Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Oligonucleotides containing a piperazino-modified 2′-amino-LNA monomer exhibit very high duplex stability and remarkable nuclease resistance†
Chenguang
Lou
a,
Birte
Vester
b and
Jesper
Wengel
*a
aBiomolecular Nanoscale Engineering Center, Department of Physics, Chemistry and Pharmacy, University of Southern Denmark, Campusvej 55, 5230 Odense M, Denmark. E-mail: jwe@sdu.dk; Tel: +45 65502510
bDepartment of Biochemistry and Molecular Biology, University of Southern Denmark, Campusvej 55, 5230 Odense M, Denmark
First published on 3rd February 2015
Abstract
Incorporation of a piperazino-modified 2′-amino-LNA monomer (PipLNA-T) into oligonucleotides conferred very high affinity and base-pairing selectivity towards complementary DNA and RNA strands. Furthermore, one PipLNA-T modification provided a robust nuclease resistance that safeguarded three neighbouring natural nucleosides from 3′-exonucleolytic degradation. These favourable properties render PipLNA-T a promising oligonucleotide modification for various biological applications.
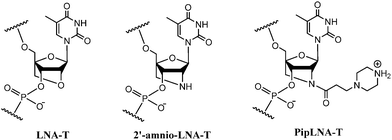
DNA encodes the genetic information in living organisms and has attracted extensive and continuous interest in the areas of biochemistry, biology and nanotechnology within the past few decades.1–4 Many modified nucleotide monomers have been incorporated into oligonucleotides (ONs) in order to optimise affinity and sequence selectivity towards complementary nucleic acids and to obtain nuclease resistance.5–9 Locked nucleic acid (LNA) monomers represent a class of nucleotides containing ribofuranose units locked into a C3′-endo (North-type) conformation,10,11 and incorporation of LNA-T and 2′-amino-LNA-T monomers (Fig. 1) into ONs provides increased duplex stability, favorable mismatch discrimination and improved resistance to nucleolytic digestion.12–16
Besides LNA, the introduction of alkylamine chains into ONs is recognised as a successful strategy to improve binding affinity, probably due to electrostatic interactions between the protonated amine functionalities and the negatively charged phosphodiester backbone.17–20 Modification of ONs with a piperazino group introduces a cationic center under physiological conditions, and provides a possibility for further functionalisation.17,19,21 Therefore, we decided to investigate a piperazino-modified 2′-amino-LNA monomer (PipLNA-T; Fig. 1) by combining the advantageous characteristics of the 2′-amino-LNA and piperazino scaffolds.
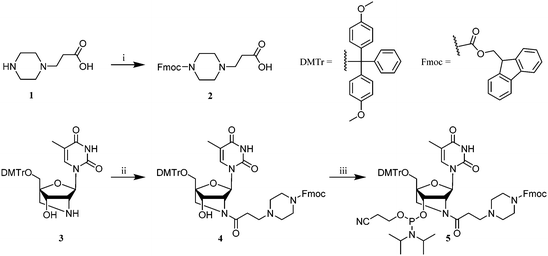
Synthesis of the PipLNA-T phosphoramidite monomer 5 was accomplished in three steps from 3-piperazinopropionic acid (1) and a DMTr-protected 2′-amino-LNA-T precursor 322 (Scheme 1).
The secondary amine of derivative 1 was reacted with N-(9-fluorenylmethoxycarbonyloxy)succinimide (Fmoc-OSu) in pyridine to produce 2 in 88% yield. The carboxylic group of 2 was subsequently reacted with 2′-amino-LNA nucleoside 3 in anhydrous DMF in the presence of 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU) and N,N-diisopropylethylamine (DIPEA) to give nucleoside 4 in 84% yield, which was finally phosphitylated by reaction with bis(diisopropylamino)(2-cyanoethoxy)phosphine to provide the desired phosphoramidite monomer 5 in 67% yield.
ON synthesis was carried out using standard solid-phase phosphoramidite chemistry on an automated DNA synthesiser where the PipLNA-T phosphoramidite monomer 5 was coupled using a hand-coupling process.23 Fmoc protection of the secondary amine of PipLNA-T not only prevents it from being irreversibly acetylated in the capping step of the standard synthesis cycle, but also allows ON deprotection under mild conditions after completion of ON synthesis.24,25 The stepwise coupling yield of 2′-amino-LNA-T and PipLNA-T were 96% and 80%, respectively. The imperfect coupling efficiency on PipLNA-T monomer may result from a close proximity of the bulky Fmoc-protected piperazino moiety and the diisopropylamino leaving group.
The synthesised ONs were purified by DMTr-ON reversed-phase HPLC (and/or ion-exchange HPLC) and their composition and purity (>90%) characterised by MAIDI-TOF MS and ion-exchange HPLC analysis, respectively.
A 9-mer sequence12,17 was chosen for evaluation of the binding affinity of PipLNA-containing ONs towards complementary DNA and RNA strands (Table 1). In comparison with all DNA (ON1) and 2′-amino-LNA-T-containing controls (ON2 and ON4), ONs containing one and three PipLNA-T modifications were studied (ON3 and ON5). The data show that a PipLNA-T monomer confers substantially higher duplex stability than thymidine (T) and 2′-amino-LNA-T in DNA–DNA contexts. The same tendency was observed for the corresponding DNA–RNA hybrids, but with even greater enhancement of stability relative to T and less dramatic difference between the PipLNA-T and the 2′-amino-LNA-T monomers. These hybridisation data are consistent with previous results showing that ONs containing 2′-amino-LNA-T exhibit higher affinity towards complementary RNA than DNA,12,26 but show a remarkable additional affinity-enhancing effect due to the incorporation of the piperazino group in monomer PipLNA-T.
| Sequence | X | Complementary DNA | Complementary RNA | |||
|---|---|---|---|---|---|---|
| Medium salt | Low salt | Medium salt | Low salt | |||
| a T m values (°C) of unmodified and modified (X = 2′-amino-LNA-T or PipLNA-T) DNA–DNA and DNA–RNA duplexes measured as an average of two independent melting temperature determinations with a deviation <0.5 °C. Numbers in brackets are ΔTm measured as the difference in Tm values between modified and unmodified duplexes. The experiments were carried out at pH 7.0 in medium salt buffer (5.8 mM NaH2PO4–Na2HPO4 buffer, containing 100 mM NaCl and 0.10 mM EDTA) and low salt buffer (6.7 mM NaH2PO4–Na2HPO4 buffer, containing 0.10 mM EDTA). The increase in melting temperature of 2′-amino-LNA-T is in a general agreement with previous results reported for ON2 and ON4.12 | ||||||
| 5′-GTGATATGC | — | ON1 | 32.0 | 18.0 | 29.5 | 16.0 |
| 5′-GTGAXATGC | 2′-Amino-LNA-T | ON2 | 35.5 (+3.5) | 37.0 (+7.5) | ||
| PipLNA-T | ON3 | 39.0 (+7.0) | 38.5 (+9.0) | |||
| 5′-GXGAXAXGC | 2′-Amino-LNA-T | ON4 | 41.0 (+9.0) | 27.5 | 49.0 (+19.5) | 36.0 |
| PipLNA-T | ON5 | 49.5 (+17.5) | 39.0 | 54.0 (+24.5) | 42.5 | |
The effect of PipLNA-T monomers on the thermal stability of duplexes is cumulative. This is demonstrated by the results that three PipLNA-T monomers provide far greater duplex stability than a single modification irrespective of either a DNA or RNA complementary strand.
In order to determine the importance of protonation of the distal nitrogen atom of the piperazino moiety (pKa ≈ 9 (ref. 27)), thermal stability of six duplexes formed by ON1, ON4 and ON5 with complementary DNA and RNA were evaluated at low salt buffer condition (6.7 mM NaH2PO4–Na2HPO4 buffer and 0.10 mM EDTA, Table 1). Smaller decreases in melting temperature were found for the duplexes involving ON5 with three PipLNA-T nucleotides than for the two control duplexes involving ON1 and ON4, suggesting that decreased duplex stability under low salt conditions can be alleviated by a stabilising effect from the positively-charged piperazino moieties of PipLNA-T. A similar effect was not observed for ON4 containing three 2′-amino-LNA-T monomers, which is consistent with the reported pKa value of 6.17 for the N2′-protonated derivative of the 2′-amino-LNA-T monomer.28
The mismatch discrimination ability of the novel PipLNA-T monomer was assessed for the central position in ON3 and ON5 in comparison to all-DNA control ON1 (Table S3, ESI†). Singly-modified ON3 showed higher base-pairing specificity for its correctly matched complementary strands than ON1, except for slightly reduced discrimination in the case of the mismatched RNA target having a cytidine monomer in the central position. For ON5 (three substitutions with PipLNA-T), the efficient mismatch discrimination ability of PipLNA-T was similar to that of DNA-T.
The ability of the PipLNA-T monomer to partake in triplex formation was evaluated using a 29-mer dsDNA target containing the wild-type HIV-1 polypurine tract sequence29 (Table S2, ESI†). Compared to a wild-type all-DNA TFO (Table S5, ESI†), incorporation of PipLNA-T greatly improved the triplex stability at both pH 6.0 and 7.0. Interestingly, no obvious improvement in triplex stability was observed when LNA-T was replaced by PipLNA-T, possibly due to the short propanoyl tether between the moderately bulky piperizino group and the nucleoside itself, restricting the conformational flexibility which could prevent the piperazino group from finding an optimal position for triplex stabilisation.
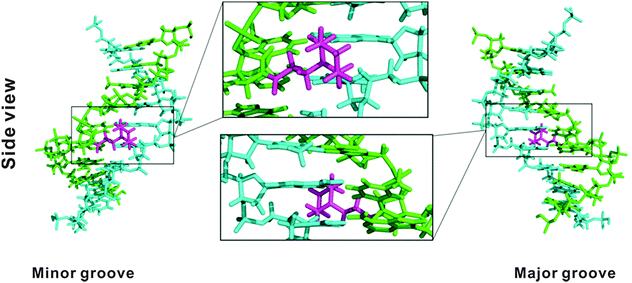
Molecular modelling30,31 (Fig. 2) on the duplex ON3:DNA with one PipLNA-T substitution indicated that the 3-(1-piperazino)propanoyl group of PipLNA-T is oriented in the minor groove of the double helix. The distances measured in the model structure between the protonated amino group of the piperazino ring and the four nearest negatively-charged oxygen anions on the phosphate backbone are ∼8–10 Å, which implies that any favourable electrostatic interaction has to be mediated through water molecule(s)32,33 in the minor groove.
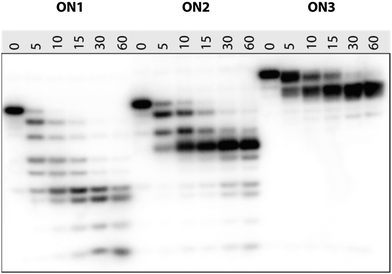
It is known that 2′-modified nucleotides usually increase nuclease resistance of ONs.34–36 In order to evaluate their resistance towards nucleolytic degradation, we studied the stability of 5′-32P labeled ON1–ON3 upon incubation with snake venom phosphordiesterase (SVPDE; 3′-exonuclease). As shown in Fig. 3, the unmodified oligonucleotide (X = T, ON1) was rapidly degraded, with no trace of full-length ON1 left after 10 min incubation. The degradation progressed over time with bands corresponding to 3- and 4-mers and 32P-labelled guanosine monophosphate remaining after 60 min. Also ON2 with the central thymidine substituted by a 2′-amino-LNA-T monomer, showed no full-length product after 15 min incubation. Intriguingly, the degradation was severely inhibited once the TGC-3′ triad was digested and the resulting 5′-GTGAXA exhibited very strong nuclease resistance with only little further digestion after 60 min incubation. This shows that a 2′-amino-LNA-T monomer is much more nuclease-resistant than its natural counterpart and that it protects the 3′-neighbouring nucleotide from 3′-exonucleolytic cleavage.
ON3 (X = PipLNA-T) showed retarded mobility in the gel compared to ON1 and ON2, probably due to a combination of reduced overall negative charge because of protonation of the piperazino moiety and increased bulkiness. Fig. 3 demonstrates that a single, centrally positioned PipLNA-T monomer when incorporated into the 9-mer ON induces significantly improved resistance to SVPDE when compared to both 2′-amino-LNA-T and DNA-T monomers. This counts with respect to full length product of which a trace remained after 15 min incubation, but more pronouncedly also by the remarkable stability of the 8-mer product resulting from cleavage of the 3′-nucleotide. This product is well preserved even after 60 min incubation. This implies that a PipLNA-T modification protects its neighbourhood up to three natural nucleosides towards its 3′-end from 3′-exonucleolytic degradation. A similar mechanism has also been proposed for C5-substituted LNA pyrimidines.20 This behaviour strongly contrasts that of a single LNA nucleotide which has been reported to confer no or only little resistance towards exonucleolytic degradation,37,38 and suggests that the PipLNA-modification may prove uniquely suited for increasing the biostability of antisense ONs.
In conclusion, a novel PipLNA-T phosphoramidite monomer has been synthesised from a known 2′-amino-LNA-T precursor and has been incorporated into oligonucleotides. The PipLNA-T monomer obeys the Watson–Crick base pairing rules, and has a pronounced stabilising effect on nucleic acid duplexes and triplexes and a strong stabilising effect against 3′-exonucleolytic degradation. These properties make PipLNA-modified oligonucleotides attractive in the context of various biomedical and nanotechnological applications.
The Villum Foundation is thanked for funding The Biomolecular Nanoscale Engineering Center. Alaa Salah Gouda and Tamer Kosbar are thanked for help with molecular modelling. Joan Hansen, Louise Johansson, Tina Grubbe Hansen and Lykke Haastrup Hansen are thanked for excellent technical assistance.
Notes and references
- E. T. Kool, Chem. Rev., 1997, 97, 1473 CrossRef CAS PubMed.
- A. Sancar, L. A. Lindsey-Boltz, K. Unsal-Kacmaz and S. Linn, Annu. Rev. Biochem., 2004, 73, 39 CrossRef CAS PubMed.
- J. L. Sessler, C. M. Lawrence and J. Jayawickramarajah, Chem. Soc. Rev., 2007, 36, 314 RSC.
- A. J. Boersma, R. P. Megens, B. L. Feringa and G. Roelfes, Chem. Soc. Rev., 2010, 39, 2083 RSC.
- J. Wengel, Acc. Chem. Res., 1999, 32, 301 CrossRef CAS.
- N. M. Bell and J. Micklefield, ChemBioChem, 2009, 10, 2691 CrossRef CAS PubMed.
- T. Yamamoto, M. Nakatani, K. Narukawa and S. Obika, Future Med. Chem., 2011, 3, 339 CrossRef CAS PubMed.
- K. R. Fox and T. Brown, Biochem. Soc. Trans., 2011, 39, 629 CrossRef CAS PubMed.
- G. F. Deleavey and M. J. Damha, Chem. Biol., 2012, 19, 937 CrossRef CAS PubMed.
- M. A. Campbell and J. Wengel, Chem. Soc. Rev., 2011, 40, 5680 RSC.
- S. Obika, S. M. A. Rahman, A. Fujisaka, Y. Kawada, T. Baba and T. Imanishi, Heterocycles, 2010, 81, 1347 CrossRef CAS PubMed.
- S. K. Singh, R. Kumar and J. Wengel, J. Org. Chem., 1998, 63, 10035 CrossRef CAS.
- K. Fluiter, M. Frieden, J. Vreijling, C. Rosenbohm, M. B. De Wissel, S. M. Christensen, T. Koch, H. Ørum and F. Baas, ChemBioChem, 2005, 6, 1104 CrossRef CAS PubMed.
- R. Owczarzy, Y. You, C. L. Groth and A. V. Tataurov, Biochemistry, 2011, 50, 9352 CrossRef CAS PubMed.
- A. S. Jørgensen, L. H. Hansen, B. Vester and J. Wengel, Bioorg. Med. Chem. Lett., 2014, 24, 2273 CrossRef PubMed.
- I. K. Astakhova and J. Wengel, Acc. Chem. Res., 2014, 47, 1768 CrossRef CAS PubMed.
- T. Bryld, T. Højland and J. Wengel, Chem. Commun., 2004, 1064 RSC.
- T. P. Prakash, A. Püschl, E. Lesnik, V. Mohan, V. Tereshko, M. Egli and M. Manoharan, Org. Lett., 2004, 6, 1971 CrossRef CAS PubMed.
- J. Skov, T. Bryld, D. Lindegaard, K. E. Nielsen, T. Højland, J. Wengel and M. Petersen, Nucleic Acids Res., 2011, 39, 1953 CrossRef CAS PubMed.
- (a) D. C. Guenther, P. Kumar, B. A. Anderson and P. J. Hrdlicka, Chem. Commun., 2014, 50, 9007 RSC; (b) P. Kumar, B. Baral, B. A. Anderson, D. C. Guenther, M. E. Østergaard, P. K. Sharma and P. J. Hrdlicka, J. Org. Chem., 2014, 79, 5062 CrossRef CAS PubMed.
- G. M. Mitev, B. L. Mellbye, P. L. Iversen and B. L. Geller, Antimicrob. Agents Chemother., 2009, 53, 3700 CrossRef CAS PubMed.
- A. S. Madsen, A. S. Jørgensen, T. B. Jensen and J. Wengel, J. Org. Chem., 2012, 77, 10718 CrossRef CAS PubMed.
- V. K. Rajwanshi, A. E. Håkansson, B. M. Dahl and J. Wengel, Chem. Commun., 1999, 1395 RSC.
- C. Lou, M. Shelbourne, K. R. Fox and T. Brown, Chem. – Eur. J., 2011, 17, 14851 CrossRef CAS PubMed.
- C. Lou, A. Dallmann, P. Marafini, R. Gao and T. Brown, Chem. Sci., 2014, 5, 3836 RSC.
- A. A. Koshkin, S. K. Singh, P. Nielsen, V. K. Rajwanshi, R. Kumar, M. Meldgaard, C. E. Olsen and J. Wengel, Tetrahedron, 1998, 54, 3607 CrossRef CAS.
- F. Khalili, A. Henni and A. L. L. East, J. Chem. Eng. Data, 2009, 54, 2914 CrossRef CAS.
- O. Plashkevych, S. Chatterjee, D. Honcharenko, W. Pathmasiri and J. Chattopadhyaya, J. Org. Chem., 2007, 72, 4716 CrossRef CAS PubMed.
- M. Faria, C. D. Wood, L. Perrouault, J. S. Nelson, A. Winter, M. R. H. White, C. Helene and C. Giovannangeli, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 3862 CrossRef CAS.
- S. J. Weiner, P. A. Kollman, D. T. Nguyen and D. A. Case, J. Comput. Chem., 1986, 7, 230 CrossRef CAS.
- MacroModel, version 9.1, Schrödinger, LLC, New York, NY, 2005 Search PubMed.
- M. A. Young, B. Jayaram and D. L. Beveridge, J. Am. Chem. Soc., 1997, 119, 59 CrossRef CAS.
- H. E. L. Williams and M. S. Searle, J. Mol. Biol., 1999, 290, 699 CrossRef CAS PubMed.
- T. P. Prakash and B. Bhat, Curr. Top. Med. Chem., 2007, 7, 641 CrossRef CAS.
- V. K. Sharma, R. K. Sharma and S. K. Singh, Med. Chem. Commun., 2014, 5, 1454 RSC.
- P. M. D. Moreno and A. P. Pego, Front. Chem., 2014, 2, 87 Search PubMed.
- S. Obika, M. Onoda, K. Morita, J.-I. Andoh, M. Koizumi and T. Imanishi, Chem. Commun., 2001, 1992 RSC.
- Y. Hari, T. Morikawa, T. Osawa and S. Obika, Org. Lett., 2013, 15, 3702 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details on synthesis of PipLNA-T monomer, oligonucleotide synthesis, purification and analysis (IE-HPLC and MAIDI-TOF MS), ultraviolet duplex melting studies, ultraviolet triplex melting studies, nuclease resistance assay, molecular modelling; copies of representative IE-HPLC curves, MAIDI-TOF mass spectra, and UV-melting curves. See DOI: 10.1039/c5cc00322a |
| This journal is © The Royal Society of Chemistry 2015 |