Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Electrochemically monitoring the antibiotic susceptibility of Pseudomonas aeruginosa biofilms†
Thaddaeus A.
Webster
,
Hunter J.
Sismaet
,
I-ping J.
Chan
and
Edgar D.
Goluch
*
Department of Chemical Engineering, Northeastern University, 360 Huntington Ave, 313 Snell Engineering, Boston, MA 02115, USA. E-mail: e.goluch@neu.edu; Tel: +1-617-373-3500
First published on 23rd September 2015
Abstract
The condition of cells in Pseudomonas aeruginosa biofilms was monitored via the electrochemical detection of the electro-active virulence factor pyocyanin in a fabricated microfluidic growth chamber coupled with a disposable three electrode cell. Cells were exposed to 4, 16, and 100 mg L−1 colistin sulfate after overnight growth. At the end of testing, the measured maximum peak current (and therefore pyocyanin concentration) was reduced by approximately 68% and 82% in P. aeruginosa exposed to 16 and 100 mg L−1 colistin sulfate, respectively. Samples were removed from the microfluidic chamber, analyzed for viability using staining, and streaked onto culture plates to confirm that the P. aeruginosa cells were affected by the antibiotics. The correlation between electrical signal drop and the viability of P. aeruginosa cells after antibiotic exposure highlights the usefulness of this approach for future low cost antibiotic screening applications.
1. Introduction
The ability to monitor the effect novel antibiotics have on bacteria is important for infection control. The conventional approach to determining antibiotic efficacy requires the creation of culture plates with the antibiotic cocktail of choice at a series of concentrations.1 After culturing (for 24 h or longer, depending on the strain), the plates are visually inspected for growth. At a certain concentration, known as the minimum inhibitory concentration (MIC), no bacterial growth is observed. This concentration is then used to design an antibiotic schedule for the patient. This effective approach suffers from the use of large amounts of reagents required to produce the culture plates. Furthermore, these screens only measure the effectiveness of the antibiotic against planktonic cell growth; not removal of biofilms, which are commonly associated with infections and significantly more difficult to treat.2,3One intriguing alternative is coupling microfluidics, to grow the bacteria, with antibiotic screens. Kim et al. (2012) utilized a microfluidic system to simultaneously expose biofilms of Escherichia coli to eight different concentrations of antibiotics on a single chip.4 The smaller volumes, inherent in microfluidic devices, along with the ability to produce multiple concentration gradients provided a faster, cheaper alternative to current antibiotic susceptibility tests. By flowing antibiotics over the grown biofilms, researchers more closely simulated in vivo conditions. Many current microfluidic studies determine biofilm viability based on the presence of fluorescent proteins during exposure to antibiotics.5,6 While these methods are certainly robust and promising, the fluorescent signal requires expensive optical equipment and genetically modified bacteria or selective labels.7–10 A cheaper and easier method of determining the relative amount of live cells in a biofilm under exposure to antibiotics can be achieved by monitoring the electrochemical response of the system. Robust bacterial biofilms produce a plethora of molecules that promote communication, defend the colony, and cause infection.11–13 Of interest are molecules that provide information about the condition of the biofilm, which can be detected by electrochemical methods.
Pseudomonas aeruginosa is an opportunistic pathogen that infects patients with compromised immune systems.14–18 This bacterial species progresses to infection rapidly, often establishing a biofilm within 24 h.19P. aeruginosa produces the blue electro-active molecule pyocyanin (PYO). PYO has been linked to several adverse human health effects including neutrophil death and reduced lung cilia beating.20–22 The detrimental results of PYO exposure enable P. aeruginosa to thrive in the infected host. PYO is able to undergo reversible redox (exchange of electrons) reactions, and its presence can be measured with standard electrochemical techniques.23–26 Additionally, the prevalence of multi-drug resistant P. aeruginosa has increased, leading to the need for new antibiotics in the doctor's tool kit.27 Colistin, a cyclic polypeptide mixture of colistin A and B, is a polymyxin antibiotic that previously fell out of use due to its toxicity.28–30 Recently, however, this antibiotic has begun to see renewed use for patients suffering from multi-drug resistant P. aeruginosa infections.31
Electrochemically monitoring the viability of P. aeruginosa cells in a microfluidic system has previously been demonstrated.32 Pires et al. (2013) combined impedance and amperometric measurements to simultaneously monitor the growth and respiration of P. aeruginosa cells.32 This approach emphasizes the potential to non-destructively observe P. aeruginosa, but it lacks the ability to measure the produced PYO itself, a potential marker of cell viability and virulence.33 A simple means of measuring excreted PYO electrochemically is accomplished by square wave voltammetry (SWV) over the range of voltages where PYO is reduced (half wave potential is approximately −250 mV vs. Ag/AgCl reference) via the following reaction: [PYO]ox + 2H+ + 2e− ↔ [PYO]red.34 The ability to measure a virulence factor as it relates to the amount of live cells in a biofilm during exposure to antibiotics, could help in determining effective treatment procedures. To this end, the present study looks at the killing of cells inside of a P. aeruginosa biofilm (grown in a microfluidic environment) via detected PYO, using SWV at a disposable three electrode cell, when exposed to different concentrations of the antibiotic colistin sulfate.
2. Experimental
2.1 Materials
P. aeruginosa strain PA14 and m-cherry Escherichia coli strain K12 were used for all antibiotic tests performed. Trypticase soy broth (BD 211768) was used as the nutrient source for all bacteria grown in these tests. Colistin sulfate (Adipogen AG-CN2-0065-G001) was dissolved in trypticase soy broth (TSB) at 1 g L−1 and used as a stock solution. When not in use, the stock solution was stored at 4 °C. Polydimethylsiloxane (Ellsworth Adhesives 184 Sil. Elast. Kit 0.5 kg) was used to prepare all microfluidic devices. Disposable three electrode cells (Zensor TE100) were used for all measurements in this study. The electrochemical cells consist of carbon working and counter electrodes with a Ag/AgCl paste reference electrode. Tubing and luer lock fittings for microfluidic connections were purchased from Amazon Supply (B001GMWZM) and Value Plastic (MTLL230). To prevent bacteria from leaving the microfluidic growth chambers, Minisart RC4 0.2 micron regenerated cellulose luer lock syringe filters (17821 K) were attached to the inlets and outlets of the devices via 19 gauge luer lock syringes (NE192PL-25). A syringe pump (Harvard Apparatus Fusion 200 211097) was used to control the flow rate of growth media and antibiotic through the microfluidic chamber. Electrochemical measurements were made using a multipotentiostat (CHI 1040C A2728).2.2 Device fabrication
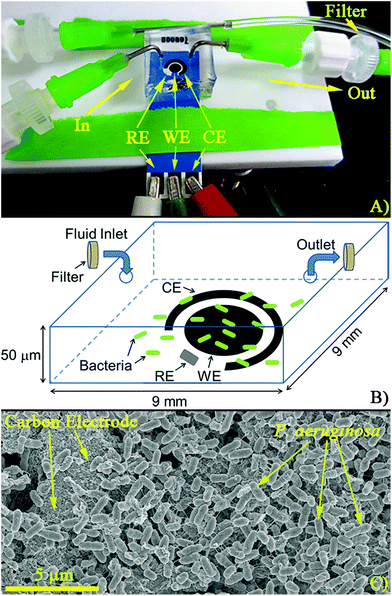
Polydimethylsiloxane (PDMS) wells were fabricated from 9 mm × 9 mm tape molds made on glass slides (3M Scotch Tape, tape thickness ≈50 μm) using a standard method.35 The PDMS wells had a final volume of approximately 4 μL and were designed to cover the entire electrochemical cell (Fig. 1A). Inlets and outlets in the wells were drilled and the resulting microfluidic devices were fabricated by irreversibly bonding PDMS to the disposable electrochemical cells using air plasma (Anatech SP-100, 5–7 s at 100 W). The microfluidic channels were filled with trypticase soy broth (TSB) at a flow rate of 10 μL min−1. To facilitate complete filling of the chamber (removal of air bubbles), an empty syringe was attached to the outlet. By pulling and releasing vacuum on the outlet, TSB was pulled through the chamber displacing any air bubbles.2.3 Cell culture and loading
For antibiotic testing, cultures of PA14 and E. coli were grown overnight in 3 mL of TSB (concentration approximately 1011 cells per mL). After overnight growth, samples were centrifuged for 3 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The supernatant was discarded and the cultures were reconstituted in 3 mL of fresh TSB. After removing the inlet syringe filter, approximately 24 μL of reconstituted cell culture was loaded into the growth chamber at a flow rate of 10 μL min−1. At this flow rate the velocity in the chamber was such that PA14 could not resist flow (P. aeruginosa speed ≈ 30–50 μm s−1), while the outlet filter prevented cells from exiting.36 After loading cells, the filter was replaced, sealing the bacteria into the growth chamber. Biofilms were then allowed to grow at room temperature overnight under stagnant conditions. Stagnant conditions were chosen to ensure that the cells had ample time to adhere to the surface and form a biofilm over the sensor. After overnight growth, flow at 0.1 μL min−1 was initiated with either TSB or colistin sulfate in TSB and the electrochemical response was monitored.6 Steady fluid flow out of the chamber through the outlet filter was always observed during the experiments once the syringe pump was actuated, indicating that the filter was not clogged. Fresh filters were used for each experiment.
000 rpm. The supernatant was discarded and the cultures were reconstituted in 3 mL of fresh TSB. After removing the inlet syringe filter, approximately 24 μL of reconstituted cell culture was loaded into the growth chamber at a flow rate of 10 μL min−1. At this flow rate the velocity in the chamber was such that PA14 could not resist flow (P. aeruginosa speed ≈ 30–50 μm s−1), while the outlet filter prevented cells from exiting.36 After loading cells, the filter was replaced, sealing the bacteria into the growth chamber. Biofilms were then allowed to grow at room temperature overnight under stagnant conditions. Stagnant conditions were chosen to ensure that the cells had ample time to adhere to the surface and form a biofilm over the sensor. After overnight growth, flow at 0.1 μL min−1 was initiated with either TSB or colistin sulfate in TSB and the electrochemical response was monitored.6 Steady fluid flow out of the chamber through the outlet filter was always observed during the experiments once the syringe pump was actuated, indicating that the filter was not clogged. Fresh filters were used for each experiment.
2.4 Electrochemical monitoring
Samples were scanned from −0.5 to 0.2 V versus the internal Ag/AgCl reference electrode on the disposable electrochemical cell (Zensor). Square wave voltammetry (SWV) was used at an amplitude voltage of 50 mV and a frequency of 15 Hz. SWV was chosen due to its increased sensitivity and its ability to monitor the electrochemical peak of PYO compared to other voltammetric and amperometric techniques.32,37 PYO concentration was approximated by a calibration curve of PYO in TSB (Fig. S1†). After loading the PDMS chambers with TSB, the sample was scanned 10 times and the average taken to get the mean response of the TSB. All subsequent measurements were then compared to this response. Three measurements were taken during the loading of the cells, with additional measurements taken every 30 min during the remainder of the tests. For each concentration of antibiotic tested, three different microfluidic setups were used. Electrochemical measurements were processed by subtracting the baseline signal. One way analysis of variation (ANOVA) was used to determine the statistical significance of resulting measurements from the control.2.5 Scanning electron microscopy (SEM) sample preparation
Samples were prepared for SEM imaging by fixing in a 2.5% glutaraldehyde (EMSDIASUM 16120) in a 0.1 M sodium cacodylate buffer (EMSDIASUM 11654). After fixing, samples were washed in cacodylate buffer, and then dehydrated in increasing concentrations of ethanol (Fisher BP2818-4 30–100%). After dehydration, ethanol was removed via critical point drying (Samdri-PVT-3D) using liquid CO2. The final step in SEM preparation was plasma sputtering (Cressington Sputter Coater 208HR) 5 nm of palladium metal onto the samples making them conductive. Once prepared for imaging, samples were loaded into a Field Emission SEM (Hitachi S-4800) and probed at an acceleration voltage and emission current of 3 kV and 10 mA, respectively.2.6 Cell staining
Cell viability after exposure to colistin sulfate was assessed using a LIVE/DEAD staining kit (EMD Chemicals Millipore 50-231-0606). Stains were prepared per manufacturer's operating procedure. After staining, 10 μL of sample was injected into an INCYTO C-chip disposable hemocytometer (DHC-N01). Cells were imaged using a fluorescence microscope, and the number of PA14 cells that were alive after exposure was determined using IMAGE J (ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/).3. Results and discussion
3.1 Monitoring cells in the chamber
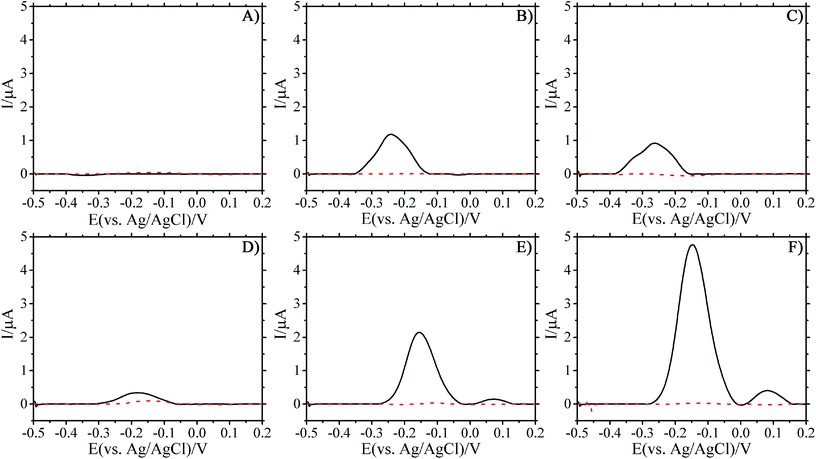
SWV were collected every 30 min from overnight cultures of PA14 in TSB, starting from the point at which they were loaded into the PDMS chambers, to determine whether electro-active molecules were being produced. P. aeruginosa continuously produces PYO as it grows, in both planktonic and biofilm phenotypes, which can be monitored electrochemically during the experiments.24 The utility of this approach is highlighted in Fig. 2 where the electrochemical response of PA14 grown in TSB is monitored over time.The lack of observable peaks during loading indicated that no detectable PYO was present initially in the fresh TSB cell suspension (Fig. 2A). As the biofilm formed under stagnant conditions, the oxidation peak height increased over time (Fig. 2A and B). SEM images of the PDMS growth chamber and the working electrode substrate showed bacteria carpeting both surfaces (see Fig. 2S and 3S in ESI†) after overnight growth under stagnant conditions. Initiating the flow of fresh TSB into the channels after overnight growth allowed the biofilm to thrive. Indeed, the electrical signal increased after TSB flow was initiated (Fig. 2D–F), indicating the increased production rate of PYO. The presence of a second peak at later time points was observed. The first peak is due to PYO, while the appearance of a second peak is ascribed to the electrochemical reaction of a second phenazine derivative that has been reported in the literature as being 5-methylphenazine-1-carboxylic acid or one of its derivatives.38,39 The change in the oxidation potential, after the initiation of flow, where the peak current was measured can be attributed to the internal Ag/AgCl pellet used as the reference for these studies. Drift due to fluid flow is an unavoidable consequence of having the reference in direct contact with the test fluid.40,41 The measured peak potential stabilized over time with constant fluid flow and the peak current at this new potential was used for calculations. The movement of the peak over time can be observed in Fig. S4–S7 in the ESI.† Measurement of the PA14 cultures with a traditional Ag/AgCl reference electrode (BASi MW-2030) showed that the PYO peak current appeared at the expected potential.
While the overall electrical signal increased over time, a decrease was observed consistently at the initiation of fluid flow. There are two possibilities for the observed result. First, the signal decrease can be an indicator of how firmly the biofilm has adhered to the surface of the microfluidic channel. The role of shear stress on cell adhesion has been studied previously; and, the results show that cells can be removed from surfaces at high shear stresses.42,43 As growth media flows through the channel it may remove bacteria if the biofilm is not firmly attached.44 The removal of bacteria in turn would lead to reduced production of PYO in the vicinity of the sensor (lowering the electrical signal). This is unlikely as the applied flow rates in this study are similar to those used by other groups and should be slow enough to avoid significant removal of the bacterial biofilm.5,6
Second, it is possible that the decrease in signal is due to PYO in solution being removed during flow, and it is only when a sufficiently large concentration of PYO is produced, to overcome convective transport, that the signal rebounds. Koley et al. (2011) demonstrated the presence of a PYO gradient (electrocline) in biofilms of P. aeruginosa using scanning electro-chemical microscopy.45 The authors showed that this electrocline extended hundreds of microns above the biofilm's surface. The change in the PYO electrocline due to fluid flow is likely responsible for the initial drop in signal when bulk fluid flow starts. Regardless, it is clear that even after the initiation of flow within the microfluidic chamber, the peak current remains indicating the cells are indeed growing within the chamber (Fig. 3 and ESI Fig. S4–S6†).
E. coli in TSB was used as a control since it is not expected to produce molecules that are redox-active in this voltage window.24,46 The lack of any discernible peak confirms that there are no electrochemical molecules produced by E. coli and that there is no contamination of the chambers by P. aeruginosa from the environment over the course of the experiment (see ESI Fig. S4–S6†). The absence of oxidation peaks from E. coli cells highlights the limitations of the proposed approach to electrochemically monitor the antibiotic susceptibility of other bacterial species. Alternatively, the ability to electrochemically measure the viability of PA14 by the production of PYO can be a useful selective marker of P. aeruginosa in patient samples.46 Furthermore the transparent nature of the PDMS used to fabricate growth chambers facilitates the use of fluorescent bacterial species and markers as reported in the literature.5,6
3.2 Effect of colistin sulfate on PYO signal
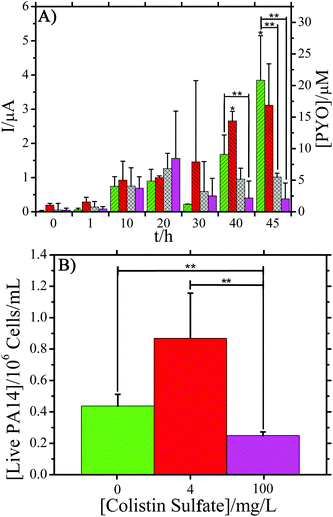
After overnight growth of P. aeruginosa, 0.100 μL min−1 flow of colistin sulfate at 4, 16, and 100 mg L−1 in TSB was initiated. These concentrations were chosen to cover the range of colistin sulfate MIC values that are reported in literature.1 SWV measurements were taken to determine what effect the reported MIC concentrations of colistin sulfate (4 and 16 mg L−1) have on PYO production. This, in turn, can be an indicator of P. aeruginosa biofilm susceptibility to colistin sulfate. Three devices per concentration of colistin sulfate were used and the average peak current reported (Fig. 3A and Fig. S4†). Error bars represent the standard deviation of the mean for three separate measurements at that time point, unless otherwise indicated. As a control, E. coli biofilms were exposed to the same concentrations of colistin sulfate. One replicate per concentration was performed for these tests. No oxidation peaks were observed for E. coli exposed to colistin sulfate signifying a lack of electrochemically active molecules (Fig. S5–S7†).24,47ANOVA was used to identify significant differences between the average peak currents of the three antibiotic concentrations and the control experiment without antibiotic (Fig. 3A). The analysis showed that the average peak current was significantly lower (P < 0.05) for PA14 exposed to 16 and 100 mg L−1 colistin sulfate concentrations when compared against the control. The average percent decrease in the maximum peak current at the end of testing for PA14 exposed to 16 and 100 mg L−1 colistin sulfate was 68% and 82%, respectively, compared to the current produced by the cells in the control experiment. The average percent decrease in the measured current, compared to the control cells, was calculated by %Decrease = 100%*(It − Ic)/Ic where It equals the average peak current at time t and Ic is the average peak current of the control P. aeruginosa cells. The decreased current response is directly related to a decrease in the measured PYO, indicating a correlation between the colistin sulfate concentration and PYO production. In contrast, the average response for cells treated with 4 mg L−1 colistin sulfate showed no significant difference when compared to biofilms exposed to only TSB, indicating that the lower MIC value was not significantly affecting the production of PYO. Importantly, Fig. 3 shows that continuous electrochemical monitoring allows the researcher to view the efficacy of an anti-pseudomonas antibiotic via a reduction in PYO production. Fig. S8† supports these results by demonstrating that PA14 exposed to ampicillin, an antibiotic that is not effective against this species, has no effect on PYO production. By reducing the amount of PYO produced by the bacteria, the host's body may be able to more effectively fight off the infection.48
The inherent resistance of PA14 to the lowest MIC value used in this study could explain why the pyocyanin response did not significantly differ from blank measurements. Liquid samples of PA14 cultured on 4 mg L−1 colistin sulfate agar plates were able to grow indicating that this concentration had no effect on planktonic cell attachment and growth (Fig. S9†). As such, it makes sense that biofilms of PA14 exposed to this concentration would not be affected and should produce similar levels of pyocyanin.
3.3 Effect of colistin sulfate on post exposure growth
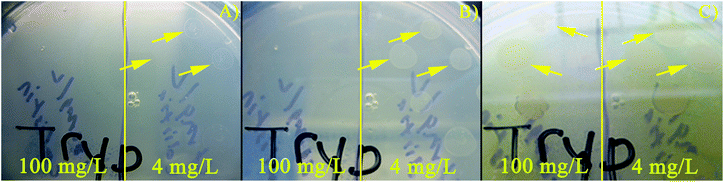
The number of living cells measured after exposure to three different concentrations of colistin sulfate were compared (Fig. 3B). After the biofilm was exposed to antibiotic in the device, the PDMS chambers were peeled off and 100 μL of fresh TSB was spotted on the biofilm and pipetted vigorously to remove material from the surface of the electrode. Removed samples were used to measure live cell counts, performed with Millipore 3P Live/Dead Stain, using a haemocytometer. Each measurement was performed in triplicate and the error bars show one standard deviation of the mean. A concentration of approximately 4 × 105 live cells per mL was measured in the biofilms not exposed to colistin sulfate. Biofilms typically have lower concentrations of live cells than agitated liquid cultures. A statistically significant reduction in the number of live PA14 cells was measured for samples exposed to 100 mg L−1 colistin sulfate compared to cells exposed to 0 and 4 mg L−1 colistin sulfate. The ∼2× reduction in the number of living cells supports the hypothesis that a reduction in the PYO signal is correlated with a reduction in the number of living cells. Recently, Connell et al. (2014) supported these findings as well by showing a correlation between the number of cells trapped in a chamber and the concentration of PYO that is present around the cells.49Fig. 4 shows the culture results at three different time points for cells exposed to 4 and 100 mg L−1 colistin sulfate in microfluidic devices. Growth was observed in samples exposed to 4 mg L−1 colistin sulfate after only 4.3 h of incubation implying that this concentration had little effect on the cells’ viability (Fig. S10†). No growth was observed for cells exposed to 100 mg L−1 colistin sulfate after 6.5 h of incubation. Growth was observed for samples collected from chambers exposed to 100 mg L−1 colistin sulfate after 74 h, indicating that the complete elimination of viable bacteria from inside the chamber was not achieved. The qualitative results of the live cell stain are consistent with the culture plate experiments. The live cell concentration in biofilms exposed to 100 mg L−1 colistin sulfate (Fig. 3B), however, is higher than expected when compared to the reduced rate of colony formation on agar plates (Fig. 4). Taken together, these results suggest that the cells exposed to this antibiotic may have reduced their metabolic activity to make them less susceptible to the antibiotic.
These results support the findings in the literature, drawing attention to the lower efficacy of reported MICs against microbial biofilms.6 It is clear from the results in Fig. 3 and 4 that a reduction in PYO production, under exposure to colistin sulfate, is correlated with a reduction in the viability of PA14. This reduction in pyocyanin and inhibited growth rate may allow a person's immune response to successfully fight off the bacterial infection.20–22
4. Conclusions
We demonstrated, for the first time, the possibility of using electrochemical sensors to monitor metabolites produced by a biofilm that is exposed to antibiotics. The time to detection using this electrochemical approach (∼45 h) is comparable to standard culture plate techniques. While simple identification of bacterial species can be accomplished within 24 hours, sensitivity tests typically require an additional 24–72 hours of incubation on several plates. Biochemical and molecular methods are available commercially that provide sensitivity information within minutes after the initial 24 hour colony formation period, but they require expensive reagents/instrumentation and additional sample processing. The analysis time of the proposed method may potentially be lowered by employing miniature microfabricated electrochemical sensors that, in turn, allow for smaller microfluidic chambers to be employed compared to those utilized in this current study. Smaller chambers would potentially decrease the time to detection due to the confinement imposed on the cells.4–6In healthcare situations, such as wound infections, biofilms form rapidly and require immediate treatment. This approach can also be utilized to study biofilms that are more mature or exposed to any number of other experimental variables. Ultimately, an electrochemical sensor for susceptibility determination may be valuable for low-resource settings or for monitoring the status of infections in vivo while they are being treated with antibiotics.
Acknowledgements
The authors would like to thank Dr Slava Epstein for providing the cultures of m-cherry E. coli used in this study. The authors would like to thank Dr Gerald Pier from the Channing Laboratory at Brigham and Women's Hospital for providing the PA14 used in this study. The authors would like to thank William Fowle of Northeastern's EM Lab for help preparing samples for SEM imaging. This study was funded in part by NSF Award #1125535.Notes and references
- J. M. Andrews, J. Antimicrob. Chemother., 2001, 48, 5–16 CrossRef CAS PubMed.
- P. K. Singh, A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh and E. P. Greenberg, Nature, 2000, 407, 762–764 CrossRef CAS PubMed.
- C. F. Schierle, M. de la Garza, T. A. Mustoe and R. D. Galiano, Wound Repair Regen, 2009, 17, 354–359 CrossRef PubMed.
- J. Kim, M. Hegde, S. H. Kim, T. K. Wood and A. Jayaraman, Lab Chip, 2012, 12, 1157–1163 RSC.
- J. Kim, H. D. Park and S. Chung, Microfluidic approaches to bacterial biofilm formation, Molecules, 2012, 17, 9818–9834 CrossRef CAS PubMed.
- K. P. Kim, Y. G. Kim, C. H. Choi, H. E. Kim, S. H. Lee, W. S. Chang and C. S. Lee, Lab Chip, 2010, 10, 3296–3299 RSC.
- L. Richter, C. Stepper, A. Mak, A. Reinthaler, R. Heer, M. Kast, H. Bruckl and P. Ertl, Lab Chip, 2007, 7, 1723–1731 RSC.
- H.-Y. N. Holman, R. Miles, Z. Hao, E. Wozei, L. M. Anderson and H. Yang, Anal. Chem., 2009, 81, 8564–8570 CrossRef CAS PubMed.
- Y. Yawata, K. Toda, E. Setoyama, J. Fukuda, H. Suzuki, H. Uchiyama and N. Nomura, J. Biosci. Bioeng., 2010, 110, 130–133 CrossRef CAS PubMed.
- Y. Yawata, K. Toda, E. Setoyama, J. Fukuda, H. Suzuki, H. Uchiyama and N. Nomura, J. Biosci. Bioeng., 2010, 110, 377–380 CrossRef CAS PubMed.
- M. B. Miller and B. L. Bassler, Annu. Rev. Microbiol., 2001, 55, 165–199 CrossRef CAS PubMed.
- M. D. P. Willcox, H. Zhu, T. C. R. Conibear, E. B. H. Hume, M. Givskov, S. Kjelleberg and S. A. Rice, Microbiology, 2008, 154, 2184–2194 CrossRef CAS PubMed.
- G. W. Lau, D. J. Hassett, H. Ran and F. Kong, Trends Mol. Med., 2004, 10, 599–606 CrossRef CAS PubMed.
- J. B. Goldberg, F1000 Biol. Rep., 2010, 2(29) DOI:10.3410/B2-29.
- J. B. Lyczak, C. L. Cannon and G. B. Pier, Microbes Infect., 2002, 2, 1051–1060 CrossRef.
- R. L. Gibson, J. L. Burns and B. W. Ramsey, Am. J. Respir. Crit. Care Med., 2003, 168, 918–951 CrossRef PubMed.
- J. Chastre and J. Fagon, Ventillator-associated pneumonia, Am. J. Respir. Crit. Care Med., 2002, 165, 867–903 CrossRef PubMed.
- J. E. Fergie, S. J. Shema, L. Lott, R. Crawford and C. C. Patrick, Clin. Infect. Dis., 1994, 18, 390–394 CrossRef CAS PubMed.
- A. Park, H. Jeong, J. Lee, K. P. Kim and C. Lee, BioChip J., 2011, 5, 236–241 CrossRef CAS.
- L. Allen, D. H. Dockrell, T. Pattery, D. G. Lee, P. Cornelis, P. G. Hellewell and M. K. B. Whyte, J. Immunol., 2005, 174, 3643–3649 CrossRef CAS.
- L. R. Usher, R. A. Lawson, I. Geary, C. J. Taylor, C. D. Bingle, G. W. Taylor and M. K. B. Whyte, J. Immunol., 2002, 168, 1861–1868 CrossRef CAS.
- R. Wilson, T. Pitt, G. Taylor, D. Watson, J. Macadermot, D. Sykes, D. Roberts and P. Cole, J. Clin. Invest., 1987, 79, 221–229 CrossRef CAS PubMed.
- Y. Wang and D. K. Newman, Environ. Sci. Technol., 2008, 42, 2380–2386 CrossRef CAS PubMed.
- D. Sharp, P. Gladstone, R. B. Smith, S. Forsythe and J. Davis, Bioelectrochemistry, 2010, 77, 114–119 CrossRef CAS PubMed.
- H. J. Sismaet, T. A. Webster and E. D. Goluch, Analyst, 2014, 139, 4241–4246 RSC.
- T. A. Webster, H. J. Sismaet and E. D. Goluch, Nano LIFE, 2013, 03, 1340011 CrossRef.
- D. Banerjee and D. Stableforth, Drugs, 2000, 60, 1053–1064 CrossRef CAS PubMed.
- C. Dai, J. Li, W. Lin, G. Li, M. Sun, F. Wang and J. Li, Toxicol. Mech. Methods, 2012, 22, 592–596 CrossRef CAS PubMed.
- B. Lin, C. Zhang and X. Xiao, J. Vet. Pharmacol. Ther., 2005, 28, 349–354 CrossRef CAS PubMed.
- M. T. Eadon, B. K. Hack, J. J. Alexander, C. Xu, M. E. Dolan and P. N. Cunningham, Physiol. Gen., 2013, 45, 877–888 CrossRef CAS PubMed.
- M. E. Evans, D. J. Feola and R. P. Rapp, Ann. Pharmacother., 1999, 33, 960–967 CrossRef CAS PubMed.
- L. Pires, K. Sachsenheimer, T. Kleintschek, A. Waldbaur, T. Schwartz and B. E. Rapp, Biosens. Bioelectron., 2013, 47, 157–163 CrossRef CAS PubMed.
- X. Mulet, G. Cabot, A. A. Ocampo-Sosa, M. A. Domínguez, L. Zamorano, C. Juan, F. Tubau, C. Rodríguez, B. Moyà, C. Peña, L. Martínez-Martínez and A. Oliver, Antimicrob. Agents Chemother., 2013, 57, 5527–5535 CrossRef CAS PubMed.
- T. A. Webster and E. D. Goluch, Lab Chip, 2012, 12, 5195–5201 RSC.
- A. B. Shrirao and R. Perez-Castillejos, Simple fabrication of microfluidic devices by replicating scotch-tape masters, Chips Tips, 2010 Search PubMed.
- T. S. Murray and B. I. Kazmierczak, J. Bacteriol., 2006, 188, 6995–7004 CrossRef CAS PubMed.
- A. J. Bard and L. R. Faulkner, Electrochemical methods fundamentals and applications, John Wiley & Sons Inc., 2011 Search PubMed.
- V. B. Wang, S. L. Chua, B. Cao, T. Seviour, V. J. Nesatyy, E. Marsili, S. Kjelleberg, M. Givskov, T. Tolker-Nielsen, H. Song, J. S. Loo and L. Lang, PLoS One, 2013, 8, e63129 CrossRef CAS PubMed.
- D. L. Bellin, H. Sakhtah, J. k. Rosenstein, P. M. Levine, J. Thimot, K. Emmett, L. E. Dietrich and K. L. Shepard, Nat. Commun., 2014, 5, 3256 CrossRef PubMed.
- M. W. Shinwari, D. Zhitomirsky, I. A. Deen, P. R. Selvaganapathy, M. J. Deen and D. Landheer, Sensors, 2010, 10, 1679–1715 CrossRef CAS PubMed.
- L. Rassaei, K. Mathwig, E. D. Goluch and S. G. Lemay, J. Phys. Chem. C, 2012, 116, 10913–10916 CrossRef CAS.
- J.-C. Ochoa, C. Coufort, R. Escudié, A. Liné and E. Paul, Chem. Eng. Sci., 2007, 62, 3672–3684 CrossRef CAS.
- Y.-P. Tsai, Biofouling, 2005, 21, 267–277 CrossRef PubMed.
- M. M. Salek, S. M. Jones and R. J. Martinuzzi, Biofouling, 2009, 25, 711–725 CrossRef PubMed.
- M. M. R. D. Koley, A. J. Bard and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 19996–20001 CrossRef PubMed.
- T. A. Webster, H. J. Sismaet, J. L. Conte, I. P. J. Chan and E. D. Goluch, Biosens. Bioelectron., 2014, 60, 265–270 CrossRef CAS PubMed.
- E. Kim, T. Gordonov, W. E. Bentley and G. F. Payne, Anal. Chem., 2013, 85, 2102–2108 CrossRef CAS PubMed.
- G. M. Denning, L. A. Wollenweber, M. A. Railsback, C. D. Cox, L. L. Stoll and B. E. Britigan, Infect. Immun., 1998, 66, 5777 CAS.
- J. L. Connell, J. Kim, J. B. Shear, A. J. Bard and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 18255–18260 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional electrochemical experimental results. See DOI: 10.1039/c5an01358e |
| This journal is © The Royal Society of Chemistry 2015 |