9412–9416 CrossRef CAS PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Curr. Protoc. Nucleic Acid Chem., 2012, 48, 12.9.1 Search PubMed.
- A. Rajendran, M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, ACS Nano, 2011, 5, 665–671 CrossRef CAS PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Chem. Rev., 2014, 114, 1493–1520 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka, P. L. T. Tran, J.-L. Mergny, R. J. Gorelick and H. Sugiyama, J. Am. Chem. Soc., 2013, 135, 18575–18585 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2013, 135, 1117–1123 CrossRef CAS PubMed.
- M. Endo and H. Sugiyama, Acc. Chem. Res., 2014, 47, 1645–1653 CrossRef CAS PubMed.
- T. Ando, T. Uchihashi and S. Scheuring, Chem. Rev., 2014, 114, 3120–3188 CrossRef CAS PubMed.
- A. Renaud de la Faverie, F. Hamon, C. D. Primo, E. Largy, E. Dausse, L. Delauriere, C. Landras-Guetta, J.-J. Toulme, M.-P. Teulade-Fichou and J.-L. Mergny, Biochimie, 2011, 93, 1357–1367 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka, P. L. T. Tran, J.-L. Mergny and H. Sugiyama, Nucleic Acids Res., 2013, 41, 8738–8747 CrossRef CAS PubMed.
- A. Rajendran, M. Endo, K. Hidaka, P. L. T. Tran, M.-P. Teulade-Fichou, J.-L. Mergny and H. Sugiyama, RSC Adv., 2014, 4, 6346–6355 RSC.
- K. Numajiri, M. Kimura, A. Kuzuya and M. Komiyama, Chem. Commun., 2010, 46, 5127–5129 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5cc01678a |
| ‡ Current address: Institute of Advanced Energy, Kyoto University, Gokasho, Uji-shi, Kyoto-fu, 611-0011, Japan. |
| This journal is © The Royal Society of Chemistry 2015 |
