Open Access Article
Open Access ArticleBoosting of activity enhancement of K+-responsive quadruplex hammerhead ribozyme†
Yudai
Yamaoki
,
Tsukasa
Mashima
,
Takashi
Nagata
* and
Masato
Katahira
*
Institute of Advanced Energy, Graduate School of Energy Science, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan. E-mail: katahira.masato.6u@kyoto-u.ac.jp; nagata.takashi.6w@kyoto-u.ac.jp
First published on 2nd March 2015
Abstract
Two second-generation quadruplex hammerhead ribozymes, whose activity enhances in response to K+via quadruplex formation of embedded r(GGA)3GG, were developed. Different strategies were applied to suppress basal activity when K+ is absent. As a result, the activity enhancement upon the addition of K+ has reached as high as 21-fold.
Nucleic acids that are rich in guanosine residues can take on a compact quadruplex structure in the presence of K+, but they are more likely to be elongated and single-stranded in the absence of K+.1 Thus, we recognized these guanine-rich RNAs as a structural switch that can be turned ‘ON’, by using K+ as a switching stimulator. Intracellular concentrations of K+ are much higher (approximately 100–150 mM) than in the extracellular environment (3.5–5.5 mM). Therefore, in principle, such RNAs rich in guanosine residues should show a rather dramatic change in structure on being delivered from the outside to the inside cells. We hypothesized that if we introduced such guanine-rich RNAs into functional RNAs such as ribozymes and aptamers, we would be able to obtain novel functional RNAs that sense differences in the K+ concentrations of cellular environments and thereby regulating their own functions.
We previously described the quadruplex structure of 5′-GGAGGAGGAGGA-3′ (R12) in the presence of K+.2 R12 folds into a compact parallel quadruplex, with G:G:G:G tetrad and G:(A):G:G:(A):G hexad planes. The R12 quadruplex forms a stable dimer through hexad–hexad stacking. However, in the absence of K+, R12 takes on an elongated single-stranded form (Fig. S1a, ESI†). We have also shown that R11, which lacks the terminal adenosine residue of R12, has similar, if not the same, structural characteristics in the presence and absence of K+.3
We previously introduced R11 into a hammerhead ribozyme (HR) and developed an ‘intelligent’ ribozyme, named here Quadruplex Hammerhead Ribozyme (QHR), whose activity is exerted in response to K+.3 In a hammerhead ribozyme (HR), two portions of the catalytic core, the 5′-HR domain (5′-GCGGUCUGAUGA-3′) and the 3′-HR domain (5′-GAAACAGG-3′), are linked via a central stem, thereby being brought into close proximity and exerting activity (Fig. S1b, ESI†).4 We replaced this stem with R11 and uridine-linkers to obtain the QHR. In QHR, the 5′-HR and 3′-HR domains are not brought into close proximity in the absence of K+ because R11 takes on the single-stranded form, but they are brought into close proximity in the presence of K+ due to the formation of the compact R11 quadruplex, by which QHR exerts activity (Fig. S1c, ESI†).3
The first-order rate constants of QHR at 25 °C in the absence of K+ and presence of 100 mM K+ were measured previously, the values being k0mMobs = ca. 0.04 min−1 and k100mMobs = ca. 0.1 min−1, respectively (Fig. S2, ESI†).3 The ratio of the first-order rate constants, k100mMobs/k0mMobs, can be considered as an index of enhancement of the enzymatic activity of QHR through quadruplex formation in response to K+, and is approximately 2.5-fold for QHR (Fig. S2, ESI†).3 It has been noticed that QHR exhibits weak residual activity even in the absence of K+. The presence of such basal activity of QHR at the concentration of 0 mM K+ is unfavorable, because it may cause a destructive attack on target and/or non-target molecules before the switch is turned ‘ON’. Suppression of the basal activity is important not only for this reason, but also to realize higher enhancement of the enzymatic activity of QHR in response to K+, namely higher switching efficiency.
We hypothesized that the origin of the above mentioned basal activity of QHR even in the absence of K+ was partial and/or transient formation of the active core structure.3 In detail, we suggested that due to joining of the 5′-HR and 3′-HR domains through R11 together with some linker residues, the fluctuation of the structure of QHR in solution or binding of QHR to the substrate may bring the 5′-HR and 3′-HR domains into close proximity, even without quadruplex formation. This may cause the basal activity. Along these lines, here we adopt two strategies to inhibit the formation of the partial and/or transient structure of the active core and suppress the basal activity of QHR in the absence of K+: for the first strategy, we split QHR in the middle of the R11 region; and for the second strategy, we introduced partially complementary RNA sequences into QHR. These second-generation QHRs not only suppressed the basal activity in the absence of K+ but also resulted in the improvement of the activity enhancement in response to K+.
In the first strategy, to keep the 5′-HR and 3′-HR domains away from each other in the absence of K+, we split QHR in the middle of the R11 region (Fig. 1a). We named the resulting subunits 5′-spQHR (5′-GCGGUCUGAUGAUUUGGAGGA-3′) and 3′-spQHR (5′-GGAGGUUGAAACAGG-3′), respectively. Since these 5′- and 3′-spQHRs are no longer covalently connected, we thought formation of the catalytic core would be prevented, expecting the basal activity to be suppressed. In the presence of K+, however, we expected the R11 regions of the 5′- and 3′-spQHRs would come close to each other, and form the same or a similar quadruplex structure through an intermolecular interaction. This is likely since four d(GGAGGA) strands reportedly fold into a compact parallel quadruplex dimer composed of two G:G:G:G tetrad planes and two G:(A):G:G:(A):G hexad ones,5 which resembles the structure of the quadruplex dimer of R12.
The enzymatic activities of a mixture of the 5′- and 3′-spQHRs in the absence of K+ and presence of 100 mM K+ were analyzed by gel electrophoresis, the resulting cleavage products being isolated and their band intensities being measured. The time course of the percentage cleaved is plotted for each case in Fig. 1b. The first-order rate constants of the mixture of the 5′- and 3′-spQHRs were k0mMobs = 1.24 × 10−3 min−1 and k100mMobs = 3.90 × 10−3 min−1 (Fig. 1c), and therefore enhancement of the activity by K+ (k100mMobs/k0mMobs) was 3.2-fold (Fig. 1d). These values indicate that the basal activity in the absence of K+ was reduced to one thirty-first, and enhancement of the activity by K+ was improved 1.4-fold as compared with in the case of the original QHR.
The first-order rate constants of the mixture of the 5′- and 3′-HRs were k0mMobs = 1.41 × 10−3 min−1 and k100mMobs = 1.19 × 10−3 min−1 (Fig. 1c). Both these values were comparable to k0mMobs of the mixture of the 5′- and 3′-spQHRs (Fig. 1c). It was clearly shown that there was no activity enhancement for the mixture of the 5′- and 3′-HRs upon the addition of K+ (Fig. 1d), which makes perfect sense because the 5′- and 3′-HRs (green in Fig. 1a) do not have a quadruplex-forming sequence in the first place.
The mixture of the 5′- and 3′-spQHRs showed a much lower k100mMobs value than the original QHR. We suggest that this observation is partially due to the fact that other combinations of the 5′- and 3′-spQHRs are possible, although 5′-spQHR/3′-spQHR is the correct combination. Thus, in principle, if a stoichiometry of one-to-one pairing is assumed, the combinations of 5′-spQHR/5′-spQHR, 5′-spQHR/3′-spQHR, and 3′-spQHR/3′-spQHR are possible, all of which may form the quadruplex structure. In theory, the population ratio should then be 5′-spQHR/5′-spQHR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5′-spQHR/3′-spQHR
5′-spQHR/3′-spQHR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3′-spQHR/3′-spQHR = 1
3′-spQHR/3′-spQHR = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Since only 5′-spQHR/3′-spQHR should be active, i.e. the others should not be active, k100mMobs of the mixture of the 5′- and 3′-spQHRs is expected to be half that of the original QHR. However, this was not the case and k100mMobs of the mixture of the 5′- and 3′-spQHRs was much lower. The reason for this may be the formation of an inactive quadruplex structure such as an anti-parallel form.
1. Since only 5′-spQHR/3′-spQHR should be active, i.e. the others should not be active, k100mMobs of the mixture of the 5′- and 3′-spQHRs is expected to be half that of the original QHR. However, this was not the case and k100mMobs of the mixture of the 5′- and 3′-spQHRs was much lower. The reason for this may be the formation of an inactive quadruplex structure such as an anti-parallel form.
Here, we constructed 5′- and 3′-spQHRs, a mixture of which exhibited suppressed basal activity in the absence of K+, and also improved the enhancement of the activity by K+, both as compared with the original QHR. To our knowledge, this is the first study to utilize intermolecular quadruplex formation to control the activity switching of a ribozyme.
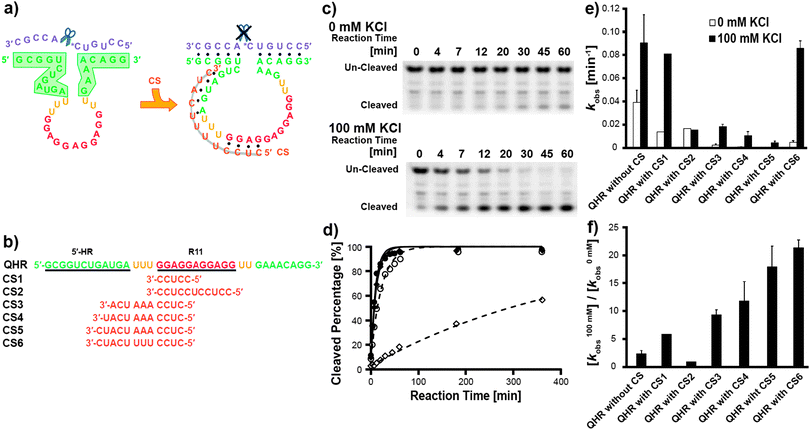
In the second strategy, we designed several RNA sequences that are complementary to different regions of QHR, and named them CSs. We attempted to find a CS that binds to QHR and forms a double-strand in the absence of K+, but in the presence of K+, dissociates from QHR with the formation of the quadruplex structure in the R11 region of QHR. In this way, we intended to keep the 5′-HR and 3′-HR domains of the QHR apart completely from each other in the absence of K+ and suppress the basal activity (Fig. 2a). When in the presence of K+, on the other hand, QHR is expected to retain full activity through the formation of a catalytic conformation upon dissociation of the CS and quadruplex formation for R11.
Fig. 2b lists the sequences of the CSs used in this study. CS1 and CS2 are 5- and 11-nucleotides (nt) long, respectively, each targeting a sequence within the R11 region of QHR. CS3, CS4, and CS5 are each 10-, 11-, and 12-nt long, respectively, all targeting sequences in the R11 region and in the 5′-HR domain of QHR. Finally, CS6 is 12-nt long, and targets the same sequence as CS5 but has a three base-pair mismatch as to QHR.
The enzymatic activities of the QHR with each CS (CS1–CS6) in the absence of K+ and presence of 100 mM K+ were analyzed by gel electrophoresis (Fig. 2c). The first-order rate constants for all combinations of QHR with CSs were obtained by applying these results (Fig. 2e). The ratios of the first-order rate constants, k100mMobs/k0mMobs, for all combinations of QHR with CSs were calculated and are shown in Fig. 2f.
CS1 is 5-nt long and is complementary to the sequence within the R11 region of QHR. The basal activity of QHR with CS1, k0mMobs, was repressed to one-third. Although this is rather mild repression, the formation of the double-strand within the R11 region was shown to be effective (Table S1, ESI† and Fig. 2e). Additionally, the enhancement of the activity of QHR with CS1 by K+ (k100mMobs/k0mMobs) was 5.9-fold. This is an improvement as compared with that of free QHR (2.3-fold), having a comparable k100mMobs value (just ca. 10% reduction) to free QHR (Table S1, ESI† and Fig. 2f).
Next, to achieve further repression of the basal activity, we constructed CS2, which is complementary to the whole sequence of the R11 region. The k0mMobs value of QHR with CS2, however, did not show further improvement as compared with that of QHR with CS1, exhibiting a similar residual activity value (Table S1, ESI† and Fig. 2e). This suggests that covering of only the R11 region and inhibition of its quadruplex formation are not sufficient to eliminate the basal activity, probably due to partial and/or transient formation of the active core structure in the HR domains, even though the quadruplex structure is not formed. On the other hand, the k100mMobs/k0mMobs value of QHR with CS2 turned out to be 0.92-fold, indicating that there was no enhancement of the activity by K+ (Table S1, ESI† and Fig. 2f). It is assumed that the double-strand formed with CS2 is more stable than the quadruplex structure in the R11 region even in the presence of K+, and thereby activity switching by K+ is restrained. Therefore, keeping an appropriate length for binding to the R11 region, while searching for an appropriate complementary sequence to other regions, such as the HR domains, may lead to the finding of a suitable CS.
In CS3, CS4, and CS5, the sequence that is complementary to the R11 region is short (4-nt long, which is one nucleotide shorter than CS1) and identical, while the sequences that are complementary to 5′-HR domain are varied (lengthened by one base in CS3, CS4, and CS5 in that order) (Fig. 2b). As we have expected, these three CSs repressed the basal activity of QHR significantly, the k0mMobs values being in the range of one-fortieth to one two-hundredth of the value of free QHR (Table S1, ESI† and Fig. 2e). This indicates that connection of complementary sequences to parts of the 5′-HR domain and that to the R11 region are highly effective for suppressing the basal activity of QHR. A probable explanation for this suppression of the basal activity is that complementary sequences in CS3, CS4, and CS5 as to parts of the 5′-HR domain of QHR have formed a double-strand and thereby inhibiting the partial and/or transient formation of the active core structure in the HR domains of QHR, which would otherwise exhibit basal activity even in the absence of the quadruplex structure.
We then determined the k100mMobs/k0mMobs values of QHR with CS3, CS4, and CS5, all of which improved in that order (9.3-, 11.8-, and 17.9-fold), and were much larger than that of free QHR, 2.3-fold (Table S1, ESI† and Fig. 2f). However, the k100mMobs values of QHR with CS3, CS4, and CS5 decreased in that order, and more importantly, all had much smaller values than that of free QHR (Table S1, ESI† and Fig. 2e). A probable reason for the latter point is that each of CS3, CS4, and CS5 forms a double-strand with QHR through a rather long stretch (regions through a part of 5′-HR, a uridine-linker, and a part of the R11 region), thereby dissociation of CSs, formation of the quadruplex structure in the R11 region, and activity switching in the presence of K+ are strongly restricted. Although the further improvement of suppression of the basal activity, k0mMobs, and activity enhancement, k100mMobs/k0mMobs, for CS3, CS4, and CS5, as compared with CS1, is encouraging, k100mMobs needs to be increased.
Finally, CS6 targets exactly the same region of QHR as CS5, except that CS6 has a three base-pair mismatch as to QHR (Fig. 2b). This base-pair mismatch was introduced to make an adjustment to obtain a sequence that retains a similar level of basal activity suppression as CS5 but is capable of dissociating from QHR upon quadruplex structure formation by R11 in the presence of K+. Although the k0mMobs of QHR with CS6 was not as low as that of QHR with CS5, QHR with CS6 exhibited effective suppression of the basal activity, the k0mMobs value being 4.77 × 10−3 min−1, which is one-eighth of the value of free QHR (Table S1, ESI† and Fig. 2e). Importantly, the k100mMobs value was 8.57 × 10−2 min−1, which is just a minor decrease from the k100mMobs value of free QHR (only ca. 5% reduction), and thereby the k100mMobs/k0mMobs value turned out to be as high as 21.4-fold (Table S1, ESI† and Fig. 2f). We suggest that the three base-pair mismatch between CS6 and QHR was suitable for CS6 to form a double-strand with QHR in the absence of K+, and also for CS6 to dissociate from QHR upon formation of the quadruplex of R11, thereby the active structure of QHR being induced in the presence of K+.
We attempted to design a CS that suppresses the basal activity of QHR in the absence of K+ and also leads to QHR high activity enhancement (k100mMobs/k0mMobs value) through quadruplex formation in response to K+. In total, we examined the effects of six CSs on the activity of QHR (Fig. 2b). Among them, CS6 exhibited good results in leaving QHR with highly suppressed residual basal activity in the absence of K+, and causing high activity enhancement for QHR in response to K+. Thus, we have successfully developed a second-generation QHR.
Several ribozymes were reportedly conjoined with aptamers, by which the activities were altered in response to small compounds.6 These ribozymes that are activated by small compounds are used as biosensors and gene regulators. Another activity switching ribozyme, that was reported, was composed of a ribozyme portion and a conjoined inhibitor portion, which is an antisense RNA against the ribozyme portion. This ribozyme–inhibitor system is activated upon the presence of a complementary RNA strand (named activator), by which the inhibitor portion is displaced. This system, which employs a cis-acting RNA duplex is somewhat similar to our QHR–CS system in the sense that it uses inhibitor of ribozyme.7 To our knowledge, only two groups have reported ribozymes connected with quadruplexes instead of an aptamer. Hartig and coworkers replaced the stem of a hammerhead ribozyme with a guanosine-rich RNA fragment, r(GGGGUGGGUGGGUGGG), with which the activity was enhanced by the addition of meso-5,10,15,20-tetrakis-(N-methyl-4-pyridyl) porphine (TMPyP4), a known quadruplex stabilizer.8 The activity enhancement of their conjoined ribozyme upon addition of TMPyP4 was about 8-fold. Perreault and coworkers incorporated two guanosine-rich RNA fragments into hepatitis D virus ribozyme.9 It is thought that one of the incorporated fragments forms a series of hydrogen bonds with part of the original hepatitis D virus ribozyme in the absence of K+, resulting in a misfolded ribozyme with no activity. In the presence of K+, however, the two fragments are suggested to fold into a quadruplex and thereby the active catalytic structure is restored. In the latter two cases, the precise structures of the quadruplexes are not fully understood.
In our case, we have precise, at atomic resolution, structural knowledge of the quadruplex that we have used, which provides us a guide to obtain the ability of activity switching to other ribozymes and aptamers. Our strategy enabling induction of the activity of a ribozyme aptamer in response to K+ is rather straightforward. Firstly, the ribozyme or aptamer is divided into two parts, and each part is linked to one end of R11 through some linker residues. At this stage, we only need to focus on obtaining such a ribozyme or aptamer conjoined with R11 that exerts as high activity as possible in the presence of K+. This can be accomplished by finding an appropriate position to divide the ribozyme or aptamer, and adjusting the length of the linker residues. At the next stage, we use two different strategies, both of which are fruits of the present study, to suppress the basal activity in the absence of K+ and to achieve highly efficient enhancement of the activity in response to K+: splitting the R11 region in the middle or introducing partially complementary RNA sequences. In summary, we have shown two methods, which might be applicable to other ribozymes and aptamers, endowing them with activity switching capabilities that can be controlled.
We thank Professors T. Morii and E. Nakata of Kyoto University for their technical support with the fluorescence gel imaging systems, and Professor Y. Tanaka of Tohoku University for the valuable discussion on the idea of splitting QHR. We are grateful to the Ministry of Education, Science, Sports and Culture of Japan for Grants-in-Aid for Scientific Research (23570146 and 26440026 to T.N.; 25115507, 25291013, 26104520, and 26650014 to M.K.).
Notes and references
- (a) S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402 CrossRef CAS PubMed; (b) D. E. Gilbert and J. Feigon, Curr. Opin. Struct. Biol., 1999, 9, 305 CrossRef CAS; (c) A. N. Lane, J. B. Chaires, R. D. Gray and J. O. Trent, Nucleic Acids Res., 2008, 36, 5482 CrossRef CAS PubMed; (d) A. T. Phan, V. Kuryavyi and D. J. Patel, Curr. Opin. Struct. Biol., 2006, 16, 288 CrossRef CAS PubMed.
- (a) T. Mashima, A. Matsugami, F. Nishikawa, S. Nishikawa and M. Katahira, Nucleic Acids Res., 2009, 37, 6249 CrossRef CAS PubMed; (b) T. Mashima, F. Nishikawa, Y. O. Kamatari, H. Fujiwara, M. Saimura, T. Nagata, T. Kodaki, S. Nishikawa, K. Kuwata and M. Katahira, Nucleic Acids Res., 2013, 41, 1355 CrossRef CAS PubMed.
- T. Nagata, Y. Sakurai, Y. Hara, T. Mashima, T. Kodaki and M. Katahira, FEBS J., 2012, 279, 1456 CrossRef CAS PubMed.
- (a) K. J. Hertel, A. Pardi, O. C. Uhlenbeck, M. Koizumi, E. Ohtsuka, S. Uesugi, R. Cedergren, F. Eckstein, W. L. Gerlach and R. Hodgson, Nucleic Acids Res., 1992, 20, 3252 CrossRef CAS PubMed; (b) T. Sakamoto, M. H. Kim, Y. Kurihara, N. Sasaki, T. Noguchi, M. Katahira and S. Uesugi, J. Biochem., 1997, 121, 288 CrossRef CAS; (c) O. C. Uhlenbeck, Nature, 1987, 328, 596 CrossRef CAS PubMed.
- A. Kettani, A. Gorin, A. Majumdar, T. Hermann, E. Skripkin, H. Zhao, R. Jones and D. J. Patel, J. Mol. Biol., 2000, 297, 627 CrossRef CAS PubMed.
- (a) M. Famulok, J. S. Hartig and G. Mayer, Chem. Rev., 2007, 107, 3715 CrossRef CAS PubMed; (b) M. Wieland and J. S. Hartig, ChemBioChem, 2008, 9, 1873 CrossRef CAS PubMed; (c) M. N. Win and C. D. Smolke, Biotechnol. Genet. Eng. Rev., 2007, 24, 311 CrossRef CAS.
- D. H. Burke, N. D. Ozerova and M. Nilsen-Hamilton, Biochemistry, 2002, 41, 6588 CrossRef CAS PubMed.
- M. Wieland and J. S. Hartig, Angew. Chem., 2006, 45, 5875 CrossRef CAS PubMed.
- J. D. Beaudoin and J. P. Perreault, RNA, 2008, 14, 1018 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and experimental details. See DOI: 10.1039/c5cc00961h |
| This journal is © The Royal Society of Chemistry 2015 |