Open Access Article
Open Access ArticleReliable microspotting methodology for peptide-nucleic acid layers with high hybridization efficiency on gold SPR imaging chips†
L.
Simon
a,
G.
Lautner‡
b and
R. E.
Gyurcsányi
*ab
aMTA-BME “Lendület” Chemical Nanosensors Research Group, Department of Inorganic and Analytical Chemistry, Budapest University of Technology and Economics, Szent Gellért tér 4, H-1111 Budapest, Hungary. E-mail: robertgy@mail.bme.hu
bDepartment of Inorganic and Analytical Chemistry, Budapest University of Technology and Economics, Szent Gellért tér 4, H-1111 Budapest, Hungary
First published on 10th June 2015
Abstract
One-step direct immobilization of peptide-nucleic acid (PNA) probes onto gold surfaces through Au–S chemistry is critical in terms of generating self-assembled monolayers with high hybridization efficiency. We found that this problem is more severe if the immobilization is performed by contact microspotting to generate PNA arrays. Therefore, here we propose a novel microspotting-based immobilization method to generate PNA arrays with high hybridization efficiency on bare gold surface plasmon resonance imaging (SPRi) chips. The essence of the approach is to spot thiol labelled PNA strands prehybridized with a short complementary DNA strand instead of conventionally used single stranded PNA (ssPNA) probes. After immobilization the complementary DNA strands could be easily removed to activate the surface confined PNA probes. The incubation time and the type of spotting needle also have a marked influence on the hybridization efficiency of the PNA layers. However, we show that if all other conditions remain the same, PNA layers from prehybridized PNA probes exhibit superior hybridization efficiency than those from ssPNA probes.
Introduction
Peptide nucleic acids (PNAs)1 are artificial nucleic acid analogs in which the nucleotide bases are attached to a peptide backbone typically formed from aminoethylglycine units. They can form Watson–Crick base pairs with complementary nucleic acid strands (DNA or RNA).2 The immediate consequence of replacing the deoxyribose phosphodiester backbone is that PNAs lack the negative charge of natural nucleic acids. This is a major advantage in hybridization assays because there is no charge repulsion between the hybridized strands. Accordingly, the hybridization of PNA strands in solution is not affected by the ionic strength and PNAs form stronger complexes with complementary nucleic acid strands than their natural counterparts. As the chemical and biochemical stabilities of PNAs are also superior to those of DNA strands,3 their drawbacks seem to be limited to their higher cost and the need for a more careful probe design to avoid self-complementarity. PNA arrays and chips have been made by using various substrates and immobilization methodologies.4–7 However, the self-assembly of PNA strands directly attached through terminal thiol groups onto gold surfaces remains one of the preferred choices for electrochemical,8–11 surface plasmon resonance12–14 and quartz crystal microbalance15 transducers. In a series of studies it was found that the direct attachment of PNAs to gold via Au–S chemistry is rather critical in terms of the efficiency of the subsequent hybridization step.16–18 In fact an early study formulated fully discouraging conclusions regarding the use of Au–S chemistry for direct attachment of PNA strands to gold as opposed to biotin–streptavidin-base coupling of biotinylated PNA strands. Using a quartz crystal microbalance with energy dissipation, a very low energy dissipation was observed during thiol-PNA immobilization suggesting that PNA is rigidly attached through several unspecific contact points on gold. Thus the strands most likely “lie down” adsorbed on the gold surface, which hampers subsequent hybridization.16 Similar observations were also made for DNA strands by neutron reflectivity at high salt concentrations showing that the DNA strands are non-specifically adsorbed onto the gold surface.19 In fact terminal attachment of the DNA strands through thiol groups resulting in high hybridization efficiency was only obtained when a post-treatment with mercaptohexanol (MH) was performed to reduce the direct contact of the DNA strands with the gold surface. Therefore, in many studies the biotin–avidin coupling13,20,21 is still preferred over direct attachment of the thiol labeled nucleic acid probes to the gold surface.Extensive studies by Martin-Gago and co-workers22 on self-assembled single stranded PNA (ssPNA) layers on gold have revealed that the formation of PNA monolayers is a concentration dependent two-step process. It starts with the adsorption of ssPNA molecules on the gold surface in a “lying down” orientation while above a certain surface coverage a phase transition occurs and the strands realign in a “standing-up” position.22,23 The concentration threshold was suggested to be at ca. 1 μM ssPNA in the aqueous solutions used for surface modification, resulting in ordered arrangements.24 However, at concentrations higher than this value the surface rapidly saturates and at 5–10 μM ssPNA becomes so compact that no DNA binding was detected by X-ray photoemission spectroscopy (XPS). The repulsive interactions generated within immobilized PNA layers, upon hybridization with ssDNA probes, are in fact at the core of cantilever bending in cantilever-based sensors.25 In contrast to other studies that demonstrated low hybridization efficiency of the PNA strands lying down on the surface, Briones et al.22 observed by XPS a close to 100% yield for hybridization. This discrepancy may be due to the different experimental conditions as they used a very high, 100 μM, concentration of complementary DNA, for their hybridization study, which is many orders of magnitude higher than those used in analytical studies. The model of “lying-down” and “standing-up” PNA molecules was confirmed later also by electrochemical means using ssPNA strands labeled at the C and N terminus with cysteine and ferrocene, respectively.26,27 Besides orientation and steric effects, the ssPNA probe density on the surface can influence the hybridization efficiency in other ways as well. While the PNA–DNA hybridization per se is not affected by the ionic strength, it was reported that in the case of compact surface confined PNA layers, electrostatic repulsion can occur between the closely bound DNA strands. This effect is independent of the type of coupling chemistry and can be eliminated either by increasing the ionic strength of the hybridization buffer or by decreasing the surface concentration of the PNA probe until the hybridization of complementary strands follows the Langmuir adsorption model.21
While apparently the optimization of the surface concentration and orientation of thiol labeled PNAs on gold for high hybridization efficiency is difficult, if successful, it offers major advantages in terms of versatility and single-step coupling. It eliminates the need for additional cross-linking reagents,10,28 and consequently reduces the cost of fabrication and the structural complexity of the attached layers. Owing to the large variability in terms of length and sequence of the immobilized strands as well as co- or post-immobilized spacers, it is unlikely that universally applicable optimum conditions for immobilization can be found, although this would be preferable for the preparation of PNA microarrays.
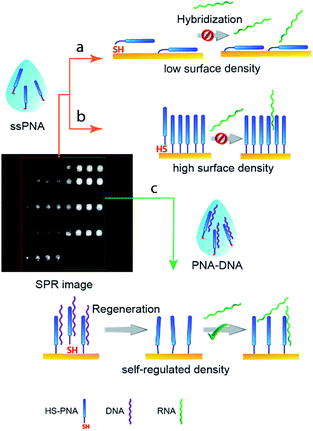
In this study we aimed at developing a reliable preparation method for PNA receptor layers by microspotting thiol-labeled PNA strands on bare gold SPR imaging chips and taking advantage of the multiplex readout for high throughput optimization. Since PNAs due to their very high affinity are ideal candidates for the determination of micro-RNAs (miRNAs) the method is demonstrated through the hybridization assay of a 22-mer miRNA (hsa-miR-208a) that was identified as a biomarker of myocardial injury.29,30 The approach we used is based on implementing a “self-regulating”21 mechanism for improved hybridization efficiency of the immobilized PNA strands (Scheme 1). Our hypothesis was that using thiol labeled PNA strands prehybridized with complementary DNA, instead of ssPNA strands, will automatically adjust the optimal surface conditions for subsequent hybridization. Moreover, we assumed that the non-specific surface adsorption of PNA strands on gold will be less of a problem if their duplex structures with DNAs are used for surface modification. The latter assumption is indirectly supported by the observation of Li and Rothberg on the differential adsorption of ss and dsDNAs on gold colloids,31i.e., single-stranded DNAs adsorb strongly while double-stranded oligonucleotides do not. The intuitive explanation for this behavior was that in the case of dsDNAs the nucleotide bases are involved in the formation of hydrogen bonds between the complementary strands and as such are not available for interaction with gold, which is, however, not the case for the flexible ssDNAs.
Experimental
Chemicals and materials
Twelve (N′–TGCTCGTCTTAT–C′) and 18-mer (N′–GCTTTTTGCTCGTCTTAT–C′) PNA strands complementary to the microRNA hsa-miR-208a as well as a random non-complementary PNA strand (NC-PNA (18-mer): N′–GCCGCTTCTTTATCTTTT–C′) with a thiol group attached to the C terminus of the PNA backbones through a spacer consisting of two ethylene glycol units (C6-AEEA, ca. 2.4 nm, see ESI, Scheme S1†) were purchased from Eurogentec (Seraing, Belgium). The 22-mer hsa-miR-208a microRNA sequence (5′–AUAAGACGAGCAAAAAGCUUGU–3′), its DNA analog (C-DNA, 5′–ATAAGACGAGCAAAAAGCTTGT–3′), and a non-complementary random 22-mer RNA (NC-RNA; 5′–AGUACUAAUUCGUCUCUGUUCU–3′) were purchased from Sigma. RNase and DNase-free water for molecular biology (DEPC-treated and sterile filtered; Sigma) and DNA LoBind centrifuge tubes (Eppendorf) were used for preparing the RNA and DNA stock solutions in a UV-cabinet for PCR operations. All other reagents such as inorganic salts and buffer components were of highest bioanalytical grade (Sigma-Aldrich). Phosphate buffer saline (PBS) solution was prepared from PBS tablets. The other buffers used for spotting, i.e., saline-sodium citrate (SSC) 3 × concentrate contained 45 mM trisodium citrate, and 450 mM NaCl at pH = 7.0 (adjusted with 1 M HCl), while borate buffered saline (BBS) contained 10 mM sodium borate, and 150 mM NaCl at pH = 10.0 (adjusted with 1 M NaOH). All aqueous solutions were prepared with ultrapure deionized water (18.2 MΩ cm resistivity, Millipore).Methods
Bare gold SPR sensor slides (HORIBA Jobin Yvon S.A.S. Palaiseau, France) were cleaned immediately before microspotting in a UV generated ozone atmosphere (Novascan Technologies, Ames, IA, USA) for 15 minutes. The immobilization of PNA strands was performed by microspotting using a BioOdyssey™ Calligrapher™ miniarrayer (Bio-Rad, Hercules, CA, USA) by means of either a solid pin (Stealth Solid Pin, 375 μm, Arrayit, Sunnyvale, CA, USA) or pins with an uptake channel, i.e., comprising a microcavity acting as a sample reservoir (SMP15 Stealth Micro Spotting Pin, with 500 μm spot diameter and 0.25 μL uptake volume). The thiolated PNA probes were spotted onto the gold surfaces from a 384 well LD-PE plate previously blocked with protein-free TBS blocking buffer (Pierce, Thermo-Fisher, Rockford, IL USA) for 1 hour, washed with DI water, and dried. In all cases the wells were filled with 20 μL of 5 μM PNA strands. At least three parallel spots were made for each probe formulation at 65% rh and with the spotting stage thermostated at 15 °C. The spotted gold SPR sensors were incubated at 15 ± 1 °C and 65% rh in the humidity chamber of the microspotter for periods between 4 and 19 h. Under these conditions the drying of the spotted droplets was avoided. The droplets were still visible before the unspotted gold surface of the chips was blocked with 1 mM mercaptohexanol (MH) in phosphate buffer saline (PBS) for 15 min. Finally, the chips were washed with 300 mL DI water and gently dried under a N2 stream.Surface plasmon resonance imaging (SPRi) measurements were carried out by using a SPRi-Plex II system (HORIBA Jobin Yvon S.A.S. Palaiseau, France) at a fixed angle. First the working angle was selected based on the recorded SPR curves and then the refractive index calibration was performed to normalize the SPR response of each spot with the signal change measured at the respective location for a given refractive index change of the solution. The binding of nucleic acid strands (DNA or RNA) to the PNA microarrays (Fig. S1†) was monitored at 25.00 °C, at a flow rate of 50 μL min−1. The activation of the immobilized phPNA strands, i.e. removal of prehybridized complementary nucleic acid strands, was performed with 100 mM NaOH solution (50 μL min−1, for 4 min). The same conditions were also used to regenerate PNA surfaces between miRNA injections. The injected volume for each nucleic acid sample and regeneration solution was 180 μL. The typical durations for baseline, association and dissociation were 12, 3.6, and 8 min, respectively. The evaluation of the interaction curves was performed using Scrubber 2 (Genoptics version).
Results and discussion
To determine the validity of our hypothesis that PNA strands in a prehybridized form (phPNA) will provide receptor layers with higher hybridization efficiency than those formed by ssPNA, the effect of various experimental parameters including the composition of the spotting buffer, the type of spotting pin (solid and microcavity-based) as well as the length of PNA probes (12-mer and 18-mer) were systematically investigated. A preliminary screening was carried out to identify the concentration of PNA probes and MH used for co- or post-immobilization (Fig. S2†). The experimental protocol featured in the Experimental section is the result of this first optimization step. Taking advantage of the multiplexing capabilities of SPR imaging, the effect of the experimental parameters was determined side-by-side for ssPNA and phPNA under rigorously identical conditions. The prehybridized probe solutions were prepared by mixing PNA with a 20 mol% excess of complementary DNA (the final concentration of phPNA was 5 μM assuming quantitative association with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry). We used DNA instead of the target miRNA because it is less susceptible to biodegradation. The microspotting of thiol labelled PNA probes onto the gold surface beside the self-assembly process is expected to be influenced also by the specific conditions of spotting. Therefore, we prepared PNA chips by using microspotting thiol labelled PNA probes in different spotting buffers using both solid and microcavity-based stealth microspotting pins. Rather surprisingly, we found that the subsequent hybridization of complementary miRNA (100 nM) as determined by SPR imaging was most severely affected by the type of microspotting pin used (see Fig. 1A and B). The reflectance change, indicative of the amount of complementary miRNA bound to the immobilized PNA spots depending also on other experimental conditions, was up to 7 times higher when using the microcavity-based stealth pin as compared to the solid pin. Since the essential difference between these two types of pins is the volume of the deposited droplet, apparently larger volumes are beneficial in terms of increasing the binding capacity of the surface confined PNA probes.
1 stoichiometry). We used DNA instead of the target miRNA because it is less susceptible to biodegradation. The microspotting of thiol labelled PNA probes onto the gold surface beside the self-assembly process is expected to be influenced also by the specific conditions of spotting. Therefore, we prepared PNA chips by using microspotting thiol labelled PNA probes in different spotting buffers using both solid and microcavity-based stealth microspotting pins. Rather surprisingly, we found that the subsequent hybridization of complementary miRNA (100 nM) as determined by SPR imaging was most severely affected by the type of microspotting pin used (see Fig. 1A and B). The reflectance change, indicative of the amount of complementary miRNA bound to the immobilized PNA spots depending also on other experimental conditions, was up to 7 times higher when using the microcavity-based stealth pin as compared to the solid pin. Since the essential difference between these two types of pins is the volume of the deposited droplet, apparently larger volumes are beneficial in terms of increasing the binding capacity of the surface confined PNA probes.
The validity of our assumption that phPNA probes will provide better hybridization efficiency than ssPNAs is clearly confirmed by the results obtained for solid pin-based immobilization (Fig. 1A). For both 12- and 18-mer PNA strands as well as for all the different spotting buffers the binding to PNA spots formed from phPNA strands was in average ca. 2 times larger than to those obtained by ssPNA immobilization.
In the case of microcavity-based stealth pins the same trend is obvious for the 18-mer PNA strands, however, in the case of the 12-mer PNAs the effect is only noticeable when using PBS as spotting buffer (Fig. 1B). Such a length dependent behaviour could be explained if shorter PNA strands would assemble more easily on the gold surface to enable subsequent hybridization, and/or if their non-specific interactions with gold are weaker than that of longer PNA strands. If so it is reasonable to expect that the hybridization efficiency will depend on the time allowed for self-assembly. Therefore, to test the feasibility of these assumptions we have reduced significantly the incubation time of the spots, i.e., the time allowed after spotting for the formation of PNA SAMs in a controlled relative humidity atmosphere. Using an incubation time of only 4 hours the difference between phPNA and ssPNA spots was also visible for the shorter 12-mer PNA strand. This suggests that the immobilization of shorter PNA strands is less critical especially if enough time is allowed for the SAM to arrange. Since the length of the PNAs and the immobilization time can vary in a wide range this may explain, at least in part, the controversy in the literature regarding the performance of PNAs immobilised through terminal HS groups to gold.
In all cases the difference between the hybridization efficiency of the two types of immobilized PNAs is the largest for strands spotted from PBS buffer. Otherwise, it is difficult to choose a single optimal spotting buffer between SSC and PBS for the different length PNAs, but overall the SSC buffer seems to offer the most consistent results. Thus the hybridization efficiency of the PNA spots depends on the volume of the spotted solution, the time allowed for immobilization and the type of spotting buffer used, but very importantly in all instances using the prehybridized form for spotting provides the best results under the given conditions. The superiority of PNA spots immobilized from phPNA using microcavity-based stealth pins was confirmed for a wide concentration range of miRNA as shown in Fig. 2.
The ratio of the sensitivities (slope of the linear range of ΔR vs. log[miRNA]) of the spots showing the highest and lowest hybridization efficiency exceeds an order of magnitude, i.e. spots of phPNA made with a microcavity-based stealth spotting pin and ssPNA with a solid pin, respectively. However, even when using microcavity-based stealth pins for spotting there is a factor of two between the sensitivity of PNA spots immobilized from phPNA and ssPNA, which is remarkable. Using the optimized spotting procedure for PNA immobilization, a miRNA amount as low as 140 fmol could be detected label-free, without amplification (Fig. S3†). There is another less obvious advantage of using phPNA for immobilization. Namely, the signal change (ΔR) during the removal of the DNA strand to activate the immobilized PNA probes is a good estimate of the ΔRmax value (Fig. 3A inset) corresponding to the saturation of the PNA receptor layer with complementary RNA strands. This value is difficult (or at least unpractical and costly) in many cases to be determined by hybridization assays due to the very high concentrations of complementary RNA required. Knowing the signal at saturation enables a more exact fitting of the binding curves (Fig. 3). The validity of the approach is based on the assumptions that (i) all PNA probes bound to the surface are in hybridized form, i.e., the equilibrium dissociation constant (KD) is sufficiently small, and (ii) the first regeneration step removes all the DNA hybridized to the PNA strands. Kinetic analysis of the PNA-DNA interactions revealed a KD value of 0.9 nM for the PNA-DNA complex that makes the first assumption reasonable. The kinetic curves for miRNA interactions (Fig. 3B) confirm the efficiency of the regeneration step using 0.1 M NaOH in terms of excellent baseline recovery. This suggests that the first regeneration step that activates the PNA probes after spotting effectively removes the hybridized DNA strands. The real time monitoring of the binding cycles further support the strong interaction between the PNA and complementary miRNA as there is no detectable loss of miRNA whatsoever in the timeframe allowed for dissociation (Fig. 3B).
The selectivity of the optimized PNA layers for hsa-miR-208a was assessed by using a 22-mer random sequence RNA at 100 nM concentration. In case of PNA arrays it must be also ensured that there is no cross-talk between the different surface confined probes, therefore, a preliminary test was performed by spotting also a random 18-mer PNA under the optimized conditions. In all cases when the spotting was made according to the optimized protocol the non-specific interactions were low (Fig. 4).
Interestingly, and most importantly, we found that the selectivity towards complementary miRNA strands is somewhat enhanced by immobilizing the probes in prehybridized form as compared to the single stranded ones.
Conclusions
While this aspect has received little awareness our study showed that thiol labelled PNA probes immobilized in one step via the Au–S bond to gold are extremely sensitive in terms of subsequent hybridization with complementary nucleic acids to the immobilization conditions. The sensitivity was clearly influenced by the length of the PNA probe. The optimization of the immobilization was more critical for longer probes. The main finding of the paper is that under all practically relevant conditions the microspotting of PNA strands in prehybridized form with complementary DNA strands results in PNA layers that are superior (or equal) in terms of binding capacity to those obtained by ssPNA microspotting. The use of prehybridized PNA strands is also beneficial in terms of determining the signal corresponding to the maximum binding capacity of the respective layers upon their activation, i.e., by measuring the signal change due to the removal of the hybridized complementary strands. The immobilization strategy is potentially applicable to a wide range of gold made transducers as those used in electrochemical, SPR, and quartz crystal microbalance sensors. The results indicate that if all other conditions are the same, PNA layers from phPNA yield better hybridization efficiency than those from ssPNA, alleviating the optimization of PNA immobilization to gold surfaces.Acknowledgements
The financial support from the Lendület program of the Hungarian Academy of Sciences (LP2013-63/2013) is gratefully acknowledged.Notes and references
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497–1500 CAS.
- M. Egholm, O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden and P. E. Nielsen, Nature, 1993, 365, 566–568 CrossRef CAS PubMed.
- V. V. Demidov, V. N. Potaman, M. D. Frank-Kamenetskil, M. Egholm, O. Buchard, S. H. Sönnichsen and P. E. Nielsen, Biochem. Pharmacol., 1994, 48, 1310–1313 CrossRef CAS.
- M. Beier and J. D. Hoheisel, Nucleic Acids Res., 1999, 27, 1970–1977 CrossRef CAS PubMed.
- J. Weiler, H. Gausepohl, N. Hauser, O. N. Jensen and J. D. Hoheisel, Nucleic Acids Res., 1997, 25, 2792–2799 CrossRef CAS PubMed.
- N. Winssinger and J. L. Harris, Expert Rev. Proteomics, 2005, 2, 937–947 CrossRef CAS PubMed.
- S. Y. Lim, W. Y. Chung, H. K. Lee, M. S. Park and H. G. Park, Biochem. Biophys. Res. Commun., 2008, 376, 633–636 CrossRef CAS PubMed.
- T. H. Degefa and J. Kwak, J. Electroanal. Chem., 2008, 612, 37–41 CrossRef CAS PubMed.
- H. Aoki and Y. Umezawa, Nucleic Acids Symp. Ser., 2002, 2, 131–132 CrossRef CAS.
- D. Ozkan, A. Erdem, P. Kara, K. Kerman, J. Justin Gooding, P. E. Nielsen and M. Ozsoz, Electrochem. Commun., 2002, 4, 796–802 CrossRef CAS.
- N. Husken, M. Gebala, W. Schuhmann and N. Metzler-Nolte, ChemBioChem, 2010, 11, 1754–1761 CrossRef PubMed.
- J. Liu, L. Tiefenauer, S. Tian, P. E. Nielsen and W. Knoll, Anal. Chem., 2006, 78, 470–476 CrossRef CAS PubMed.
- K. K. Jensen, H. Ørum, P. E. Nielsen and B. Nordén, Biochemistry, 1997, 36, 5072–5077 CrossRef CAS PubMed.
- N. Prabhakar, K. Arora, S. K. Arya, P. R. Solanki, M. Iwamoto, H. Singh and B. D. Malhotra, Analyst, 2008, 133, 1587–1592 RSC.
- J. Wang, Anal. Chem., 1997, 69, 5200–5202 CrossRef CAS.
- F. Hook, A. Ray, B. Norden and B. Kasemo, Langmuir, 2001, 17, 8305–8312 CrossRef.
- G. Jágerszki, R. E. Gyurcsányi, L. Höfler and E. Pretsch, Nano Lett., 2007, 7, 1609–1612 CrossRef PubMed.
- C. Ananthanawat, T. Vilaivan and V. P. Hoven, Sens. Actuators, B, 2009, 137, 215–221 CrossRef PubMed.
- R. Levicky, T. M. Herne, M. J. Tarlov and S. K. Satija, J. Am. Chem. Soc., 1998, 120, 9787–9792 CrossRef CAS.
- Y. Sato, K. Fujimoto and H. Kawaguchi, Colloids Surf., B, 2003, 27, 23–31 CrossRef CAS.
- D. F. Yao, J. Kim, F. Yu, P. E. Nielsen, E. K. Sinner and W. Knoll, Biophys. J., 2005, 88, 2745–2751 CrossRef CAS PubMed.
- C. Briones, E. Mateo-Marti, C. Gomez-Navarro, V. Parro, E. Roman and J. A. Martin-Gago, Phys. Rev. Lett., 2004, 93, 208103 CrossRef CAS.
- E. Mateo-Marti, C. Briones, E. Roman, E. Briand, C. M. Pradier and J. A. Martin-Gago, Langmuir, 2005, 21, 9510–9517 CrossRef CAS PubMed.
- S. Ghosh and R. Mukhopadhyay, J. Colloid Interface Sci., 2011, 360, 52–60 CrossRef CAS PubMed.
- S. Ghosh, S. Mishra and R. Mukhopadhyay, J. Mater. Chem. B, 2014, 2, 960–970 RSC.
- A. Paul, R. M. Watson, P. Lund, Y. J. Xing, K. Burke, Y. F. He, E. Borguet, C. Achim and D. H. Waldeck, J. Phys. Chem. C, 2008, 112, 7233–7240 CAS.
- N. Husken, M. Gebala, F. La Mantia, W. Schuhmann and N. Metzler-Nolte, Chem.–Eur. J., 2011, 17, 9678–9690 CrossRef PubMed.
- D. Kambhampati, P. E. Nielsen and W. Knoll, Biosens. Bioelectron., 2001, 16, 1109–1118 CrossRef CAS.
- E. Wang, Y. Nie, Q. Zhao, W. Wang, J. Huang, Z. Liao, H. Zhang, S. Hu and Z. Zheng, Eur. J. Cardio. Thorac. Surg., 2013, 8, 165 Search PubMed.
- X. Ji, R. Takahashi, Y. Hiura, G. Hirokawa, Y. Fukushima and N. Iwai, Clin. Chem., 2009, 55, 1944–1949 CAS.
- H. Li and L. Rothberg, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14036–14039 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Typical SPR images and layout of the PNA arrays, results of prescreening of the spotting conditions and unprocessed SPR curves. See DOI: 10.1039/c5ay01239b |
| ‡ Present address: University of Michigan at Ann Arbor. |
| This journal is © The Royal Society of Chemistry 2015 |