Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhanced cellular uptake of engineered spider silk particles†
Martina B.
Elsner
a,
Heike M.
Herold
a,
Susanne
Müller-Herrmann
a,
Hendrik
Bargel
a and
Thomas
Scheibel
*abcde
aLehrstuhl Biomaterialien, Universitätsstraße 30, Universität Bayreuth, Bayreuth D-95447, Germany. E-mail: Thomas.scheibel@bm.uni-bayreuth.de
bBayreuther Zentrum für Kolloide und Grenzflächen (BZKG), Universitätsstraße 30, Universität Bayreuth, Bayreuth D-95447, Germany
cBayreuther Zentrum für Bio-Makromoleküle (bio-mac), Universitätsstraße 30, Universität Bayreuth, Bayreuth D-95447, Germany
dBayreuther Zentrum für Molekulare Biowissenschaften (BZMB), Universitätsstraße 30, Universität Bayreuth, Bayreuth D-95447, Germany
eBayreuther Materialzentrum (BayMAT), Universitätsstraße 30, Universität Bayreuth, Bayreuth D-95447, Germany
First published on 13th January 2015
Abstract
Drug delivery systems allow tissue/cell specific targeting of drugs in order to reduce total drug amounts administered to an organism and potential side effects upon systemic drug delivery. Most drug delivery systems are polymer-based, but the number of possible materials is limited since many commercially available polymers induce allergic or inflammatory responses or lack either biodegradability or the necessary stability in vivo. Spider silk proteins represent a new class of (bio)polymers that can be used as drug depots or drug delivery systems. The recombinant spider silk protein eADF4(C16), which can be processed into different morphologies such as particles, films, or hydrogels, has been shown to fulfil most criteria necessary for its use as biomaterial. Further, eADF4(C16) particles have been shown to be well-suited for drug delivery. Here, a new method was established for particle production to reduce particle size and size distribution. Importantly, cellular uptake of these particles was shown to be poor in HeLa cells. Therefore, variants of eADF4(C16) with inversed net charge or incorporated cell penetrating peptides and receptor interacting motifs were tested, showing much better cellular uptake. Interestingly, uptake of all silk variant particles was mainly achieved by clathrin-mediated endocytosis.
Introduction
In principle drug delivery systems allow the achievement of constant drug levels at targeted locations within the body. Two generally different systems have been developed: depot systems located specifically within the tissue of choice or mobile systems that convey drugs embedded in a carrier to targeted tissues/cells. Hydrogels or films can be used as depots,1 whereas mostly particulate systems are used as drug delivery devices. Amongst the materials employed as carrier systems are inorganic (nano)particles, lipid vehicles, and most commonly synthetic polymers like poly(lactide), poly(lactic-co-glycolic acid), or poly(glycolic acid) and natural polymers such as gelatin, alginate, chitosan or silk proteins.2–7 Polymeric systems are the preferred material because many polymers show good biocompatibility, can be chemically modified according to the desired application, and are suitable for entrapment of therapeutic agents, allowing controlled release of the encapsulated drug over days or even months.8,9 However, synthetic polymers often need organic solvents and harsh formulation conditions for processing. Natural polymers, in contrast, are produced under mild environmental conditions (i.e. aqueous buffer systems) and show much higher biocompatibility.10Amongst natural polymers, silk proteins constitute a promising new material due to their biocompatibility and biodegradability. Recently, materials made of recombinantly produced spider silk proteins or silkworm fibroin have been shown to be well tolerated by cells.11–16 In this respect, particles made of eADF4(C16) have been previously used as drug delivery vehicles. eADF4(C16) is based on the repetitive core domain of the spidroin ADF4 of the European garden spider Araneus diadematus and can be processed into different morphologies including films,17,18 hydrogels,19 non-woven mats,20 capsules,21,22 and particles.23,24 Particles are produced with adjustable particle size by salting out, upon varying either protein concentration or mixing intensity with the salting-out reagent.24 Diameters of particles were in the range of 250 nm to 3 μm. Generally, cellular uptake of silk particles is rare13,25,26 probably due to the size of the particles, lack of binding ligands or the negative net/surface charge of some silk proteins.
Here, we used three approaches to improve cellular uptake of eADF4(C16) particles. Firstly, we established a new technique to process spider silk proteins into particles with small diameters by using ionic liquids as a starting solvent. Secondly, silk proteins were functionalized with cell penetrating peptides (CPP) to enhance the cellular uptake of particles. CPPs are short (up to 30 amino acids), mostly cationic peptides, able to cross cellular membranes and, thereby, transport cargo (particles, DNA, RNA, proteins, liposomes) into cells.27–33 The transport across membranes occurs via energy-dependent and/or independent mechanisms, with the exact mechanism of how the particles cross membranes being largely unresolved. The first discovered CPP was part of the trans-activator of transcription (Tat) protein of the HI-Virus in 198934,35 (amino acid sequence GRKKRRQRRRPPQ). Nowadays, beyond protein-derived CPPs, designed peptides such as poly-arginines (Rn) are also in use.36–47 Furthermore, both Tat- and R8-peptides possess nuclear targeting properties which can be advantageous in some cases of drug delivery.42,48 In this study Tat- and R8G-peptides as well as RGD49 were employed as CPPs to functionalize eADF4(C16). Thirdly, since the net- and surface charge of particles also plays a role in cellular uptake, all glutamic acid residues of polyanionic eADF4(C16) were mutated to lysine ones, yielding the polycationic eADF4(κ16).50
Endocytosis plays an important role in various unspecific and specific functions (e.g. nutrient uptake, signal transduction, regulation mechanism of cell migration, shape and volume, transcellular transport) of a cell. For the identification of internalization of substances, particles, etc. pharmacological inhibitors are commonly used. The choice of an inhibitor for uptake studies is not easy because many are not very specific or cause side effects. Ivanov51 compared the commonly used inhibitors for different endocytotic pathways. Based on this comparison dansylcadaverine and di-methyl-amiloride are the favourable inhibitors of clathrin-mediated endocytosis and macropinocytosis due to specificity and low side effects. These inhibitors were here used to determine the cellular uptake route of the individual spider silk particles.
Experimental
Genetic modification of eADF4(C16)
eADF4(C16) is based on 16 repeats of the consensus sequence of spidroin ADF4 of the European garden spider (Araneus diadematus) (C-module: GSSAAAAAAAASGPGGY GPENQGPSGPGGYGPGGPG), and a T7-tag fused to the aminoterminus for detection purposes.52 Fusions were made using tags as described in Wohlrab et al.49 For each tag, DNA cassettes were created by annealing two synthetic oligonucleotides (R8-peptide tag: gatccatgggccgtcgccgtcgtcgccgtcgccgtggctaatgaA and agctttcattagccacggcgacggcgacgacggcgacggcccatg; Tat aminoterminal tag: CATGGGCCGCAAAAAACGCCG CAGCGCCGTCGCGGCTAATGAAA and AGCTTTCATTAGCCGCGACGGCGCT GACGGCGTTTTTTGCGGCC; Tat carboxyterminal tag: CATGGGCCGCAAAAAACGCCGTCAGCGCCGTCGCCCGGGCTAATGAAA and AGCTTTCAT TAGCCCGGGCGACGGCGCTGACGGCGTTTTTTGCGGCC). The resulting amino acid sequences and the modified proteins are shown in Fig. 1. DNA sequences of the tags were inserted into the cloning vector pCS-eADF4(C16) by seamless cloning as described by Huemmerich et al.52 Successful cloning was confirmed by sequencing.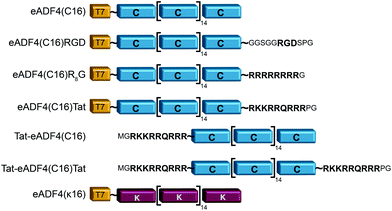 | ||
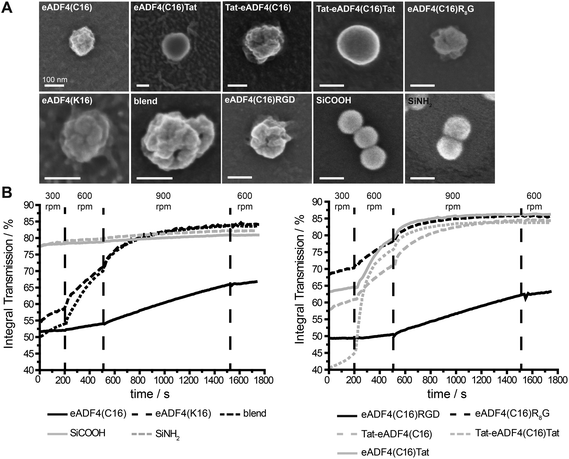
| Fig. 1 Schematic design of eADF4 variants used in this study. All constructs were created by genetic engineering.49,50 | ||
Production of recombinant spider silk proteins
Recombinant ADF4 derivates were produced in E. coli as described previously.52 Proteins were purified after lysis using ultrasonication, centrifugation of the cell debris, heat denaturation of E. coli proteins (80 °C, 20 min) and their removal by a second centrifugation step. The soluble spider silk proteins remaining in the supernatant were precipitated using 20% ammonium sulfate at 25 °C and lyophilized. Isoelectric point (pI) and molecular weight (MW) of the recombinant proteins were calculated using ExPASy ProtParam (Table 1). eADF4(C16), eADF4(C16)RGD, eADF4(C16)R8G, eADF4(C16)Tat and eADF4(κ16) contain an aminoterminal T7-tag, whereas Tat-eADF4(C16) and Tat-eADF4(C16)Tat do not have this T7-tag due to the cloning and expression system. Due to the T7-tag eADF4(C16) has a higher molecular weight than Tat-eADF4(C16), and eADF4(C16)Tat has a higher molecular weight than Tat-eADF4(C16) and Tat-eADF4(C16)Tat.| pI | Molecular weight/Da | |
|---|---|---|
| eADF4(κ16) | 9.70 | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683 683 |
| Tat-eADF4(C16)Tat | 6.36 | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
| eADF4(C16)R8G | 4.57 | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 005 005 |
| eADF4(C16)Tat | 4.57 | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 174 174 |
| Tat-eADF4(C16) | 4.46 | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 696 696 |
| eADF4(C16)RGD | 3.64 | 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 583 583 |
| eADF4(C16) | 3.48 | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 698 698 |
Spider silk particle formation
Lyophilized proteins were dissolved in 1-ethyl-3-methyl-imidazolium acetate (EMiM[acetate]) and stirred for 1 h at 95 °C. Particle formation was initiated by mixing 0.1 mg ml−1 of protein in EMiM[acetate] with 5 fold v/v excess of 2 M potassium phosphate, pH 8.23,55 After incubation for 1 h at 25 °C, the particles were centrifuged (15 min, 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C) and washed three times with ultrapure water.
000g, 4 °C) and washed three times with ultrapure water.
Dynamic light scattering (DLS) and zeta potential measurements
Spider silk and control particles (sicastar®-redF, surface modified with COOH or NH2 groups, micromod, Rostock, Germany) were analyzed for their particle size (Z-average and width) and distribution indices using dynamic light scattering (ZetaSizer NanoZS, Malvern Instruments, Worcestershire, UK). The samples were measured (n = 6; eADF4(c16)Tat n = 4) in MQ-H2O at a protein concentration of 0.5 mg ml−1 at 25 °C. The electrophoretic mobilities of all spider silk particles were measured in 10 mM KCl, pH 5.5 at 25 °C (ZetaSizer NanoZS). The zeta potential was calculated according to the theory of Smoluchowski56 based on the measured electrophoretic mobilities.Scanning electron microscopy
20 μl of particle suspension were pipetted on Thermanox™ plastic cover slips and air-dried. Samples were sputtered with a 2 nm layer of platinum (Sputter coater 208 HR, Cressington, Watford, UK) and analyzed using a Leo 1530 VP Gemini SEM (Zeiss, Germany) at 2–3 kV.Cell samples were fixed with 2.5% v/v glutaraldehyde, 80 mM HEPES, 3 mM CaCl2, pH 7.3, and washed twice with fixation buffer without glutaraldehyde for 15 min. After washing two times with ultrapure water, the samples were dehydrated by incubation for 20 min in 25%, 50%, 70%, 95%, each, and three times in 100% acetone. Afterwards, all samples were critically point dried (transitional medium CO2, Balzers CPD 020), sputtered with a 2 nm layer of platinum and analyzed using a Leo 1540 CrossBeam VP Gemini SEM.
Colloidal stability analysis
Colloidal stability of spider silk and control particles was analyzed in 10 mM KCl, pH 5.5 using a LUMiFuge®114 (L.U.M. GmbH, Berlin, Germany) with a rotation frequency of 300, 600 and 900 rpm and different time intervals of 200, 300, and 1000 s. Particle suspensions were placed in tubes in horizontal positions on the disc of the LUMiFuge®114. Transparency of the suspensions was measured in the area between menisci and sediment in duplicates, three times for each protein using fresh preparations each time. Transmission was measured every 10 s over 1800 s, and the integral of transmission between meniscus and bottom of the vial was plotted against time.Cell culture
HeLa cells (ACC-57, German collection of microorganism and cell cultures DSMZ, Leibnitz Institute, Braunschweig, Germany) were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Biochrom, Berlin, Germany) containing 10% v/v fetal bovine serum (Biochrom, Berlin, Germany), 1% v/v GlutaMAX (Gibco, Grand Island, USA) and 0.1% gentamicin sulfate (Sigma-Aldrich, Seelze, Germany). The viability of the cells was confirmed by trypan blue staining (Sigma-Aldrich, Seelze, Germany) before seeding the cells for experiments. Cells were cultured in a CO2-incubator (Haereus, Hanau, Germany) at 95% humidity, 5% CO2 and 37 °C.Analysis of cell proliferation (cytotoxicity), proliferation rate and doubling time
For cell proliferation analysis, cells were seeded on treated 96-well cell culture plates (Nunc, Langenselbold, Germany) with a density of 5000 cells cm−2 for 9 days. Cells were pre-incubated with spider silk or control particles (9.6 ng μl−1) for 24 h at 37 °C. Medium was changed daily followed by analysis of cell vitality using the CellTiter Blue assay. Cells were washed two times with phosphate buffered saline (PBS). Fresh media containing 10% v/v CellTiter Blue reagent® (Promega, Madison, USA) was added, and cells incubated for 2.5 h at 37 °C. Transformation of the blue fluorescent dye resazurin into red fluorescent resorufin (λex = 530 nm; λem = 590 nm) was measured using a plate reader (Mithras LB 940, Berthold, Bad Wildbad, Germany) with 530 nm excitation and 600 nm emission filters and a counting time of 0.5 s. For each particle type, cell culture experiments were repeated 3 times with 3 replicates.Proliferation rate (μ) and doubling time (Td) were calculated using a first order Monod-type kinetic model with the assumption that the mortality rate can be neglected (eqn (1)).
| X(t) = X0eμt | (1) |
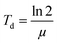 | (2) |
Uptake studies and identification of the uptake mechanism using flow cytometry
HeLa cells were cultured on treated 6-well cell culture plates (Nunc, Langenselbold, Germany) with a density of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−2 in the presence of spider silk or control fluorescent particles for 6, 24, 48 and 72 h. Cells were washed with PBS twice, treated with trypan blue, washed again with PBS and pelleted using 0.05% Trypsin/EDTA and centrifugation (300g, 5 min, 25 °C). Cells were resuspended in fresh media, and uptake was measured using flow cytometry (Cytomics FC500, Beckman-Coulter, Krefeld, Germany). Cells only appeared fluorescent upon internalization of rhodamine-labeled particles (labeling via N-Hydroxysuccinimid ester chemistry19,57). Endocytotic inhibitors were used for identification of the underlying uptake mechanism. Cells were seeded at a density of 50
000 cells cm−2 in the presence of spider silk or control fluorescent particles for 6, 24, 48 and 72 h. Cells were washed with PBS twice, treated with trypan blue, washed again with PBS and pelleted using 0.05% Trypsin/EDTA and centrifugation (300g, 5 min, 25 °C). Cells were resuspended in fresh media, and uptake was measured using flow cytometry (Cytomics FC500, Beckman-Coulter, Krefeld, Germany). Cells only appeared fluorescent upon internalization of rhodamine-labeled particles (labeling via N-Hydroxysuccinimid ester chemistry19,57). Endocytotic inhibitors were used for identification of the underlying uptake mechanism. Cells were seeded at a density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−2 with spider silk or control silica particles for 6 and 24 h. 100 μM dansylcadaverin (DC) for inhibition of clathrin-mediated endocytosis and 100 μM di-methyl-amiloride (DMA) for inhibition of macropinocytosis were added to cells 30 min prior to particle addition.
000 cells cm−2 with spider silk or control silica particles for 6 and 24 h. 100 μM dansylcadaverin (DC) for inhibition of clathrin-mediated endocytosis and 100 μM di-methyl-amiloride (DMA) for inhibition of macropinocytosis were added to cells 30 min prior to particle addition.
Cell staining for fluorescence microscopy
HeLa cells were cultured on μ-slides (8-well, ibidi GmbH, Martinsried, Germany) at a cell density of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−2 and incubated for 6, 24, 48 and 72 h with rhodamine-labeled particles at 37 °C. The cells were fixed with 4% w/v paraformaldehyde for 15 min at 25 °C, washed with PBS and permeabilized with 0.1% v/v Triton X-100 for 3 min. After two additional PBS washing steps, cells were incubated with HCS CellMask™ Blue stain (Invitrogen, Darmstadt, Germany) for 30 min and washed again with PBS. All samples were kept in PBS. Fluorescence microscopy was performed using a LifeCell microscope (DMI6000, Leica, Wetzlar, Germany).
000 cells cm−2 and incubated for 6, 24, 48 and 72 h with rhodamine-labeled particles at 37 °C. The cells were fixed with 4% w/v paraformaldehyde for 15 min at 25 °C, washed with PBS and permeabilized with 0.1% v/v Triton X-100 for 3 min. After two additional PBS washing steps, cells were incubated with HCS CellMask™ Blue stain (Invitrogen, Darmstadt, Germany) for 30 min and washed again with PBS. All samples were kept in PBS. Fluorescence microscopy was performed using a LifeCell microscope (DMI6000, Leica, Wetzlar, Germany).
Results and discussion
Spider silk protein modification
Recombinant spider silk protein eADF4(C16) has been well investigated in recent years.19,22–24,49,55,58–62 Here, the protein was modified with different tags for stimulating internalization of respective protein particles by HeLa cells (Fig. 1). A Tat-peptide was fused either to the carboxy-, the aminoterminus or both termini of eADF4(C16), and an R8- or an RGD-peptide was fused with the aminoterminus of eADF4(C16).49 To study particle-charge-dependence of cellular uptake, the recently established positively charged eADF4(κ16) was used.50 In this variant, all glutamic acid residues of eADF4(C16) are replaced by lysine ones rendering the protein polycationic under neutral conditions (in contrast to polyanionic eADF4(C16)). All modifications were analyzed and confirmed by MALDI-TOF MS (data not shown).Particle production and characterization
Particles were produced by salting out of the protein dissolved in EMiM[acetate]. In contrast to previously published methods23 the ionic liquid EMiM[acetate] was used to minimize the particle diameter and distribution. In addition to “single-protein” particles also eADF4(C16) and eADF4(κ16) blend particles were produced.All spider silk particles had spherical morphologies (Fig. 2A, Fig. S1†), indicating that modifying the sequence as well as blending had no influence on particle formation. Particles were also analyzed concerning size and surface potential (Table 2). Commercially available silica particles with a theoretical diameter of 100 nm were used as control. Their measured diameter was 113.5 ± 0.5 nm. Spider silk particles had diameters between 239 and 294 nm except for eADF4(κ16) (324 nm) and blend particles (393 nm). Importantly, the previously reported broad particle size distribution could be significantly reduced.24 Many studies investigated the uptake of particles produced from different materials such as polystyrene,63,64 chitosan based polymers,65 silica,66 or poly(lactic-co-glycolic acid) (PLGA).67 Independently from the material, the optimal particle size for uptake was in the range of 100–200 nm.
| (Spider silk) particles | Particle size/nm | Particle size distribution index | Zeta potential/mV | Electrophoretic mobility/10−8 m2 s−1 V−1 |
|---|---|---|---|---|
| eADF4(κ16) | 325 ± 38 | 0.193 ± 0.091 | 13.2 ± 5.3 | 1.04 ± 0.42 |
| Blend | 393 ± 135 | 0.545 ± 0.152 | −5.7 ± 7.5 | −0.45 ± 0.58 |
| Tat-eADF4(C16)Tat | 282 ± 61 | 0.165 ± 0.077 | −8.4 ± 5.3 | −0.66 ± 0.42 |
| eADF4(C16)R8G | 242 ± 19 | 0.123 ± 0.027 | −17.1 ± 3.3 | −1.35 ± 0.26 |
| eADF4(C16)Tat | 294 ± 50 | 0.166 ± 0.136 | −23.0 ± 7.0 | −1.80 ± 0.55 |
| Tat-eADF4(C16) | 239 ± 17 | 0.106 ± 0.055 | −23.5 ± 3.6 | −1.85 ± 0.28 |
| eADF4(C16)RGD | 263 ± 7 | 0.157 ± 0.074 | −24.8 ± 2.7 | −1.88 ± 0.36 |
| eADF4(C16) | 242 ± 22 | 0.123 ± 0.087 | −26.7 ± 2.6 | −2.09 ± 0.20 |
| SiNH2 | 113 ± 1 | 0.014 ± 0.001 | −55.0 ± 1.0 | −4.32 ± 0.08 |
| SiCOOH | 114 ± 2 | 0.020 ± 0.015 | −55.9 ± 2.3 | −4.39 ± 0.18 |
eADF4(C16), eADF4(C16)RGD, Tat-eADF4(C16) and eADF4(C16)Tat particles showed zeta potentials between −20 and −30 mV, similar to the previously published zeta potential of eADF4(C16).60 Zeta potentials of Tat-eADF4(C16)Tat and eADF4(C16)R8G were determined to be −8.4 and −17.4 mV, and those of blends varied between −13.2 and +1.8 mV due to the fact that each particle had different amounts of eADF4(C16) and eADF4(κ16) (especially on its surface). Particles with a zeta potential below −20 mV typically show little agglomeration in suspension.68 Control silica particles showed a surface potential of −55 mV and were therefore quite stable.68
Next, all particle suspensions were analyzed concerning their colloidal stability using a LUMiFuge®114 (Fig. 2B). Control silica particles were colloidally stable (no change in curve behavior) as suggested by their zeta potentials, whereas spider silk particles behaved differently. eADF4(C16) and eADF4(C16)RGD showed some sedimentation behavior at 300 and 600 rpm and started to sediment above 900 rpm, fitting with the measured zeta potentials of −26.7 and −24.8 mV. As expected, eADF4(C16)R8G, eADF4(C16)Tat and Tat-eADF4(C16) showed less colloidal stability, and eADF4(κ16) and blend particles were even less colloidally stable, with Tat-eADF4(C16)Tat particles being the least stable ones.
Cytotoxicity
Cytotoxicity of spider silk and control silica particles was analyzed using the CellTiter Blue® assay over 9 days after particle addition (24 h). HeLa cells cultured in the presence of spider silk or control silica particles showed indistinguishable growth behavior in comparison to HeLa cells grown in the absence thereof. Proliferation rates (μ) and doubling times (Td) also showed no significant differences (Table 3). Therefore, none of the particles seemed to have any cytotoxic effect on HeLa cells.| (Spider silk) particles | μ max/d−1 | Doubling time/h |
|---|---|---|
| No particles | 0.38 ± 0.03 | 43.9 ± 3.3 |
| eADF4(κ16) | 0.36 ± 0.03 | 47.0 ± 4.7 |
| Blend | 0.38 ± 0.01 | 44.1 ± 0.9 |
| Tat-eADF4(C16)Tat | 0.37 ± 0.02 | 45.3 ± 3.1 |
| eADF4(C16)R8G | 0.37 ± 0.01 | 45.2 ± 1.5 |
| eADF4(C16)Tat | 0.36 ± 0.01 | 46.8 ± 0.9 |
| Tat-eADF4(C16) | 0.37 ± 0.01 | 45.0 ± 1.2 |
| eADF4(C16)RGD | 0.37 ± 0.01 | 44.5 ± 0.7 |
| eADF4(C16) | 0.40 ± 0.01 | 42.1 ± 0.7 |
| SiNH2 | 0.38 ± 0.02 | 44.3 ± 2.3 |
| SiCOOH | 0.39 ± 0.04 | 43.0 ± 4.2 |
Particle internalization
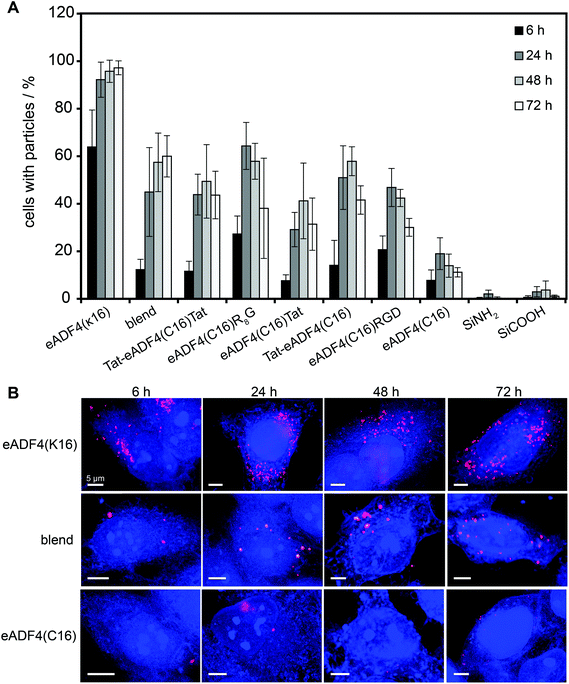
HeLa cells were incubated with spider silk or control particles for 6, 24, 48 and 72 h at 37 °C and analyzed using flow cytometry (Fig. 3A) or fluorescence microscopy (Fig. 3B, shown for eADF4(C16), eADF4(κ16) and blends). After 6 h of incubation, all spider silk particles were indistinguishably internalized by HeLa cells (between 10 and 30% of cells showed internalized particles) except for eADF4(κ16) particles where 64% of the cells contained particles (Fig. 3A). After 24 h of incubation, 92% of the cells showed internalized eADF4(κ16) particles, after 48 h 95% and after 72 h 97%, with all of the cells containing more than one internalized particle (Fig. 3B). Similar internalization numbers have been previously reported for particles produced from silk fibroin and albumin.25 As expected, polyanionic eADF4(C16) particles were internalized by only 19% of the HeLa cells after 24 h, with the amount of cells not increasing after 48 and 72 h.Blend particles were internalized from about 45% of the cells after 24 h, 57% after 48 h and 60% after 72 h, thus, the number of cells with internalized blend particles was between that of cells containing eADF4(C16) and eADF4(κ16) particles. Further, each cell (5000 cells measured per set) contained on average more blend particles than eADF4(C16) particles, but less than eADF4(κ16) particles. This result was expected since cells usually show a negative surface potential (HeLa cells approximately −50 mV69), and, therefore, charge–charge interactions with positively charged particles are pronounced.
The number of cells containing particles made of all CPP-fusion proteins was increased in comparison to that of eADF4(C16) alone. No significant differences were obtained when the Tat peptide was fused to the amino- or carboxyterminus or to both. Interestingly, the uptake of the CPP- and RGD-fusion particles was similar to that of blend particles.
Identification of the internalization mechanism of spider silk particles
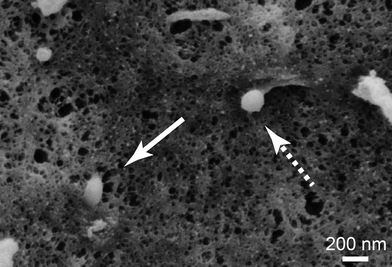
To get a closer look at the internalization mechanism of spider silk particles, SEM visualization of cell surfaces was performed. The particles could be seen in different stages of internalization (Fig. 4). Furthermore, HeLa cells were cultured in the presence of different endocytotic inhibitors (dansylcadaverine (DC): inhibitor of clathrin-mediated endocytosis; di-methyl-amiloride (DMA): inhibitor of macropinocytosis51,70–72) before adding the silk particles. After particle addition, the cells were incubated for 6 and 24 h and analyzed using flow cytometry (Table 4 and Table S1†).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−2 were incubated in the presence of particles for 24 h in the absence and presence of endocytosis inhibitors (100 μM of dansylcadaverine (DC) or di-methyl-amiloride (DMA)) at 37 °C. Inhibitors were added 30 min before particle addition
000 cells cm−2 were incubated in the presence of particles for 24 h in the absence and presence of endocytosis inhibitors (100 μM of dansylcadaverine (DC) or di-methyl-amiloride (DMA)) at 37 °C. Inhibitors were added 30 min before particle addition
| (Spider silk) particles | Cells with particles/% | ||
|---|---|---|---|
| − | + DC | + DMA | |
| eADF4(κ16) | 92.2 ± 7.4 | 47.3 ± 12.5 | 72.9 ± 7.1 |
| blend | 44.9 ± 18.7 | 16.6 ± 6.7 | 29.0 ± 17.5 |
| eADF4(C16)R8G | 64.5 ± 9.9 | 23.4 ± 5.9 | 63.7 ± 14.9 |
| Tat-eADF4(C16) | 51.0 ± 13.4 | 11.1 ± 0.6 | 26.7 ± 15.7 |
| eADF4(C16)RGD | 46.9 ± 8.0 | 19.9 ± 10.5 | 33.7 ± 8.4 |
| eADF4(C16) | 19.0 ± 6.7 | 8.5 ± 3.4 | 7.7 ± 4.9 |
| SiNH2 | 2.1 ± 1.6 | 3.2 ± 2.3 | 2.9 ± 1.9 |
| SiCOOH | 3.0 ± 2.2 | 3.1 ± 1.6 | 2.2 ± 2.2 |
The number of cells containing eADF4(C16) particles was reduced from 7.8% to 5.4% after 6 h and from 19% to 8.5% after 24 h in the presence of DC. Further, particles made of eADF4(C16) were the only ones being internalized by less cells in the presence of DMA after 6 h (2.4%) compared to internalization by cells in the presence of DC. After 24 h of incubation, the number of cells containing eADF4(C16) particles in the presence of DMA (7.7%) was again similar to that of cells in the presence of DC. The number of cells containing eADF4(C16) particles was reduced from 7.8% to 5.4% after 6 h and from 19% to 8.5% after 24 h in the presence of DC. Further, particles made of eADF4(C16) were the only ones being internalized by less cells in the presence of DMA after 6 h (2.4%) compared to internalization by cells in the presence of DC. After 24 h of incubation, the number of cells containing eADF4(C16) particles in the presence of DMA (7.7%) was again similar to that of cells in the presence of DC.
eADF4(κ16) particle internalization was severely inhibited with DC after 6 h (27.3%) and 24 h (47.3%) whereas in the presence of DMA the internalization was less inhibited after 6 h (44%) and 24 h (73%). Accordingly, numbers of cells with blend, Tat-eADF4(C16), eADF4(C16)R8G or eADF4(C16)RGD particles decreased moderately in the presence of DMA in comparison to that in the presence of DC (Table 4) indicating that macropinocytosis plays a minor and clathrin-mediated endocytosis a major role in silk particle uptake in the used experimental setup.
Generally, three different groups can be identified which differ in particle internalization. The first group is eADF4(C16) particles with indistinguishable inhibition of internalization in the presence of both inhibitors. The second group contains Tat-eADF4(C16), eADF4(C16)RGD, and blend particles with a decreased cellular uptake in the presence of DC and a slightly less decreased uptake in the presence of DMA indicating that clathrin-mediated endocytosis is the main route of particle uptake with macropinocytosis playing a medium role. Unexpectedly, macropinocytosis played a more important role in case of eADF4(C16)RGD, despite most well-known receptors involved in receptor-mediated endocytosis (e.g. LDL receptor73) being internalized via clathrin-mediated endocytosis. Here, the amino acid sequence of the RGD peptide (GRGDSP) is optimized for the recognition by α5β1- and αvβ3-integrin.49,74 These integrin receptors are internalized by clathrin-dependent and –independent as well as caveolin-mediated endocytosis75–80 which would explain the involvement of marcopinocytosis in eADF4(C16)RGD particle internalization. The third group represents eADF4(κ16) and eADF4(C16)R8G particles which were internalized by most cells. Here, uptake is severely inhibited in the presence of DC, whereas inhibition of uptake is only moderate in the presence of DMA, indicating that here macropinocytosis only plays a minor role.
The fact that eADF4(κ16) and eADF4(C16)R8G internalization is higher than that of other modified particles (Fig. 3) makes them promising candidates for drug delivery vehicles.
Conclusions
A new route for producing spider silk particles has been established using the ionic liquid EMiM[acetate] as a starting solvent. Particles made therefrom show small diameters and are quite homogenous with a narrow particle size distribution. Beneath previously employed engineered spider silk proteins several variants with different (cell penetrating) peptide tags were used to produce particles. The tags and blending of proteins had no influence on particle formation but on the electrophoretic mobility, zeta potential and therefore on the colloidal stability. Interestingly, the uptake efficiency of spider silk particles by HeLa cells was highest in the presence of a poly-arginine tag (zeta potential −17.1 mV) or when using the polycationic mutant (without tag) eADF4(κ16) (zeta potential 13.2 mV) in which all naturally occurring glutamic acid residues have been replaced by lysine ones. Both the number of cells containing spider silk particles as well as the number of internalized particles was increased (Fig. 3B). Uptake mostly occurred through clathrin-mediated endocytosis for all used spider silk particles as determined by using dansylcadaverine. In combination with the recently reported encapsulation efficiency of low molecular weight drugs, low molecular weight proteins or even nucleic acids by eADF4(κ16) particles,50 engineered silk particles show high potential as novel drug delivery systems.Acknowledgements
We gratefully thank Nicolas Helfricht for the assistance with zeta potential measurements, Jasmin Schmidt for help/discussion on colloidal stability, Marino Zerial and Undine Schubert for experimental help and inspiring discussion, Johannes Diehl and Andreas Schmidt for fermentation of eADF4(C16)R8G, eADF4(C16)RGD and eADF4(κ16), and Stefanie Wohlrab, Martin Humenik, Christian Borkner and Eileen Lintz for fruitful discussions and comments on the manuscript.References
- R. Langer and N. A. Peppas, AIChE J., 2003, 49, 2990–3006 CrossRef CAS.
- Y. B. Choy, F. Cheng, H. Choi and K. K. Kim, Macromol. Biosci., 2008, 8, 758–765 CrossRef CAS PubMed.
- C. Berkland, K. Kim and D. W. Pack, J. Controlled Release, 2001, 73, 59–74 CrossRef CAS PubMed.
- J. Herrmann and R. Bodmeier, J. Controlled Release, 1995, 36, 63–71 CrossRef CAS.
- E. Wenk, A. J. Wandrey, H. P. Merkle and L. Meinel, J. Controlled Release, 2008, 132, 26–34 CrossRef CAS PubMed.
- S. Hofmann, C. T. Wong Po Foo, F. Rossetti, M. Textor, G. Vunjak-Novakovic, D. L. Kaplan, H. P. Merkle and L. Meinel, J. Controlled Release, 2006, 111, 219–227 CrossRef CAS PubMed.
- S. Freiberg and X. X. Zhu, Int. J. Pharm., 2004, 282, 1–18 CrossRef CAS PubMed.
- S. Grund, M. Bauer and D. Fischer, Adv. Eng. Mater., 2011, 13, B61–B87 CrossRef.
- A. H. Faraji and P. Wipf, Bioorg. Med. Chem., 2009, 17, 2950–2962 CrossRef CAS PubMed.
- S. K. Nitta and K. Numata, Int. J. Mol. Sci., 2013, 14, 1629–1654 CrossRef CAS PubMed.
- K. Numata, B. Subramanian, H. A. Currie and D. L. Kaplan, Biomaterials, 2009, 30, 5775–5784 CrossRef CAS PubMed.
- A. S. Gobin, R. Rhea, R. A. Newman and A. B. Mathur, Int. J. Nanomed., 2006, 1, 81–87 CrossRef CAS PubMed.
- B. Subia, S. Chandra, S. Talukdar and S. C. Kundu, Integr. Biol., 2014, 6, 203–214 RSC.
- L. Meinel and D. L. Kaplan, Adv. Drug Delivery Rev., 2012, 64, 1111–1122 CrossRef CAS PubMed.
- B. Kundu, R. Rajkhowa, S. C. Kundu and X. Wang, Adv. Drug Delivery Rev., 2013, 65, 457–470 CrossRef CAS PubMed.
- A. B. Mathur and V. Gupta, Nanomed., 2010, 5, 807–820 CrossRef CAS PubMed.
- U. Slotta, M. Tammer, F. Kremer, P. Koelsch and T. Scheibel, Supramol. Chem., 2006, 18, 465–471 CrossRef CAS.
- K. Spiess, S. Wohlrab and T. Scheibel, Soft Matter, 2010, 6, 4168–4174 RSC.
- K. Schacht and T. Scheibel, Biomacromolecules, 2011, 12, 2488–2495 CrossRef CAS PubMed.
- A. Leal-Egaña, G. Lang, C. Mauerer, J. Wickinghoff, M. Weber, S. Geimer and T. Scheibel, Adv. Eng. Mater., 2012, 14, B67–B75 CrossRef.
- K. D. Hermanson, D. Huemmerich, T. Scheibel and A. R. Bausch, Adv. Mater., 2007, 19, 1810–1815 CrossRef CAS.
- C. Blüm, A. Nichtl and T. Scheibel, Adv. Funct. Mater., 2014, 24, 763–768 CrossRef.
- U. K. Slotta, S. Rammensee, S. Gorb and T. Scheibel, Angewandte Chemie International Edition, 2008, 47, 4592–4594 Search PubMed.
- A. Lammel, M. Schwab, U. Slotta, G. Winter and T. Scheibel, ChemSusChem, 2008, 1, 413–416 CrossRef CAS PubMed.
- B. Subia and S. C. Kundu, Nanotechnology, 2013, 24, 035103 CrossRef CAS PubMed.
- F. P. Seib, G. T. Jones, J. Rnjak-Kovacina, Y. Lin and D. L. Kaplan, Adv. Healthcare Mater., 2013, 2, 1606–1611 CrossRef CAS PubMed.
- E. Gros, S. Deshayes, M. C. Morris, G. Aldrian-Herrada, J. Depollier, F. Heitz and G. Divita, Biochim. Biophys. Acta, 2006, 1758, 384–393 CrossRef CAS PubMed.
- B. Gupta, T. S. Levchenko and V. P. Torchilin, Adv. Drug Delivery Rev., 2005, 57, 637–651 CrossRef CAS PubMed.
- G. P. Dietz and M. Bahr, Mol. Cell. Neurosci., 2004, 27, 85–131 CrossRef CAS PubMed.
- M. Mae and U. Langel, Curr. Opin. Pharmacol., 2006, 6, 509–514 CrossRef PubMed.
- T. Lehto, K. Kurrikoff and U. Langel, Expert Opin. Drug Delivery, 2012, 9, 823–836 CrossRef CAS PubMed.
- P. Jarver, K. Langel, S. El-Andaloussi and U. Langel, Biochem. Soc. Trans., 2007, 35, 770–774 CrossRef CAS PubMed.
- B. R. Meade and S. F. Dowdy, Adv. Drug Delivery Rev., 2008, 60, 530–536 CrossRef CAS PubMed.
- M. Green, M. Ishino and P. M. Loewenstein, Cell, 1989, 58, 215–223 CrossRef CAS PubMed.
- E. Vives, P. Brodin and B. Lebleu, J. Biol. Chem., 1997, 272, 16010–16017 CrossRef CAS PubMed.
- S. Futaki, T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda and Y. Sugiura, J. Biol. Chem., 2001, 276, 5836–5840 CrossRef CAS PubMed.
- P. Guterstam, F. Madani, H. Hirose, T. Takeuchi, S. Futaki, S. El Andaloussi, A. Graslund and U. Langel, Biochim. Biophys. Acta, 2009, 1788, 2509–2517 CrossRef CAS PubMed.
- C. Y. Jiao, D. Delaroche, F. Burlina, I. D. Alves, G. Chassaing and S. Sagan, J. Biol. Chem., 2009, 284, 33957–33965 CrossRef CAS PubMed.
- K. S. Kawamura, M. Sung, E. Bolewska-Pedyczak and J. Gariepy, Biochemistry, 2006, 45, 1116–1127 CrossRef CAS PubMed.
- D. J. Mitchell, D. T. Kim, L. Steinman, C. G. Fathman and J. B. Rothbard, J. Pept. Res., 2000, 56, 318–325 CrossRef CAS PubMed.
- I. Nakase, M. Niwa, T. Takeuchi, K. Sonomura, N. Kawabata, Y. Koike, M. Takehashi, S. Tanaka, K. Ueda, J. C. Simpson, A. T. Jones, Y. Sugiura and S. Futaki, Mol. Ther., 2004, 10, 1011–1022 CrossRef CAS PubMed.
- I. Nakase, Y. Konishi, M. Ueda, H. Saji and S. Futaki, J. Controlled Release, 2012, 159, 181–188 CrossRef CAS PubMed.
- G. Ter-Avetisyan, G. Tuennemann, D. Nowak, M. Nitschke, A. Herrmann, M. Drab and M. C. Cardoso, J. Biol. Chem., 2009, 284, 3370–3378 CrossRef CAS PubMed.
- S. A. Nasrollahi, S. Fouladdel, C. Taghibiglou, E. Azizi and E. S. Farboud, Int. J. Dermatol., 2012, 51, 923–929 CrossRef CAS PubMed.
- D. M. Copolovici, K. Langel, E. Eriste and U. Langel, ACS Nano, 2014, 8, 1972–1994 CrossRef CAS PubMed.
- F. Madani, S. Lindberg, U. Langel, S. Futaki and A. Graslund, J. Biophys., 2011, 2011, 414729 Search PubMed.
- N. Schmidt, A. Mishra, G. H. Lai and G. C. Wong, FEBS Lett., 2010, 584, 1806–1813 CrossRef CAS PubMed.
- S. Yigit, O. Tokareva, A. Varone, I. Georgakoudi and D. L. Kaplan, Macromol. Biosci., 2014, 14, 1291–1298 CrossRef CAS PubMed.
- S. Wohlrab, S. Müller, A. Schmidt, S. Neubauer, H. Kessler, A. Leal-Egaña and T. Scheibel, Biomaterials, 2012, 33, 6650–6659 CrossRef CAS PubMed.
- E. Doblhofer and T. Scheibel, J. Pharm. Sci., 2014 DOI:10.1002/jps.24300.
- A. I. Ivanov, in Exocytosis and Endocytosis, ed. A. I. Ivanov, Humana Press, Totowa, 2008, vol. 440, ch. 2, pp. 15–33 Search PubMed.
- D. Huemmerich, C. W. Helsen, S. Quedzuweit, J. Oschmann, R. Rudolph and T. Scheibel, Biochemistry, 2004, 43, 13604–13612 CrossRef CAS PubMed.
- E. Gasteiger, A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel and A. Bairoch, Nucleic Acids Res., 2003, 31, 3784–3788 CrossRef CAS PubMed.
- E. Gasteiger, C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel and A. Bairoch, in The Proteomics Protocols Handbook, ed. J. M. Walker, Humana Press Inc., Totowa, 2005, pp. 571–607 Search PubMed.
- K. Spiess, A. Lammel and T. Scheibel, Macromol. Biosci., 2010, 10, 998–1007 CrossRef CAS PubMed.
- R. J. Hunter, Zeta Potential in Colloid Science: Principles and Applications, Academic Press Inc., San Diego, 1981 Search PubMed.
- M. Humenik and T. Scheibel, ACS Nano, 2014, 8, 1342–1349 CrossRef CAS PubMed.
- M. Humenik, M. Magdeburg and T. Scheibel, J. Struct. Biol., 2014, 186, 431–437 CrossRef CAS PubMed.
- C. Blüm and T. Scheibel, BioNanoScience, 2012, 2, 67–74 CrossRef.
- N. Helfricht, M. Klug, A. Mark, V. Kuznetsov, C. Blum, T. Scheibel and G. Papastavrou, Biomater. Sci., 2013, 1, 1166–1171 RSC.
- M. P. Neubauer, C. Blüm, E. Agostini, J. Engert, T. Scheibel and A. Fery, Biomater. Sci., 2013, 1, 1160–1165 RSC.
- A. Leal-Egana and T. Scheibel, J. Mater. Chem., 2012, 22, 14330–14336 RSC.
- S. A. Kulkarni and S. S. Feng, Pharm. Res., 2013, 30, 2512–2522 CrossRef CAS PubMed.
- R. Firdessa, T. A. Oelschlaeger and H. Moll, Eur. J. Cell Biol., 2014, 93, 323–337 CrossRef CAS PubMed.
- C. B. He, Y. P. Hu, L. C. Yin, C. Tang and C. H. Yin, Biomaterials, 2010, 31, 3657–3666 CrossRef CAS PubMed.
- J. Zhu, L. Liao, L. Zhu, P. Zhang, K. Guo, J. Kong, C. Ji and B. Liu, Talanta, 2013, 107, 408–415 CrossRef CAS PubMed.
- K. Y. Win and S. S. Feng, Biomaterials, 2005, 26, 2713–2722 CrossRef CAS PubMed.
- R. H. Müller, Zetapotential und Partikelladung in der Laborpraxis, Wissenschaftlicher Verlagsgesellschaft mbH Stuttgart, Stuttgart, 1996 Search PubMed.
- I. Szabo, S. Brutsche, F. Tombola, M. Moschioni, B. Satin, J. L. Telford, R. Rappuoli, C. Montecucco, E. Papini and M. Zoratti, EMBO J., 1999, 18, 5517–5527 CrossRef CAS PubMed.
- R. J. Davis and M. P. Czech, J. Biol. Chem., 1985, 260, 2543–2551 CAS.
- S. Schutze, T. Machleidt, D. Adam, R. Schwandner, K. Wiegmann, M. L. Kruse, M. Heinrich, M. Wickel and M. Kronke, J. Biol. Chem., 1999, 274, 10203–10212 CrossRef CAS PubMed.
- M. Koivusalo, C. Welch, H. Hayashi, C. C. Scott, M. Kim, T. Alexander, N. Touret, K. M. Hahn and S. Grinstein, J. Cell Biol., 2010, 188, 547–563 CrossRef CAS PubMed.
- H. Jeon and S. C. Blacklow, Annu. Rev. Biochem., 2005, 74, 535–562 CrossRef CAS PubMed.
- U. Hersel, C. Dahmen and H. Kessler, Biomaterials, 2003, 24, 4385–4415 CrossRef CAS PubMed.
- T. Pellinen, S. Tuomi, A. Arjonen, M. Wolf, H. Edgren, H. Meyer, R. Grosse, T. Kitzing, J. K. Rantala, O. Kallioniemi, R. Fassler, M. Kallio and J. Ivaska, Dev. Cell, 2008, 15, 371–385 CrossRef CAS PubMed.
- F. Shi and J. Sottile, J. Cell Sci., 2008, 121, 2360–2371 CrossRef CAS PubMed.
- B. G. Galvez, S. Matias-Roman, M. Yanez-Mo, M. Vicente-Manzanares, F. Sanchez-Madrid and A. G. Arroyo, Mol. Biol. Cell, 2004, 15, 678–687 CrossRef CAS PubMed.
- T. Nishimura and K. Kaibuchi, Dev. Cell, 2007, 13, 15–28 CrossRef CAS PubMed.
- L. Liu, B. He, W. M. Liu, D. Zhou, J. V. Cox and X. A. Zhang, J. Biol. Chem., 2007, 282, 31631–31642 CrossRef CAS PubMed.
- P. T. Caswell, S. Vadrevu and J. C. Norman, Nat. Rev. Mol. Cell Biol., 2009, 10, 843–853 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4bm00401a |
| This journal is © The Royal Society of Chemistry 2015 |