Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optimization of enzyme immobilization on magnetic microparticles using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) as a crosslinking agent
F.
Kazenwadel
,
H.
Wagner
,
B. E.
Rapp
and
M.
Franzreb
*
Karlsruhe Institute of Technology, Institute of Functional Interfaces, Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany. E-mail: Matthias.Franzreb@kit.edu
First published on 6th November 2015
Abstract
Enzyme immobilization is a versatile tool in biotransformation processes to enhance enzyme activity and to secure an easy separation of catalysts and products and the reusability of enzymes. A simple and commonly used method for crosslinking enzymes to a solid support is the zero-length crosslinking agent 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). This work shows the optimization of the EDC-crosslinking protocol for two enzymes, glucose oxidase (GOx) and horseradish peroxidase (HRP), to functionalized magnetic microparticles. For GOx the optimization of the immobilization parameters pH-value and the enzyme to particle ratio results in activity yields of up to 36%, which is in the usual range for undirected enzyme immobilisations. In contrast, for HRP the activity yield does not exceed 6% even after optimization of the protocols. The main reasons for this unusually low activity yield are the presence of multiple HRP isoforms in the enzyme solution used for immobilisation and the observed tendency of HRP to be inactive even in the case of simple physisorption to the particle surface.
Introduction
The use of enzymes as catalysts offers a number of advantages compared to classical organic-chemical synthesis. As enzymes are highly substrate- and product-specific and are able to perform stereoselective reactions, the formation of unwanted side products can be reduced or even prevented and cost-intensive purification processes can be simplified.1 Moreover, as enzyme reactions mostly occur in aqueous media, preferring mild reaction conditions, their application can be a substantial contribution to green and sustainable production processes.2 The high selectivity and performance of enzymes also allow their use in diagnostic and sensing applications, as for example for blood glucose detection, excellently reviewed by Yoo and Lee in 2010.3 However, main drawbacks of using proteins as biocatalysts are their high costs and their susceptibility against harsh reaction conditions.4 Furthermore, in order to purify the product, enzymes have to be depleted from the reaction solution, whereby they often get inactivated. This problem can be solved by immobilizing the biocatalysts to solid supports which enables the easy separation from the product stream post-synthesis. In addition, an immobilization often causes enhanced stability and activity of enzymes5,6 especially concerning temperature, pH values or substrate specificity.7,8 However, in some cases the activity can also be reduced or even destroyed, depending on the respective technique.8 Many different immobilization procedures were reported, differing in their specificity, efficiency, simplicity and purpose.9 For example the sophisticated use of genetically encoded tags results in an exact site specific immobilization,10 while approaches that do not need genetic manipulation, for example by using “click chemistry”, lead to a more random distribution and orientation of the proteins on the support.11 One simple and fast way of immobilizing proteins to functionalized solid supports or among each other12 is the use of crosslinking agents. Chemical crosslinking means linking two or more molecules by a stable covalent bond. Beside the use in immobilization purposes, chemical crosslinking is also employed for the stabilization of the protein structure13 or the identification of unknown interaction partners.14 Crosslinking agents normally consist of a chemical backbone which is mainly defined by its spacer arm length. An exception are the so-called zero-length crosslinkers that only serve as activators, but do not introduce spacer atoms while connecting molecules.15 The functional groups at the ends of the crosslinking molecule directly react with specific side chains of superficial amino acids of the target protein. Such functional groups are primary amines, carboxylic groups, sulfhydryls or carbonyls.16 The classification of interlinking molecules depends on the nature of their functional groups. If they carry the same functional group on each arm, they are called homobifunctional crosslinkers (e.g. glutaraldehyde17). Crosslinkers are classified as heterobifunctional if they carry two different functional groups. They can be further classified by their physicochemical properties such as solubility in organic solvents or water, their behaviour when applied to living cells or their reaction mechanism. Some crosslinking agents contain sites for subsequent controlled cleavage of the immobilized target enzyme after the immobilization process.18 The use of crosslinking molecules in enzyme immobilization involves one main disadvantage: as enzymes are attached to the respective surface in a random orientation, activity often decreases immensely or even gets lost. The use of short crosslinkers results in a small distance between the surface and molecule, leading to a restricted flexibility of the molecule that can be accompanied by a decrease in activity and/or selectivity.19 However, the use of chemical crosslinkers for enzyme immobilization also offers some crucial advantages: the main benefit is the simplicity of the method. Neither enzymes nor supports have to be modified costly before the reaction. This enables the use of a broad range of different materials. As most crosslinking reactions of enzymes can be performed in aqueous media under mild reaction conditions, the process is environmentally sustainable and little hazardous. A wide variety of crosslinking molecules exists that can address a broad range of functional groups, providing an appropriate molecule for each immobilization application.Besides this variety of potential crosslinkers, different supports can be applied when proteins are to be immobilized. To obtain optimal results carriers should provide a large specific surface for biocatalyst attachment while being easily separable from the reaction solution. Magnetic carriers combine both requirements as nano- or micro-particles provide large specific surfaces and can be separated rapidly and reliably by applying an external magnetic field.20 Magnetic particles can be purchased with a wide variety of different chemical functionalities and can be easily further modified.
Table 1 shows an overview of publications reporting the use of chemical crosslinkers to immobilize enyzmes to magnetic carriers. The immobilization method is specified, naming the crosslinking method and the pH-value used in the coupling process. Furthermore, the isoelectric point and molecular size of the target-protein are listed and yields in binding and activity upon immobilization compared to free enzymes, if published, are cited. Publications range from the early 1970s to the current research state, pointing out that chemical crosslinking has a long history in the immobilization of enzymes to magnetic carriers. Most enzymes are technical enzymes that are needed for the conversion of substrates used in industrial processes, such as invertase or lipase. Other enzymes are immobilized to serve as sensoric tools, as for example cholesterol esterase. Crosslinking agents mainly contain carbodiimides, but also glutaraldehyde or epoxy groups are used for the crosslinking of amino groups. In addition in some cases stable non-covalent binding can be found. pH-values during the immobilization process are for this discussion expected to be optimized for the respective enzyme and range from 4.0 to 8.0. The pI-values of the enzymes used also vary from 3.8 (invertase from yeast) to 10.8 (bovine trypsin). It is often recommended to adjust the pH-value during the coupling process close to the isoelectric point of the enzyme in order to avoid superficial loading that may lead to a repulsion of the enzyme from the carriers. The pH-value is adjusted near the pI (±0.5–1) in 4 of the cited publications, and the rest differs for more than 1.5 pH magnitudes. Looking at Table 1 it is hard to find general trends. However, it becomes obvious that if low enzyme amounts per g of magnetic particles are offered, the resulting binding yields are high and vice versa. Most articles report initial conditions of less than 50 mg enzyme per g of particles and binding yields of 70–100%, demonstrating the effectiveness of using crosslinking agents for the immobilization of enzymes. Choosing an optimum, it has to be considered that while increasing the specific activity of the immobilisates by increasing the enzyme amount bound, the activity of the immobilized enzyme might decrease because of steric hindrance.21 Activity yields, saying how much of the initially offered activity of free enzymes finally is detectable on the immobilisates, are much less predictable. If published, they range between less than 10% to more than 100%. The same variety holds true, even if we restrict our analysis to the cases in which the same particles as in our study were used (magnetic polyvinylalcohol particles of the company Perkin-Elmer chemagen). Assuming that all protocols are optimized, these findings show that not all enzymes are identically well suited for immobilization.
| Literature | Carrier | Protein | Crosslinking agent | pH | pI | Enzyme offered | Mass | Binding yield | Activity yield |
|---|---|---|---|---|---|---|---|---|---|
| van Leemputten & Horisberger (1974)8 | Aminoalkylsilylated magnetite (Fe3O4) | Trypsin | Glutaraldehyde | 8.0 | 10.8 | 50 mg enzyme per g particles | 23.3 kDa | 72% | 46%, 8% |
| Invertase | 3.8 | 40 mg enzyme per g particles | 270 kDa | 11% | 8.7% (+1% sucrose) | ||||
| Bahar & Celebi (1999)24 | Magnetic poly(styrene) particles | Glucoamylase | Aldehyde-groups | 4.0 | 4.2 | 8 mg enzyme per g particles | 72 kDa | 70% | 70% |
| Liao & Chen (2001)25 | Fe3O4 magnetic nanoparticles (10.6 nm) | Yeast alcohol dehydrogenase | Carbodiimide | 6.0 | 5.4–5.8 | 0.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) 1 (w/w) |
141–151 kDa | 100% | 62% |
| Akgöl et al. (2001)26 | Magnetic PVA microspheres | Invertase | 1,1′-Carbonyldiimidazole | 7.0 | 3.8 | N/A | 270 kDa | 7.18 mg g per particles | 74% |
| Zheng et al. (2003)21 | Magnetic poly(VAc–DVB) microspheres | Candida cylindracea lipase | Adsorptive | 7.0 | 4.5 | N/A | 43 kDa | 8–35 mg per g particles | 6750 IU per g carrier |
| Wang & Lee (2003)27 | Fe3O4 magnetic nanoparticles | Trypsin | Carbodiimide | — | 10.8 | 17 mg enzyme per g particles | 23.3 kDa | 86% | N/A |
| Avidin | Cyanamid | 10.5 | 66 kDa | 100% | N/A | ||||
| Bozhinova et al. (2004)28 | Magnetic PVA microparticles | E.coli penicillin amidase | Epoxygroups glutaraldehyde | 7.5 | 4.3–7.0 | 30 mg enzyme per g particles | 70 kDa | 10–93% | 50–100% |
| Kouassi et al. (2005)29 | Magnetic nanoparticles | Cholesterol oxidase | Carbodiimide | 7.4 | 5.1–5.4 | 7–10 mg enzyme per g particles | 34 kDa | 98–100% | >100% |
| Bruno et al. (2005)30 | Magnetic POS–PVA particles | Mucor miehei lipase | Glutaraldehyde | 7.0 | 3.8 | N/A | 32 kDa | N/A | 65% |
| N. Schultz (2007)31 | Magnetic PVA microparticles | Candida antarctica lipase A (CALA) | Carbodiimide | 7.0 | 7.5 | 16.7 mg enzyme per g particles | 45 kDa | 30% | 8% |
| Huang et al. (2008)7 | Fe3O4 magnetic particles (12.7 nm) | Candida rugosa lipase | Carbodiimide | 6.0 | 4.5 | <33 mg enzyme per g particle | 43 kDa | 100% | 141% |
| Magario et al. (2008)32 | Magnetic PVA microparticles | Naringinase | Carbodiimide | 7.0 | ∼5 | 3.7 mg enzyme per g particles | 90 kDa | 82% | 36% |
| Ricco et al. (2014)33 | Magnetic nanoparticles | Almond beta-glucosidase | Carbodiimide | 4.6 | 7.3 | 400–1000 mg enzyme per g particle | 110 kDa | 18–24% | N/A |
| Morhardt et al. (2014)34 | Magnetic PVA microparticles | Chymotrypsine | SMCC | 7.2 | 8.75 | 2.9–95.5 mg enzyme per g particles | 25 kDa | 75–100% | 9–45% |
| Adsorptive | 5.5 | ||||||||
| Carbodiimide | 5.3 | 75–100% | |||||||
| Kazenwadel et al. (2015) | Magnetic PVA microparticles | Glucose oxidase | Carbodiimide | 4.0 | 4.2 | 5–15 mg enzyme per g particles | 160 kDa | 67% | 34% |
| Horseradish peroxidase | |||||||||
| 4.0 | 3.0–9.0 | 44 kDa | 100% | 6.5% |
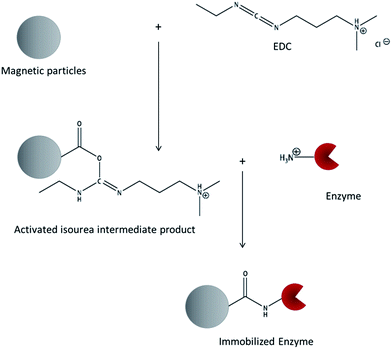
Table 1 also shows that 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) is one of the most commonly used crosslinking molecules. As it only activates carboxy-groups and mediates the linkage with superficial primary amino groups without the introduction of any spacer molecules, it can be classed among the so-called zero-length crosslinking agents. A neutral pH value around 7.0 during the immobilization process is required according to the standard protocols. The reaction is performed in two steps (see Fig. 1): first, the carboxy-groups of the carrier are activated by adding EDC. An active acyl-isourea intermediate product is formed. Second, the enzyme is added and a peptide bond is formed between the carboxy-groups on the surface and the superficial lysine amino side chains of the enzyme.16
In this article, the optimization of enzyme crosslinking to magnetic particles using EDC as a zero-length crosslinking agent is shown. By optimizing the protocol concerning coupling pH-value and enzyme loading, the activity of the used model enzymes glucose oxidase (GOx, Aspergillus niger, see ref. 22 for detailed information) and peroxidase (HRP, Armoracia rusticana, see ref. 23 for detailed information) could be enhanced significantly. Nevertheless, the achieved activity yields remained low, especially in the case of HRP. Possible reasons for this will be discussed and the reversible physical adsorption/desorption of the enzyme on the particle surface is identified as one critical process.
Materials and methods
Magnetic microparticles (M-PVA C22, average diameter of 1–3 μm, carboxy-group density of 950 μmol COOH g−1) were obtained from Perkin Elmer chemagen (Baesweiler, Germany). Buffer components, D-(+)-glucose and the enzymes glucose oxidase and horseradish peroxidase (lyophilized powder, grade VI) were purchased from Sigma Aldrich. TMB-substrate solutions were purchased from Sigma Aldrich (ELISA: extra slow, ready to use) and KPL (solution A, without hydrogen peroxide). PierceTM bicinchoninic acid (BCA)-assay kit (Life Technologies) was used for the determination of protein concentration. All substances were of analytical grade.Enzyme immobilization
Optimizations were based on the protocol established by Morhardt et al. in 2014.34 Magnetic microparticles (60 g L−1) were washed in 0.1 M 2-(N-morpholino)ethanesulfonic acid, pH 5.3 and activated by adding 50 mg ml−1 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and mixing at 11 °C for 35 min. After separation and washing of the particles, enzyme solutions (5 to 30 mg enzyme per g particles) in different buffers with varying pH-values (0.02 M citrate buffer, pH 4.0; 0.01 M MES-buffer pH 5.3; 0.01 M sodium phosphate buffer, pH 6.0 and 7.0) were added and incubated for 2 h at 25 °C while mixing (see the reaction scheme, Fig. 1). Samples without the addition of EDC served as controls for physical protein adsorption. Particles were washed using 0.8 M NaCl. Enzyme–particle-complexes were washed and suspended in 0.1 M sodium phosphate buffer, pH 6.0.Control experiments for the detection of enzyme inactivation during the immobilization process were performed by omitting the addition of magnetic particles and by replacing the EDC-activated carboxy-terminated magnetic particles by native M-PVA-C22 particles. Adsorption was measured by detecting the protein concentration in supernatants after coupling. The specific activity of the immobilisates [U per mg particles] was calculated by dividing the activity of the particle suspension [U ml−1] by the particle concentration [mg ml−1]. Enzyme activity [U per mg enzyme] was determined by dividing the specific activity of the immobilisates [U per mg particles] by the enzyme to particle ratio of the particles [mg enzyme per mg particle]. The binding yield [%] was determined by dividing the bound enzyme mass by the applied enzyme mass. At the end, the yield in activity [%] was determined by dividing the total activity of the immobilized enzyme by the total activity of the free enzyme initially offered for immobilisation.
Activity assay for enzyme–particle-complexes
The particle solutions were diluted and 50 μl were placed in the wells of a 96-well-plate. The HRP-activity assay was started by adding 200 μl of ELISA-TMB-solution (Sigma Aldrich). The GOx-activity assay was started by adding 10 μl 0.1 mg ml−1 HRP-solution, 100 μl TMB-solution (KPL) and 100 μl 72 g L−1D-glucose solution. The absorption change as a function of time was recorded at 653 nm (εTMB = 39 ml μmol−1 cm−1) and volumetric enzyme activity was calculated using the Lambert–Beer–Law.Recycling experiments
The enzyme immobilisates produced under optimized conditions were diluted in 0.1 M NaPO4-buffer pH 6.0 yielding a 60 μg ml−1 solution and 50 μl (=3 μg) were placed in the well of a 96-well plate. Afterwards an activity assay for enzyme immobilisates was performed as described above. As the last step of each cycle, the particles were separated, washed and resuspended in 0.1 M NaPO4-buffer to a volume of 50 μl. It was of main importance not to touch the particles with the pipette tip, as they easily adsorb to all kind of surfaces, leading to a decreased mass of applied particles in the assay. The procedure was repeated 6 times for adsorbed and covalently bound enzymes.Results
Optimization of the pH-value in the enzyme coupling step
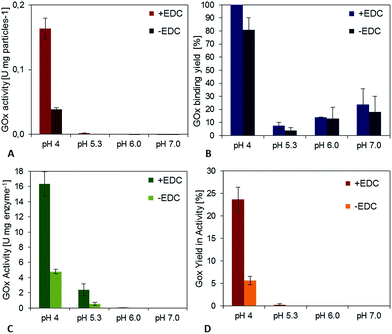
The activation step was optimized by Morhardt et al. in 201434 using a Design-of-Experiments (DoE) approach. As this step is independent from the enzymes used, it was adopted without further changes. In contrast, an optimization of the coupling step was performed, because of its dependency on the enzyme used. First experiments showed that the duration (between 2 h and 25 h) and temperature (between 11 °C and 25 °C) do not have a significant effect on binding and activity (data not shown), so these parameters were kept constant during the trials (2 h, 25 °C).The pH-value during the enzyme attachment was varied and resulting activity of the particle suspension was measured. For GOx it could be clearly shown that a decrease of the pH-value from 7.1 to 4.0 led to a significant enhancement of the specific activity of the immobilisates (Fig. 2A). It also resulted in an enhancement of the activity of control particles to which enzymes were physically adsorbed. However, particles with chemically bound enzymes (+EDC) showed more than 3.2-fold activity compared to the adsorption controls (−EDC).
In order to find a reason for this behavior, the amount of bound enzymes was determined by probing the protein concentration in the supernatants of coupling and washing steps and the yield of enzymes that bound to the particles was calculated (Fig. 2B). At a pH value of 7.0 only about 23% of the added enzyme bound to the particles, while at a pH value of 4.0 no enzyme in the supernatant could be detected after the coupling process. Interestingly, the binding yield does not increase steadily when probing pH-values between 7 and 4 during immobilization, but shows a clear minimum with binding yields of approximately 14% (pH 6.0) and 8% (pH 5.3). The difference in the GOx binding yield comparing chemically crosslinked (+EDC) and physically adsorbed samples (−EDC) was not significant. Together with the finding that enzyme activity is 3.2-fold lower for adsorbed samples, this indicates that enzymes were stabilized during the immobilization process and activity was preserved using covalent binding. This effect can also be accentuated by calculating the activity of the immobilized GOx [U per mg enzyme] for adsorbed and covalently bound samples (Fig. 2C). While there is practically no or only a little activity detectable for the pH-values 5.3, 6 and 7, the activity of crosslinked GOx at pH 4.0 is 4-fold higher compared to physically adsorbed enzymes, highlighting the importance of a covalent bond for the activity of glucose oxidase. For the immobilization of 10 mg GOx to functionalized PVA-coated magnetic particles, a total yield in the activity of approximately 24% could be achieved for covalently bound GOx and 5.5% for adsorbed GOx for pH 4, while when starting the trials using the published protocol (pH 7.0) hardly any activity could be detected (Fig. 2D).
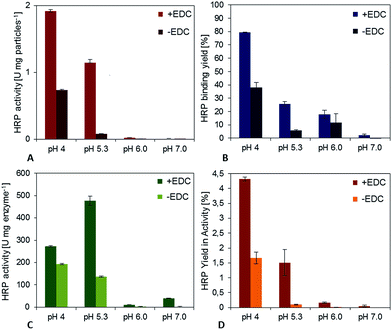
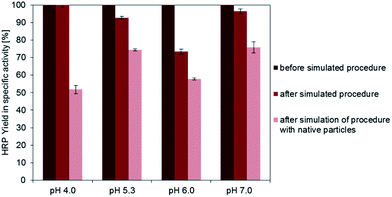
The second model enzyme, horseradish peroxidase, showed in many aspects a comparable behavior (Fig. 3). At a pH value of 7.0, a specific activity of the immobilisates of approximately 0.09 U per mg particles could be detected for covalent binding (+EDC). After decreasing the pH-value to 4.0, the specific activity could be increased significantly to 1.9 U per mg particles. The binding yield for pH 7 was only 2.5%, but it could be enhanced to 80%, when the pH-value in the coupling step was regulated to 4.0. An adjustment of the pH-value to 6.0 led to a binding yield of 18%, an adjustment to pH 5.3 to 58%. Adsorptive binding (−EDC) occurred for all pH-values (approximately 30–50% compared to covalent binding). However, the activity of physically bound enzymes was not as high as for covalent bound HRP, underlining the importance of a chemical bond for enzyme stabilization while immobilization also in this case. Fig. 3D shows the activity of immobilized enzyme. Although enzyme immobilized at pH 5.3 shows the highest activity related to immobilized mass, yield in activity is highest for pH 4.0, as more enzymes can be bound to the particle surface. The yield of immobilized activity is less than 4.5% for 10 mg HRP immobilized covalently on functionalized magnetic particles at a pH value of 4.0. For physically adsorbed enzymes it is approximately 1.5%, which is less than half compared to the crosslinking approach.
These results prove that for both enzymes the recommended immobilization conditions of pH 7 are not ideal and that the yields in immobilized mass and activity could be significantly enhanced by adjusting the pH-value in the coupling process. As it can be derived from the binding yield trend, the pH-value influences enzyme binding efficiency and thus the specific activity of enzyme immobilisates. This may be due to charging phenomena at the surface of the enzyme that depend on the isoelectric point (pI) and lead to different physical adsorption on the surface at different ambient pH-values. In the literature it is often recommended to adjust the coupling pH-value near the isoelectric point of the enzyme, in order to avoid superficial protein loading and thus repulsion from charged surfaces. This recommendation holds true for glucose oxidase, which showed a binding maximum at pH 4.0, while the isoelectric point of the enzyme is reported to be 4.2 (Sigma Aldrich, Material Data Sheet). However, for horseradish peroxidase, there is not such an explicit result. This is because the HRP used for this work is extracted from horseradish roots and contains at least 7 different isoforms, whose pI-values vary from 3.0–9.0 (Sigma Aldrich, Material Data Sheet). In view of this variety it becomes obvious that no real optimum can be found, which would result in high binding and activity yields of all HRP isoforms offered for immobilization.
Effect of the enzyme to particle ratio during immobilisation
As the density of immobilized enzymes on a surface can play an important role in enzyme activity, different enzyme to particle ratios were investigated for HRP and GOx. For glucose oxidase (Table 2), specific activity remains comparatively small for the lowest enzyme to particle ratio, 0.08 U per mg particles for 5 mg GOx per g particles and 0.16 U per mg particles for 10 mg GOx per g particles. After increasing enzyme to particle ratio to 15 mg GOx per g particles, activity was enhanced 3-fold compared to 10 mg GOx per g particles. By further increase it remained constant. However, the binding yield (Fig. 5B) at a loading of 15 mg GOx per g particles was only about 70% of the added enzyme bound to the particles. If the loading was further increased, the binding yield decreased to approximately 50% for 20 mg GOx per g particles and 30% for 30 mg GOx per g particles. Thus, regarding the relationship between the overall activity and binding yield, an enzyme to particle ratio of 15 mg GOx per g particles was found to be optimal, as the yield in activity is maximal with more than 34% and relatively small amounts of enzymes are washed off with the supernatant. This might indicate that for GOx only a certain amount of enzyme could bind to the particle surface, leading to a constant maximum overall activity while the rest of the added enzyme was washed off during the immobilization process. That is why higher loadings were not considered to be economical because of the loss of free enzymes. For GOx that was physically adsorbed to the particles, high binding yields (at least 80% of the enzyme to particle ratios reached for covalently bound enzyme) could be observed for all initial enzymes to particle mass ratios (data not shown). However, the enzyme activity [U per mg enzyme] and specific activity [U per mg particles] were significantly lower, respectively, showing the importance of covalent binding for the stabilization of glucose oxidase during immobilization.| Initial enzyme to particle mass ratio [mg enzyme per g particles] | Specific activity [U per mg particles] | Activity yield [%] | Binding yield [%] |
|---|---|---|---|
| 5 | 0.08 | 14.0 | 100.0 |
| 10 | 0.16 | 17.2 | 100.0 |
| 15 | 0.49 | 34.1 | 67.0 |
| 20 | 0.50 | 33.1 | 51.8 |
| 30 | 0.53 | 25.3 | 32.1 |
For horseradish peroxidase (Table 3), a maximum in specific activity (2.6 U per mg particles) could be proven for 20 mg HRP per g particles. After further increasing the loading, the activity yield dropped to 1.87 U per mg particles for 30 mg HRP per g particles. This effect might be due to the enzyme density that may be too high at 30 mg HRP per g particles to ensure good substrate accessibility and flexibility of the enzyme to move during the conversion process. When comparing the yield in activity compared to free enzymes a maximum of 6.5% could be found for the lowest loading of 5 mg HRP per g particles. By increasing the initial enzyme to particle mass ratio, the binding yield decreases to 1.5%. In order to further investigate the yield of bound enzyme mass, supernatants were investigated. It could be seen that only for the lowest enzyme to particle ratio (5 mg HRP per g particles) all enzymes bound to the surface. By further increasing the loading, the binding yield decreased (approximately 79% for 10 mg HRP per g particles, 41% for 15 mg HRP per g particles, 32% for 20 mg HRP per g particles and 21% for 30 mg HRP per g particles). Binding yields for physically adsorbed HRP were significantly lower than covalently bound enzymes in all cases. In addition, an adsorption of enzymes resulted in much lower activity values. Although a loading of 5 mg HRP per g particles resulted in the highest activity and binding yields, the overall activity was relatively small. This is why for further experiments, a loading of 10–15 mg HRP g particles was considered to be optimal, as only smaller amounts of enzymes were washed off while specific activity was maximal.
| Initial enzyme to particle mass ratio [mg enzyme per g particles] | Specific activity [U per mg particles] | Activity yield [%] | Binding yield [%] |
|---|---|---|---|
| 5 | 1.26 | 6.5 | 100.0 |
| 10 | 1.91 | 4.3 | 79.4 |
| 15 | 2.31 | 4.0 | 41.0 |
| 20 | 2.60 | 3.4 | 32.2 |
| 30 | 1.89 | 1.5 | 21.9 |
Recycling experiments
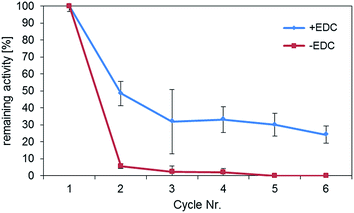
In order to determine the importance of the formation of a covalent bond during immobilization of HRP, recycling experiments were performed (Fig. 4).The samples which were chemically crosslinked showed much better reusability compared to the samples, in which HRP was only physically adsorbed. In the latter case, the activity was almost completely lost after the first reuse of the particles. In contrast, the activity of the chemically crosslinked samples could be preserved for at least 6 cycles at a value of about more than 30% of the original activity. The activity loss of about 70% during recycling might be due to a fraction of physically adsorbed enzymes that desorbed during the activity testing cycles. Although examples of stable adsorptive binding of enzymes to magnetic particles can be found in the literature,21 at least for HRP these findings point out the importance of a stable chemical bond in enzyme immobilization. The partially higher standard deviations of the recycling experiments are mainly caused by the small amount of enzyme immobilisates used in the experiments. As particles easily adsorb to the pipette tip during the recycling process, particle mass may decrease over the course of the cycles. However, as was shown by our group, working with larger volumes and higher particle concentrations reuse of magnetic enzyme immobilisates results in consistent values over twenty cycles.34
Reasons for enzyme inactivation upon immobilization
With respect to the total activity of free enzymes used in the immobilization experiments activity yields up to 6.5% for HRP (15 per mg g particles at pH 4.0) and up to 35% for GOx (15 per mg g particles at pH 4.0) were reached. In order to better understand the reason for the enzyme inactivation, the influence of the immobilization procedure itself was investigated by performing the process without particles and by adding carboxy-functionalized, but not EDC-activated particles. Activity values were determined before and after the process and the activity of free enzymes [U per mg enzyme] was calculated. In order to take into account the enzyme loss by adsorption to the magnetic particles, the protein concentration was measured before and after the process and the measured activity was related to the actual enzyme concentration. For GOx no inactivating effect by the particles or the pH-value during the process could be detected (data not shown). This indicates that the lower activity yields measured for physically immobilized GOx are not due to a short term contact with the particle surface or the different pH-values applied. Therefore the observed partial inactivation of adsorbed GOx seems to have its origin in a long term attachment to the surface accompanied by potential changes in the enzyme structure.For HRP it could be proven that, except for pH 4.0, there was an inactivation caused by the immobilization process conditions even without contact to magnetic particles, indicating a susceptibility of enzyme activity to higher pH-values during the coupling process (Fig. 5). The percentaged inactivation is further increased if particles are added to the solution. This might be due to a reversible adsorption and desorption or short term contact with the particle surface, which might affect enzyme stability. Also in the experiments with varying pH-values and enzyme to particle ratios it could be proven that adsorptive bound HRP does not show a comparably high specific activity as the covalent bound one.
Conclusion
Although it is known that enzymes enormously differ in their properties and characteristics, standard protocols are commercially available for immobilization recommending certain parameters. However, in this work it could be shown that this is only true for enzyme independent reaction steps, as for example the activation step of the carboxy-functionalized carrier using EDC. However, as proteins differ in their characteristics like size, form and surface charge properties, the coupling step needs to be adjusted for each enzyme–carrier-pair. In this work, the immobilization of horseradish peroxidase (HRP) and glucose oxidase (GOx) to carboxy-functionalized polyvinylalcohol-coated magnetic microparticles was investigated. An optimization of the pH-value and the ratio between enzyme and carrier mass resulted in a significant improvement of the resulting activity and binding yields. For HRP using a recommended protocol34 led to an activity yield of less than 0.1%. After optimizing the protocol it could be enhanced to 6.5%. For GOx, an initial activity yield of approximately 1% was reached that could be improved to approximately 35% after tuning the parameters pH and added enzyme mass in the coupling process.In the case of HRP it could be shown that already the coupling process itself leads to a partial inactivation. This is most probably due to the pH-enzyme to particle ratio values higher than 4.0 and the addition of magnetic particles that leads to a consistent adsorption and desorption of proteins what might affect enzyme activity.
Acknowledgements
This research work is part of the project “Molecular Interaction Engineering: From Nature's Toolbox to Hybrid Technical Systems”, which is funded by BMBF, funding code 031A095.Notes and references
- K. R. Jegannathan and P. H. Nielsen, J. Cleaner Prod., 2013, 42, 228–240 CrossRef CAS.
- A. Liese, K. Seelbach and C. Wandrey, Industrial biotransformations, Wiley-VCH, Weinheim, 2nd completely rev. and extended edn, 2006 Search PubMed.
- E. H. Yoo and S. Y. Lee, Sensors, 2010, 10, 4558–4576 CrossRef PubMed.
- R. DiCosimo, J. McAuliffe, A. J. Poulose and G. Bohlmann, Chem. Soc. Rev., 2013, 42, 6437–6474 RSC.
- A. M. Klibanov, Anal. Biochem., 1979, 93, 1–25 CrossRef CAS PubMed.
- C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2007, 40, 1451–1463 CrossRef CAS.
- S. H. Huang, M. H. Liao and D. H. Chen, Biotechnol. Prog., 2003, 19, 1095–1100 CrossRef CAS PubMed.
- E. Vanleemp and M. Horisber, Biotechnol. Bioeng., 1974, 16, 385–396 CrossRef.
- L. S. Wong, F. Khan and J. Micklefield, Chem. Rev., 2009, 109, 4025–4053 CrossRef CAS PubMed.
- M. Miyazaki, J. Kaneno, S. Yamaori, T. Honda, M. P. P. Briones, M. Uehara, K. Arima, K. Kanno, K. Yamashita, Y. Yamaguchi, H. Nakamura, H. Yonezawa, M. Fujii and H. Maeda, Protein Pept. Lett., 2005, 12, 207–210 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- R. A. Sheldon, Biochem. Soc. Trans., 2007, 35, 1583–1587 CrossRef CAS PubMed.
- S. S. Wong and L. J. C. Wong, Enzyme Microb. Technol., 1992, 14, 866–874 CrossRef CAS PubMed.
- X. Tang and J. E. Bruce, Methods Mol. Biol., 2009, 492, 283–293 CAS.
- Z. Grabarek and J. Gergely, Anal. Biochem., 1990, 185, 131–135 CrossRef CAS PubMed.
- Bioconjugate Techniques, ed. G. T. Hermanson, Elsevier, 2nd edn, 2008 Search PubMed.
- I. Migneault, C. Dartiguenave, M. J. Bertrand and K. C. Waldron, BioTechniques, 2004, 37, 790–802 CAS.
- H. J. Schramm and T. Dulffer, Adv. Exp. Med. Biol. A, 1977, 86, 197–206 Search PubMed.
- R. Kluger and A. Alagic, Bioorg. Chem., 2004, 32, 451–472 CrossRef CAS PubMed.
- M. Franzreb, M. Siemann-Herzberg, T. J. Hobley and O. R. T. Thomas, Appl. Microbiol. Biotechnol., 2006, 70, 505–516 CrossRef CAS PubMed.
- G. Zheng, B. Shu and S. Yan, Enzyme Microb. Technol., 2003, 32, 776–782 CrossRef.
- R. Wilson and A. P. F. Turner, Biosens. Bioelectron., 1992, 7, 165–185 CrossRef CAS.
- N. C. Veitch, Phytochemistry, 2004, 65, 249–259 CrossRef CAS PubMed.
- T. Bahar and S. S. Celebi, J. Appl. Polym. Sci., 1999, 72, 69–73 CrossRef CAS.
- M. H. Liao and D. H. Chen, Biotechnol. Lett., 2001, 23, 1723–1727 CrossRef CAS.
- S. Akgol, Y. Kacar, A. Denizli and M. Y. Arica, Food Chem., 2001, 74, 281–288 CrossRef CAS.
- T. H. Wang and W. C. Lee, Biotechnol. Bioprocess Eng., 2003, 8, 263–267 CrossRef CAS.
- D. Bozhinova, B. Galunsky, G. Yueping, M. Franzreb, R. Koster and V. Kasche, Biotechnol. Lett., 2004, 26, 343–350 CrossRef CAS PubMed.
- G. K. Kouassi, J. Irudayaraj and G. McCarty, J. Nanobiotechnol., 2005, 3, 1 CrossRef PubMed.
- L. M. Bruno, J. S. Coelho, E. H. M. Melo and J. L. Lima, World J. Microbiol. Biotechnol., 2005, 21, 189–192 CrossRef CAS.
- N. Schultz, Application of Magnetic Separation Technology for the Recovery and Reuse of Immobilized Lipase of Candida Antarctica A-Type (CALA), Dissertation, 2007 Search PubMed.
- I. Magario, A. Neumann, O. Vielhauer, C. Syldatk and R. Hausmann, Biocatal. Biotransform., 2009, 27, 237–245 CrossRef CAS.
- R. Ricco, C. M. Doherty and P. Falcaro, J. Nanosci. Nanotechnol., 2014, 14, 6565–6573 CrossRef CAS PubMed.
- C. Morhardt, B. Ketterer, S. Heissler and M. Franzreb, J. Mol. Catal. B: Enzym., 2014, 107, 55–63 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2015 |