 Open Access Article
Open Access ArticlePromiscuous hydroxylases for the functionalization of polycyclic tetramate macrolactams – conversion of ikarugamycin to butremycin†‡
Christian
Greunke§
,
Janine
Antosch§
and
Tobias A. M.
Gulder
*
Biosystems Chemistry, Department of Chemistry and Center for Integrated Protein Science Munich (CIPSM), Technische Universität München, Lichtenbergstraße 4, 85747 Garching, Germany. E-mail: tobias.gulder@ch.tum.de
First published on 20th February 2015
Abstract
Polycyclic tetramate macrolactams (PTMs) are a structurally, biomedically and biosynthetically intriguing class of bacterial metabolites. By combining parts of the machineries of different PTM biosynthetic pathways, we demonstrate for the first time the substrate promiscuity of a class of PTM tailoring enzymes, thereby facilitating the (bio)synthesis of butremycin.
In the course of evolution, Nature has developed a large set of diverse natural products with unique structures and biological functions. An interesting group of such metabolites are the PTMs that are produced by bacteria. Depending on their individual structures, PTMs exhibit a number of valuable biological activities, ranging from antimicrobial to cytotoxic.1–6 All PTMs contain a tetramic acid moiety incorporated into a macrolactam ring that is further modified with a fused carbocyclic system. Both overall size of the macrolactam and oxidation levels of the individual carbon atoms vary in different PTMs, as does the number of rings and their exact cyclization pattern in the carbocyclic portion. Overall, this leads to the structural diversity of this natural product class, with HSAF (1)1 and the frontalamides (2),2 alteramide A (3),3 and ikarugamycin (4)4 being typical examples (Fig. 1).
 | ||
| Fig. 1 Structures of the PTMs HSAF (1), the frontalamides (2), alteramide (3), and ikarugamycin (4). | ||
Triggered by initial work on the structure and biosynthesis of 1 by Du et al.7a and of 2 by the Clardy lab,2 PTM biosynthetic pathways have meanwhile attracted significant attention. In particular the pioneering work by Du on HSAF assembly in Lysobacter7 and our contribution towards understanding the biosynthesis of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 that was later validated by Zhang et al.9 have revealed a number of interesting features of PTM biosynthesis. These include the unusual catalytic activities of the thioesterase domain in the mixed polyketide–non-ribosomal peptide (PKS–NRPS) biosynthetic machinery,7b,c the origin of the C-3 hydroxyl group found in many PTMs that was shown to be introduced into 1 by late-stage hydroxylation,7d the unprecedented iterative use of a single PKS module for incorporation of twelve C2-units into a bacterial metabolite,7e,8,9 and the remarkably low number of enzymes overall involved in the biosynthesis of the highly complex cyclic structures by reductive ring formation, only three in case of 4.8,9 In addition, Zhao et al. showed the applicability of their DNA assembler methodology towards unlocking a cryptic PTM biosynthetic pathway from Streptomyces griseus.10
8 that was later validated by Zhang et al.9 have revealed a number of interesting features of PTM biosynthesis. These include the unusual catalytic activities of the thioesterase domain in the mixed polyketide–non-ribosomal peptide (PKS–NRPS) biosynthetic machinery,7b,c the origin of the C-3 hydroxyl group found in many PTMs that was shown to be introduced into 1 by late-stage hydroxylation,7d the unprecedented iterative use of a single PKS module for incorporation of twelve C2-units into a bacterial metabolite,7e,8,9 and the remarkably low number of enzymes overall involved in the biosynthesis of the highly complex cyclic structures by reductive ring formation, only three in case of 4.8,9 In addition, Zhao et al. showed the applicability of their DNA assembler methodology towards unlocking a cryptic PTM biosynthetic pathway from Streptomyces griseus.10
Owing to the broad range of biological activities of different PTMs and the simplicity of PTM biosynthetic pathways in terms of the low number of tailoring enzymes needed to shape these complex molecules, the engineered biosynthesis of PTMs by swapping PTM genes between different pathways would be highly desirable. However, such an approach has not yet been reported. The success of such experiments is highly dependent on the level of substrate promiscuity of the employed enzyme. Within this work, we set out to test this by utilizing PTM-pathway derived sterol desaturases (SD), the catalysts responsible for C-3 hydroxylation in the biosynthesis of HSAF (1),7d to functionalize ikarugamycin (4), a compound with a significantly altered cyclization pattern when compared to 1 (cf.Fig. 1).
For our experiments we decided to use the SD gene of a bacterial strain with a putative PTM biosynthetic gene cluster resembling that of the HSAF pathway in Lysobacter enzymogenes C3.7 The SD gene of the commercially available Lysobacter capsici DSM 19286 was selected as a good candidate (see Fig. 2A/B). The production of 1 by L. capsici was confirmed by its cultivation and chemical analysis of organic extracts of the culture broth. Having thus indirectly proven the presence of a biocatalytically active SD protein that hydroxylates the HSAF skeleton in L. capsici, we set out to clone the respective candidate gene into a suitable overexpression vector. To allow later purification of the recombinant SD protein, the vectors pHIS811 and pGS-21a12 were used, both encoding N-terminal poly(His)-tags facilitating affinity chromatography, with pGS-21a additionally encoding a GST tag to enhance protein solubility. The putative SD gene of the PTM biosynthetic gene cluster in L. capsici was amplified from genomic DNA by PCR using two different sets of primers (see ESI‡). This led to the production of a 810 bp copy of the SD gene as predicted by GLIMMER13 and to a longer 1002 bp copy that includes an alternative, more upstream start codon of the original sequence and better resembles the length of other annotated SDs in published genomic data. The thus obtained constructs were transferred into E. coli BL21(DE3) for recombinant protein production studies. Unfortunately, all expression experiments with the four constructs revealed only very low overproduction levels and exclusive formation of insoluble inclusion bodies. Best overproduction levels were still achieved with the 810 bp construct of SD in pGS-21a (pSDsLcap).
 | ||
| Fig. 2 Schematic representation of (A) the PTM biosynthetic pathway in Lysobacter enzymogenes C3 encoding HSAF (1)7 and the putative PTM pathways in (B) L. capsici DSM 19286 and (C) Saccharophagus degradans DSM 17024. Genes putatively coding for: red: iterative PKS–NRPS system; green: oxidoreductase; blue: alcohol dehydrogenase; grey: sterol desaturase (SD). | ||
We next examined the utility of pSDsLcap to provide the hydroxylating SD biocatalyst in vivo. The plasmid was transferred into our recently reported E. coli BAP114 based ikarugamycin (4) heterologous production system8 by electroporation. Both the transcription of the ikarugamycin (4) pathway genes as well as of the SD gene from L. capsici were controlled by lacI. Fermentation of the resulting strain in ZY autoinduction media15 revealed the exclusive production of the 3-hydroxylated derivative of 4 (see Fig. 3D), a compound very recently isolated as the natural product butremycin (5) from Micromonospora sp. K310.16 Ikarugamycin (4) was not detectable in these experiments at all, revealing its highly effective, complete conversion to 5 catalysed by SD in vivo.17 Because of the identical chromatographic behaviour of 5 produced by our approach compared to 5 from Micromonospora sp., formation of the identical diastereomer, i.e., with 3S-configuration, can be concluded.18
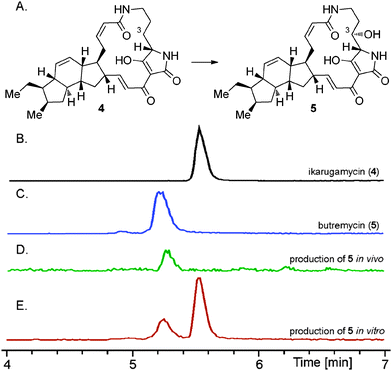 | ||
| Fig. 3 (A) Conversion of ikarugamycin (4) to butremycin (5). HPLC-MS-analyses of (B) a standard of ikarugamycin (4), (C) a standard of butremycin (5), (D) the coexpression of Ika_ABC8 in E. coli BAP114 together with SD from L. capsici, (E) the in vitro transformation of 4 using the supernatant derived from a culture of an E. coli BL21(DE3) SD (S. degradans) overexpression strain. EICs at 479.3 (4) and 495.2 (5) are shown. | ||
We next turned our attention to the possibility to hydroxylate purified ikarugamycin (4) using recombinant SD, as this methodology may easily be expanded to functionalize other PTM molecules without the need of a complete, heterologously reconstituted PTM pathway or a genetically transformable natural producer. Indeed, when feeding 4 to E. coli BL21(DE3) carrying pSDsLcap, a small portion of 4 was converted into 5 (with approx. 90% of 4 remaining, data not shown).
To improve turnover rates, which might have been hampered in the E. coli feeding experiments due to problems of 4 passing the bacterial cell wall, the in vitro modification of 4 was re-investigated. As the SD encoded by L. capsici did not yield sufficient quantities of soluble protein for purification (see above), the SD gene of the putative PTM biosynthetic gene cluster of Saccharophagus degradans DSM 17024, a close homolog of the respective Lysobacter genes, was selected as an alternative (cf.Fig. 2C). It was readily cloned into pHIS8 and pGS-21a (see ESI‡) and the resulting constructs tested in protein overproduction assays in E. coli BL21(DE3). Using the respective pGS-21a construct (pSDSde), overproduction of soluble protein was indeed achieved. Purification of the recombinant SD was conducted using Ni-affinity chromatography. However, significant degradation of SD during the isolation procedure was observed. The SD enriched fraction was nevertheless assayed for its hydroxylating activity. In these experiments, 4 was incubated with SD for 12 hours, leading to approx. 30% conversion to 5 (Fig. 3E). The overall turnover was not increased when extending the reaction time, hinting at a complete degradation of SD under the assay conditions. To potentially circumvent the problems associated with decomposition of recombinant SD during purification, the in vitro hydroxylation of 4 using cell lysates of the E. coli BL21(DE3)::pSDSde expression system was tested. These experiments gave the best results so far for the in vitro preparation of 5, leading to overall approx. 50% C-3 hydroxylation of 4 to 5. This hydroxylation step could be forced to completion by simple extractive work-up of the mixture of 4 and 5 and its repeated metabolization with fresh cell lysate.
The members of the PTM class of natural products are characterized by highly complex molecular structures that arise from remarkably streamlined biosynthetic machineries. Besides the unusual iterative PKS–NRPS system there are only a very small number of tailoring enzymes involved in PTM biosynthesis. Generating new molecules by adding or swapping individual biosynthetic genes between pathways thus seems to be a rewarding task. A precondition for success of such experiments, however, is a relaxed substrate specificity of the respective encoded enzymes. We herein prove for the first time that a class of PTM tailoring enzymes, usually annotated as sterol desaturases (SD), is capable of transforming “unnatural” substrates derived from a different PTM pathway. This led to the development of a heterologous production system of butremycin (5) utilizing the ikarugamycin biosynthetic genes together with a SD from L. capsici. In addition, even purified 4 can be converted into 5 utilizing SD alone, both in vivo and in vitro. As this class of SDs from PTM pathways seems to have highly specific activity for C-3 hydroxylation of PTMs, as also shown for the previously uncharacterized SD from Saccharophagus degradans, we propose to rename these enzymes according to their function as PTM hydroxylases. The results presented here provide diverse methods for late-stage C-3 hydroxylation of PTM molecules. The utility of the PTM hydroxylases for the functionalization of other substrates is currently being tested in our laboratory. Our work should furthermore set the stage for other PTM pathway gene swapping experiments that are likewise being investigated by our group.
We thank Dr Markus Nett (HKI Jena) for Lysobacter capsici and Dr Kwaku Kyeremeh (University of Ghana) for an analytical sample of butremycin from Micromonospora sp. K310. J.A. and C.G. thank the Fonds der Chemischen Industrie (FCI) and the Friedrich Naumann Foundation for Freedom, respectively, for their PhD scholarships. This work was generously funded by the DFG (GU1233/2-1 and the Center for Integrated Protein Science Munich CIPSM).
Notes and references
- P. R. Graupner, S. Thornburgh, J. T. Mathieson, E. L. Chapin, G. M. Kemmitt, J. M. Brown and C. E. Snipes, J. Antibiot., 1997, 50, 1014 CrossRef.
- J. A. V. Blodgett, D.-C. Oh, S. Cao, C. R. Currie, R. Kolter and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11692 CrossRef CAS PubMed.
- H. Shigemori, M.-A. Bae, K. Yazawa, T. Saaki and J. Kobayashi, J. Org. Chem., 1992, 57, 4317 CrossRef CAS.
- K. Jomon, Y. Kuroda, M. Ajisaka and H. Sakai, J. Antibiot., 1972, 25, 271 CrossRef CAS.
- S. Cao, J. A. V. Blodgett and J. Clardy, Org. Lett., 2010, 12, 4652 CrossRef CAS PubMed.
- S. Kanazawa, N. Fusetani and S. Matsunaga, Tetrahedron Lett., 1993, 34, 1065 CrossRef CAS.
- (a) F. Yu, K. Zaleta-Rivera, X. Zhu, J. Huffman, J. C. Millet, S. D. Harris, G. Yuen, X.-C. Li and L. Du, Antimicrob. Agents Chemother., 2007, 51, 64 CrossRef CAS PubMed; (b) L. Lou, G. Qian, Y. Xia, J. Hang, H. Chen, K. Zeleta-Rivera, Y. Li, Y. Shen, P. H. Dussault, F. Liu and L. Du, J. Am. Chem. Soc., 2011, 133, 643 CrossRef CAS PubMed; (c) L. Lou, H. Chen, R. L. Cerny, Y. Li, Y. Shen and L. Du, Biochemistry, 2012, 51, 4 CrossRef CAS PubMed; (d) Y. Li, J. Huffman, Y. Li, L. Du and Y. Shen, MedChemComm, 2012, 3, 982 RSC; (e) Y. Li, H. Chen, Y. Ding, Y. Xie, H. Wang, R. L. Cerny, Y. Shen and L. Du, Angew. Chem., Int. Ed., 2014, 53, 7524 CrossRef CAS PubMed.
- J. Antosch, F. Schaefers and T. A. M. Gulder, Angew. Chem., Int. Ed., 2014, 53, 3011 CrossRef CAS PubMed.
- G. Zhang, W. Zhang, Q. Zhang, T. Shi, L. Ma, Y. Zhu, S. Li, H. Zhang, Y.-L. Zhao, R. Shi and C. Zhang, Angew. Chem., Int. Ed., 2014, 53, 4840 CrossRef CAS PubMed.
- Y. Luo, H. Huang, J. Liang, M. Wang, L. Lu, Z. Shao, R. E. Cobb and H. Zhao, Nat. Commun., 2013, 4, 2894 Search PubMed.
- J. M. Jez, J. L. Ferrer, M. E. Bowman, R. A. Dixon and J. P. Noel, Biochemistry, 2000, 39, 890 CrossRef CAS PubMed.
- The expression vector pGS-21a is commercially available from GenScript.
- A. L. Delcher, K. A. Bratke, E. C. Powers and S. L. Salzberg, Bioinformatics, 2007, 23, 673 CrossRef CAS PubMed.
- B. A. Pfeifer, S. J. Admiraal, H. Gramajo, D. E. Cane and C. Khosla, Science, 2001, 291, 1790 CrossRef CAS PubMed.
- F. W. Studier, Protein Expression Purif., 2005, 41, 207 CrossRef CAS PubMed.
- K. Kyeremeh, K. S. Acquah, A. Sazak, W. Houssen, J. Tabudravu, H. Deng and M. Jaspers, Mar. Drugs, 2014, 12, 999 CrossRef CAS PubMed.
- It is important to note that the heterologous ikarugamycin production system and our natural producer of 4, Streptomyces sp. Tü 6239, do not produce any detectable amounts of butremycin (5).
- The characterization of 5 by coelution experiments was performed on a variety of different HPLC phases (XBridge C18, Chromolith RP18e, Eurospher II C18A and C8, and the chiral phase Nucleocell Delta-RP), with no chromatographic differences detectable.
Footnotes |
| † This work is dedicated to our mentor and distinguished colleague Prof. Dr Dr h.c. mult. Gerhard Bringmann. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c5cc00843c |
| § These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |
