Open Access Article
Open Access ArticleLabel-free detection of Phytophthora ramorum using surface-enhanced Raman spectroscopy†
Sezin
Yüksel‡
abc,
Lydia
Schwenkbier‡
abc,
Sibyll
Pollok
ad,
Karina
Weber
*abc,
Dana
Cialla-May
*abc and
Jürgen
Popp
abc
aLeibniz Institute of Photonic Technology Jena (IPHT), Albert-Einstein-Straße 9, 07745 Jena, Germany. E-mail: karina.weber@ipht-jena.de; Tel: +49 (0)3641-948390
bFriedrich Schiller University Jena, Institute of Physical Chemistry and Abbe Center of Photonics, Helmholtzweg 4, 07743 Jena, Germany. E-mail: dana.cialla-may@uni-jena.de; Tel: +49 (0)3641-206309
cInfectoGnostics Forschungscampus Jena, Zentrum für Angewandte Forschung, Philosophenweg 7, 07743 Jena, Germany
dErnst-Abbe-Hochschule Jena, University of Applied Sciences, Carl-Zeiss-Promenade 2, 07745 Jena, Germany
First published on 4th September 2015
Abstract
In this study, we report on a novel approach for the label-free and species-specific detection of the plant pathogen Phytophthora ramorum from real samples using surface enhanced Raman scattering (SERS). In this context, we consider the entire analysis chain including sample preparation, DNA isolation, amplification and hybridization on SERS substrate-immobilized adenine-free capture probes. Thus, the SERS-based detection of target DNA is verified by the strong spectral feature of adenine which indicates the presence of hybridized target DNA. This property was realized by replacing adenine moieties in the species-specific capture probes with 2-aminopurine. In the case of the matching capture and target sequence, the characteristic adenine peak serves as an indicator for specific DNA hybridization. Altogether, this is the first assay demonstrating the detection of a plant pathogen from an infected plant material by label-free SERS employing DNA hybridization on planar SERS substrates consisting of silver nanoparticles.
Introduction
In recent decades a vast number of invasive plant pathogens have spread across European and North American countries. Members of the genus Phytophthora are among these. They belong to the most important and aggressive plant pathogens worldwide and pose serious threats to plants in natural and landscaped environments as well as in plant cultivation.1 One prominent species is Phytophthora ramorum2 which is responsible for the dramatic die back of oaks in North America (sudden oak death) and the Larix decline in the United Kingdom.3 In order to prevent the spread of this pathogen across borders, its reliable and specific detection is mandatory. To date Phytophthora diagnosis has been mainly realized by microbiological or PCR-based techniques.4–8 However, its specific and reliable detection directly in the field from real samples and without much effort remains an on-going challenge. In this context, DNA hybridization assays have been developed which are based on the immobilization of specific capture probes and their interaction with complementary target sequences.9,10 DNA hybridization can be detected by fluorescent dyes, radioactivity or enzyme induced color changes.11–13 Recently, surface enhanced Raman spectroscopy (SERS) was highlighted as an attractive analytical tool for the identification of molecular interactions. In terms of high sensitivity and cost effectiveness, it represents an emerging and promising field in bioanalytical research.14–17 While applying SERS, it is important to consider the interaction between light and molecules as well as metallic nanostructures. The latter are eminent for the amplification of the Raman signal,18,19 which is increased by several orders of magnitude.20,21 Thus, SERS combines high molecular specificity, attributed to the Raman effect, with a high sensitivity.22–24 Therefore it is an excellent tool for both quantitative and qualitative analysis, offering almost unlimited possibilities for multiplexing.25–27 Moreover, SERS-based DNA detection provides several advantages compared to classical fluorescence techniques. Fluorescence detection requires expensive dyes and complex conjugation chemistry, resulting in a limited number of specific labels. The SERS approach enables the selection of various Raman labels, without bleaching or quenching problems.28 Furthermore, short data processing times point to the direction of implementing SERS in the development of rapid and low-cost detection platforms for pathogen diagnosis.14In the last few decades a wide range of SERS-based DNA hybridization assays have been explored. Commonly, the hybridization between specific capture probes and the corresponding target molecules is indicated by a change in the signal of the dye label.27,29–39 More recently, an elegant label-free detection scheme for specific DNA hybridization was introduced by Halas and co-workers.40 Usually the DNA spectrum is represented by the four nucleobases: adenine (A), guanine (G), cytosine (C) and thymine (T). Adenine exhibits a more prominent Raman profile than the other nucleobases and strongly dominates the SERS spectra of DNA.33,41,42 Thus, the adenine signal can serve as an endogenous marker for SERS-based DNA detection. However, in general both capture probes and target sequences consist of multiple adenine moieties, which hamper the SERS-based detection of DNA hybridization. This drawback was circumvented by the substitution of adenine by 2-aminopurine (2-AP) in order to generate adenine-free capture probes.40 2-AP serves as an adenine analogue or isomer, exhibiting identical hybridization characteristics.43–45 It forms a canonical Watson–Crick base pair with thymine and enables the study of nucleic acid structures and their dynamics. Thus, only the SERS spectra of the hybridized target DNA, containing adenine in the sequence, provide the related adenine characteristics. Inspired by this unique feature, we expanded the principle of label-free SERS-based detection to Phytophthora ramorum plant pathogens in infected Rhododendron leaves. This illustrates, to the best of our knowledge for the first time, the application of label-free SERS towards PCR products from real samples. To this end, we considered a combination of plant sampling, DNA isolation, amplification, hybridization and SERS-based DNA detection to reflect the complete analysis chain.
Experimental section
Plant infection and DNA extraction of Phytophthora ramorum
The genomic DNA (gDNA) of P. ramorum BBA9/95 was extracted from artificially infected Rhododendron leaves (see detached leaf assay, described in detail elsewhere46) by homogenizing the samples with a mortar and pestle.9 The lysate was placed into a micro reaction tube and DNA extraction was performed using the magnetic bead based innuPREP MP Basic Kit according to the recommendations of the manufacturer (Analytik Jena AG, Jena, Germany).Amplification of target DNA by PCR
Linear-after-the-exponential polymerase chain reaction (LATE-PCR) was carried out to amplify a fragment within the yeast GTP-binding protein (Ypt1) target gene region. The conditions for LATE-PCR are described elsewhere10 (Table 1). DNA of inoculated Rhododendron leaves was used in a concentration of 50 ng per reaction. The amplified Ypt1-fragments had a length of approximately 450 bp.| Sequence 5′–3′ | Modification | ||
|---|---|---|---|
| A* indicates the substitution of adenine by 2-aminopurine. | |||
| Capture probes | P. ramorum | CCC CCC A*CT TTC CGT GGG TGA* GTT TCC TTT | 5′-SH internal 2-AP |
| P. lateralis | CGG GA*G A*TT TTT TCC CGC TTT CCT TGG GGT A*A*G | 5′-SH internal 2-AP | |
| Primers | YPh1F_LATE | CAT CTC GAC CAT KGG TGT GGA CTT T | w/o |
| YPh2R | ACG TTC TCM CAG GCG TAT CT | ||
| ssDNA complementary to P. ramorum | P. ramorum | AAA GGA AAC TCA CCC ACG GAA AGT GGG GGG | w/o (SERS) 5′-FITC (fluorescence) |
SERS substrate preparation
As the SERS substrate, we applied enzymatically generated silver nanoparticles (EGNPs). In extensive studies they were proven as low-cost, highly reproducible and stable bottom-up SERS active substrates.47,48These nanoparticles were generated by an enzyme-induced growth process on glass substrates, described in detail elsewhere.47 Briefly, a biotin-labeled single-stranded DNA was immobilized onto planar substrates. A streptavidin horseradish peroxidase (HRP) conjugate bound to the biotin and catalyzed the enzymatic generation of nanoparticles from a silver solution. For this purpose the EnzMet™ kit from Nanoprobes Inc., (Yaphank, NY, USA) was used.
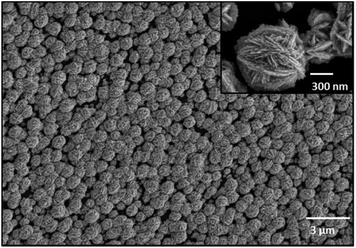
Finally, an array of individual EGNP deposits was produced, where closely packed ‘desert-rose-like’ silver nanostructures with a particle size of approximately 400 nm could be observed (Fig. 1). The strongest electromagnetic field enhancement was located at the sharp and spiky features of the silver intertwined plates.
Scanning electron microscopy (SEM)
The scanning electron microscopy (SEM) images were recorded with a high resolution field emission scanning electron microscope JEOL JSM-6300F (Tokyo, Japan), applying 5 keV accelerating voltage with an accumulation time of 300 s.Immobilization of adenine-free capture probes and DNA hybridization on EGNP array
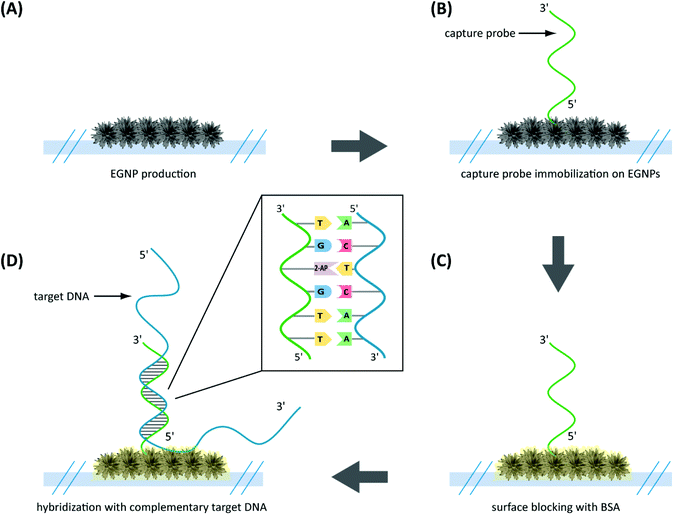
The capture probes, primers and complementary ssDNA were purchased from Eurofins MWG Operon (Ebersberg, Germany; Table 1).For either fluorescence or SERS-based detection, the capture probes were immobilized in triplicate directly on top of the EGNP array via their thiol groups (Fig. 2). Thereto they were dissolved in 5× PBS buffer to a final concentration of 20 μM and spotted on the EGNPs with a Nanoplotter 2.1 (GeSim, Germany). After UV-linking at 254 nm for 5 min the substrates were washed with 1× PBS for 2 min. A 0.5% (w/v) solution of bovine serum albumin (BSA) (Carl Roth, Karlsruhe, Germany) in 1× PBST was used to block unspecific binding sites. For the DNA hybridization either 20 μl of the LATE-PCR product or 1 μM of complementary ssDNA (Table 1) were dissolved in 3 × SSC/0.5% SDS and incubated on the substrates for two hours at 58 °C in a humidity chamber. The subsequent washing steps were performed at room temperature (2 × SSC/0.1% SDS, 0.2 × SSC and finally with 0.1 × SSC, 5 min each). Afterwards, fluorescence or SERS detection was performed.
Fluorescence microscopy
Fluorescence images were recorded with the light microscope Axio Imager Z1 (Carl Zeiss Jena GmbH, Jena, Germany). The samples were measured using a 20× objective with an exposure time of 435 ms.SERS
SERS spectra were recorded using a confocal Raman microscope (WITec GmbH, Ulm, Germany) equipped with a 488 nm excitation laser line. For the irradiation of the samples a 100× Olympus objective (NA 0.9) with a laser power of 35 μW was employed. The same objective was applied for recording the backscattered light with a spectrometer, equipped with 600 lines per mm grating and a 1024 × 127 pixel CCD camera cooled to 208 K. Average SERS spectra were calculated from ten different measurement points with an integration time of 10 s.Results and discussion
The study focused on the reliable and label-free detection of P. ramorum plant pathogen from infected Rhododendron leaves. We applied a label-free SERS-based detection approach to record the specific DNA interaction on a planar SERS substrate (Fig. 3). The assay started with the sampling of the infected plant material and the isolation of pathogenic gDNA, which was used as a template for the amplification of the Ypt1 target region by LATE-PCR. This target DNA was then used for hybridization with immobilized capture probes that possessed 2-AP instead of adenine on the EGNP array. Thus, we took advantage of the strong spectral feature of adenine in the SERS spectrum of the target DNA. The presence of adenine exclusively in the target DNA sequence served as an endogenous marker for the label-free SERS-based detection of the hybridization event. Furthermore, the entire analysis chain was considered in order to prove the near-future application of SERS for implementation in an on-site detection system.DNA isolation and amplification
The preparation of the samples is the first critical step in the successful detection of pathogens. As an example for a proof-of-concept analysis we chose Rhododendron leaves of P. ramorum-infected plants. The DNA extraction was performed by combining a mortar and pestle for effective cell disruption in conjunction with nucleic acid release.9,49 This manually operated homogenization is applicable for softer tissues such as Rhododendron leaves and provided an incredibly facile way to release gDNA from the plant material. Subsequently, magnetic particle-based DNA isolation enabled easy handling as well as short processing times. Thus, the first step of DNA extraction and purification was realized in a straightforward manner for a potential field application.LATE-PCR allowed the amplification of the yeast GTP-binding protein (Ypt1) target gene of P. ramorum. This particular region is located within a single-copy gene, which implies the presence of only one copy per genome. Therefore, a prior DNA amplification was mandatory for proper detection. Accordingly, LATE-PCR was carried out to generate sufficient amounts of single-stranded target DNA.10 The successful generation of ssDNA was indicated by the appearance of two bands in the analytical gel. The faster migrating DNA represented single-stranded DNA (450 nt) and the higher molecular weight band of 450 bp was double-stranded (see ESI Fig. S1†). By applying asymmetric PCR post-amplification treatment could be omitted.
Fluorescence microscopic detection of target DNA hybridization
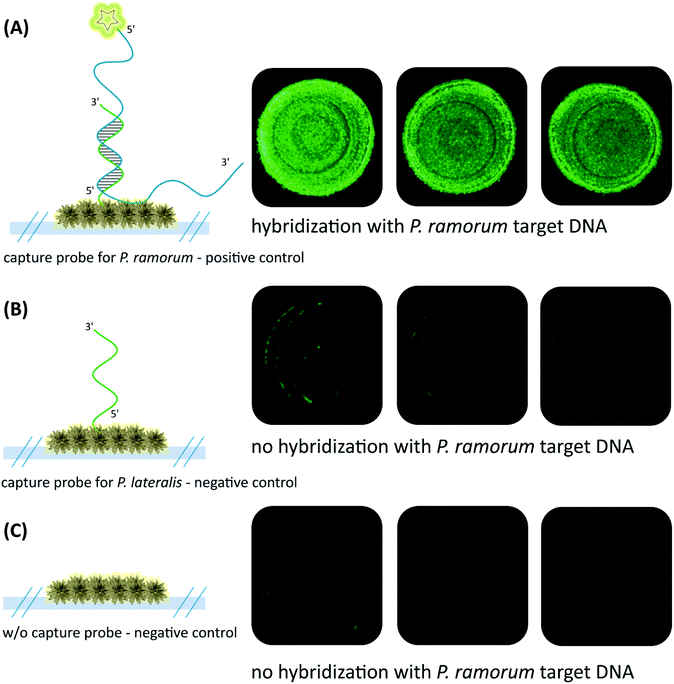
In order to verify the functionality of the hybridization assay on the SERS substrate, fluorescence microscopy was performed as a reference method. For this purpose two different capture probes, one for the target species P. ramorum (Fig. 4A) and one for the closely related species P. lateralis (Fig. 4B), were immobilized on the corresponding EGNP spots. Moreover, EGNPs without any capture probes were analyzed after identical processing steps for blocking, hybridization and washing (Fig. 4C).Subsequently, hybridization was accomplished using FITC-labeled single-stranded target DNA of P. ramorum with a length of 30 nucleotides (Table 1). The selection of this particular Phytophthora species relied upon previous studies which focused on the specificity and sensitivity of the Ypt1 target gene region.46 The highest signals were recorded for P. ramorum-specific capture probes that entirely matched the target DNA sequence (Fig. 4A). In contrast, only weak signals were detected for the P. lateralis capture probes (Fig. 4B). In comparison with the background fluorescence (Fig. 4C) those signals were negligible. Consequently, the functionality of the hybridization assay for the specific detection of P. ramorum is proven. The adenine isomer, 2-AP, in the capture probes exhibited the same hybridization characteristics.
SERS detection of target DNA hybridization
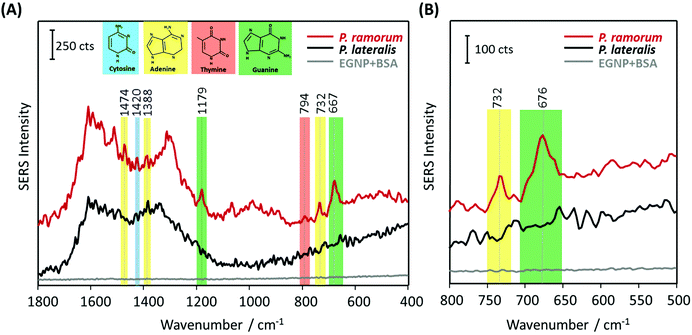
In the next step, the label-free SERS detection of P. ramorum DNA was investigated. First, DNA hybridization was accomplished using a short single-stranded target DNA of P. ramorum with a length of 30 nucleotides. After that we applied the PCR product isolated from real plant samples and recorded the corresponding SERS spectra.Hybridization experiments were performed with the 30 nucleotide single-stranded target DNA of P. ramorum. Two different adenine-free capture probes, one for the target species P. ramorum and one for the non-target species P. lateralis were immobilized on the SERS substrate and hybridization was conducted. In Fig. 5, the respective SERS spectra of the hybridized P. ramorum capture probes (indicated by a red line), the P. lateralis capture probes (indicated by a black line) and the background of the SERS substrate (indicated by a grey line) are displayed as mean values. In the recorded spectra for P. ramorum and P. lateralis capture probes, the broad carbon spectra are visible between 1200 and 1600 cm−1. This burning effect of the surface is caused by an enhanced electromagnetic field due to applying a relatively long exposure time. Owing to the high carbon background, the Raman vibrational modes of cytosine and adenine were barely detectable between 1200 and 1600 cm−1. In the SERS spectra of P. ramorum, where hybridization with the target DNA occurred (see Fig. 5A, red line), one characteristic band of ν(C–C) for cytosine at 1420 cm−1 could be observed.41,50 Moreover, the vibrational modes of guanine at 1179 cm−1 and 667 cm−1 were assigned to the ν(C–C) and ring breathing vibrations. The typical Raman bands of adenine were marked with yellow colored frames. The two Raman modes of adenine at 1474 cm−1 and 1388 cm−1 are referred to as ν(C–N) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) stretching modes respectively. Additionally, the most prominent peak of adenine at 732 cm−1 is related to the aromatic ring breathing. Fig. 5B shows the spectral zoom of the region of interest between 500 and 800 cm−1. Accordingly, P. ramorum (Fig. 5B, red line) was identified by the dominant peak of adenine at 732 cm−1. In contrast, the SERS spectra of the non-target species P. lateralis do not show any characteristic adenine-related modes, indicating that no hybridization occurred (Fig. 5B, black line). Thus, the specific binding of the P. ramorum target DNA to the matching capture probe was monitored by the presence of adenine.
N) stretching modes respectively. Additionally, the most prominent peak of adenine at 732 cm−1 is related to the aromatic ring breathing. Fig. 5B shows the spectral zoom of the region of interest between 500 and 800 cm−1. Accordingly, P. ramorum (Fig. 5B, red line) was identified by the dominant peak of adenine at 732 cm−1. In contrast, the SERS spectra of the non-target species P. lateralis do not show any characteristic adenine-related modes, indicating that no hybridization occurred (Fig. 5B, black line). Thus, the specific binding of the P. ramorum target DNA to the matching capture probe was monitored by the presence of adenine.
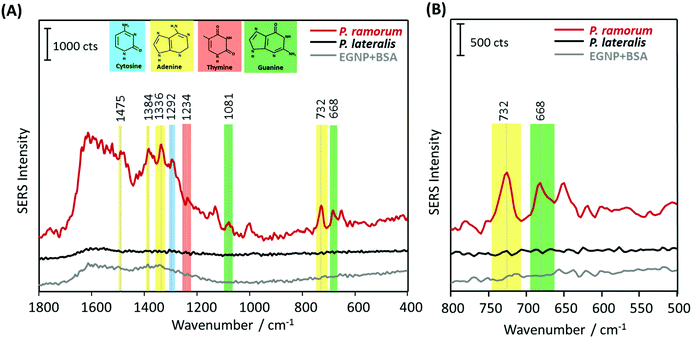
In order to demonstrate the applicability of the label-free technique for bioanalytical purposes in terms of pathogen detection, we adapted the SERS approach for DNA isolated from real plant samples, infected with the pathogen P. ramorum. The implementation of SERS as a novel tool for pathogen detection resulted in a plethora of publications in the last few years. However, the main challenge of this promising technique is the confirmation of its applicability for on-site use with real samples, e.g. from infected plant tissue. Therefore, the main objective of the present study was to demonstrate the application of label-free SERS detection for the plant pathogen DNA, extracted from Phytophthora infected Rhododendron leaves. Fig. 6 displays the mean SERS spectra of the DNA-hybrid between P. ramorum-specific capture probes and P. ramorum target DNA, which was amplified by PCR as 450 nucleotide fragments (indicated by a red line), the non-target P. lateralis (black line) and the background of the SERS substrate (grey line). In accordance with the findings for the short target DNA fragment, typical vibrational bands at 1081 cm−1 and 668 cm−1 were observed for guanine in the SERS spectra of P. ramorum (see Fig. 6A, red line). Also the less strong Raman bands of thymine at 1234 cm−1 and cytosine at 1292 cm−1 were depicted in the SERS spectra of the target DNA. However, focusing on adenine as the endogenous marker, very dominant Raman modes at 1384 cm−1 and 732 cm−1 were monitored for the DNA extracted and amplified from the infected Rhododendron. Moreover, the characteristic Raman mode of adenine at 1336 cm−1 could be detected due to the presence of a higher amount of the respective nucleobase in the target DNA strand. Similar to previous results the dominant peak at 732 cm−1 indicating the presence of adenine served as a significant marker band in comparison with the non-target P. lateralis (see Fig. 6B). Altogether, by substituting adenine with 2-AP in the capture probes, the specific hybridization of the P. ramorum target DNA could be successfully monitored even while applying this very long PCR product. A proper discrimination between P. ramorum (target) and P. lateralis (non-target) is possible. Thus, the label-free SERS detection of DNA was exemplarily demonstrated for the important plant pathogen P. ramorum.
Reproducibility of the SERS spectra
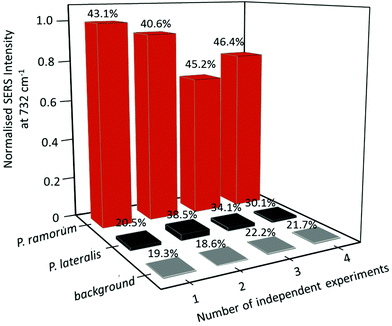
Despite the fact that SERS-based DNA detection offers great potential regarding a higher sensitivity and specificity, it is still in competition with more common techniques such as fluorescence spectroscopy. One drawback for SERS measurements is the lack of good spectral reproducibility as well as precise batch-to-batch and day-to-day recordings. For this reason the current study also addressed the batch-to-batch reproducibility of the SERS signals occurring in the case of DNA hybridization. Independent experiments were performed using various DNA extracts, conducting individual PCR runs and hybridization assays. Fig. 7 depicts the normalized integrated SERS intensity for various batches. The adenine peak at 732 cm−1 was integrated for the SERS spectra of the matching target (P. ramorum), the non-matching control (P. lateralis) and the background of the SERS substrate and then normalized to the peak of the highest intensity. In comparison with the capture probes and the background, the integrated SERS intensity at 732 cm−1 showed the highest values, having a relative standard deviation (RSD) around 40% (see Fig. 7). The high relative standard deviation can be explained by the different orientations of the DNA on the metallic surface and by an inhomogeneous coverage of BSA on silver nanoparticles. Due to the surface blocking, the SERS intensity can vary during point-to-point measurements. Hence, the results are not sufficient for quantitative detection. However, monitoring the DNA hybridization event without employing labels in SERS based detection is demonstrated. It is clearly visible that the presence of adenine in the SERS spectra of the target DNA served as an indicator for the DNA hybridization.Conclusions and outlook
In recent years, SERS has been applied for a variety of analyses in the life science sector. Due to its high sensitivity and specificity, it offers great potential for the development of novel biosensors. In this study we expanded its application for the label-free detection of important plant pathogens using the example of Phytophthora ramorum. To the best of our knowledge, this is the first report on the label-free detection of the PCR product, isolated from infected plant tissue. Here, DNA-based detection of P. ramorum was achieved by covering the whole analytical chain. The sample preparation was realized by an easy tissue disruption procedure combined with magnetic bead-based nucleic acid isolation. The subsequent DNA amplification was performed by LATE-PCR, which enabled the generation of single-stranded target DNA. Thus, a post-PCR processing step for the generation of ssDNA could be omitted. In order to improve the detection and achieve a putative in-field application, novel isothermal amplification techniques could be implemented.51,52 Moreover, the SERS-based DNA detection, relying on DNA–DNA-hybrid formation, could be performed in a handheld Raman device, that allows the applicability at the point-of-need due to its portability.In summary, a reliable, reproducible and thermally stable SERS assay has been developed for the detection of specific DNA hybridization between immobilized capture probes and the sequence-matching target DNA. We have highlighted the application of SERS to identify the adenine-containing target DNA isolated and amplified from infected Rhododendron leaves, in conjunction with adenine-free capture probes.
Acknowledgements
We thank Stephan König and Sabine Werres from the Julius Kühn–Institute Braunschweig, Germany for capture probe design, preparation of artificially inoculated Rhododendron leaves and extraction and reprocessing of the Phytophthora DNA. The project “PhytoChip-Validierung” (28-1-54.081-10) is financed by the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE) under the innovation support program. Funding of the project “JBCI 2.0” (03IPT513Y) within the framework “InnoProfile Transfer – Unternehmen Region”, the Federal Ministry of Education and Research, Germany (BMBF) is gratefully acknowledged.References
- O. K. Ribeiro and K. Lamour, Phytophthora: Global Perspect., 2013, 2, 1 Search PubMed.
- S. Werres, R. Marwitz, W. Veld, A. De Cock, P. J. M. Bonants, M. De Weerdt, K. Themann, E. Ilieva and R. P. Baayen, Mycol. Res., 2001, 105, 1155–1165 CrossRef CAS.
- C. Brasier and J. Webber, Nature, 2010, 466, 824–825 CrossRef CAS PubMed.
- G. J. Bilodeau, C. A. Levesque, A. W. A. M. de Cock, C. Duchaine, S. Briere, P. Uribe, F. N. Martin and R. C. Hamelin, Phytopathology, 2007, 97, 632–642 CrossRef CAS PubMed.
- F. N. Martin, Phytophthora: Global Perspect., 2013, 2, 19 Search PubMed.
- P. A. O'Brien, N. Williams and G. E. S. Hardy, Crit. Rev. Microbiol., 2009, 35, 169–181 CrossRef PubMed.
- L. Schena and D. E. L. Cooke, J. Microbiol. Methods, 2006, 67, 70–85 CrossRef CAS PubMed.
- L. Schena, J. M. Duncan and D. E. L. Cooke, Plant Pathol., 2008, 57, 64–75 CAS.
- S. Julich, M. Riedel, M. Kielpinski, M. Urban, R. Kretschmer, S. Wagner, W. Fritzsche, T. Henkel, R. Moeller and S. Werres, Biosens. Bioelectron., 2011, 26, 4070–4075 CrossRef CAS PubMed.
- L. Schwenkbier, S. Koenig, S. Wagner, S. Pollok, J. Weber, M. Hentschel, J. Popp, S. Werres and K. Weber, Microchim. Acta, 2014, 181, 1669–1679 CrossRef CAS.
- D. Cialla, K. Weber, R. Boehme, U. Huebner, H. Schneidewind, M. Zeisberger, R. Mattheis, R. Moeller and J. Popp, Beilstein J. Nanotechnol., 2011, 2, 501–508 CrossRef CAS PubMed.
- M. R. Hartman, R. C. H. Ruiz, S. Hamada, C. Xu, K. G. Yancey, Y. Yu, W. Han and D. Luo, Nanoscale, 2013, 5, 10141–10154 RSC.
- G. M. Santos, F. Zhao, J. Zeng, M. Li and W. C. Shih, J. Biophotonics, 2015, 9999 Search PubMed.
- D. Cialla, S. Pollok, C. Steinbruecker, K. Weber and J. Popp, Nanophotonics, 2014, 3, 383–411 CrossRef CAS.
- X. Guo, J. Biophotonics, 2012, 5, 483–501 CrossRef CAS PubMed.
- S. Mahajan, J. Richardson, T. Brown and P. N. Bartlett, J. Am. Chem. Soc., 2008, 130, 15589–15601 CrossRef CAS PubMed.
- T. Vo-Dinh, H.-N. Wang and J. Scaffidi, J. Biophotonics, 2010, 3, 89–102 CrossRef CAS PubMed.
- M. Baia, L. Baia, S. Astilean and J. Popp, Appl. Phys. Lett., 2006, 88 Search PubMed.
- S. Schluecker, Angew. Chem., Int. Ed., 2014, 53, 4756–4795 CrossRef CAS PubMed.
- E. C. Le Ru and P. G. Etchegoin, Annu. Rev. Phys. Chem., 2012, 63, 65–87 CrossRef CAS PubMed.
- M. Moskovits, J. Raman Spectrosc., 2005, 36, 485–496 CrossRef CAS.
- D. Cialla, U. Huebner, H. Schneidewind, R. Moeller and J. Popp, ChemPhysChem, 2008, 9, 758–762 CrossRef CAS PubMed.
- K. K. Hering, R. Moeller, W. Fritzsche and J. Popp, ChemPhysChem, 2008, 9, 867–872 CrossRef CAS PubMed.
- N. Pavillon, K. Bando, K. Fujita and N. I. Smith, J. Biophotonics, 2013, 6, 587–597 CrossRef CAS PubMed.
- Y. Lai, S. Sun, T. He, S. Schlucker and Y. Wang, RSC Adv., 2015, 5, 13762–13767 RSC.
- S. Niebling, H. Y. Kuchelmeister, C. Schmuck and S. Schluecker, Chem. Sci., 2012, 3, 3371–3377 RSC.
- K. K. Strelau, A. Brinker, C. Schnee, K. Weber, R. Moeller and J. Popp, J. Raman Spectrosc., 2011, 42, 243–250 CrossRef CAS.
- A. J. Driscoll, M. H. Harpster and P. A. Johnson, Phys. Chem. Chem. Phys., 2013, 15, 20415–20433 RSC.
- J. L. Abell, J. M. Garren, J. D. Driskell, R. A. Tripp and Y. Zhao, J. Am. Chem. Soc., 2012, 134, 12889–12892 CrossRef CAS PubMed.
- K. Faulds, W. E. Smith and D. Graham, Analyst, 2005, 130, 1125–1131 RSC.
- K. Gracie, E. Correa, S. Mabbott, J. A. Dougan, D. Graham, R. Goodacre and K. Faulds, Chem. Sci., 2014, 5, 1030–1040 RSC.
- D. Graham, B. J. Mallinder, D. Whitecombe, N. D. Watson and W. E. Smith, Anal. Chem., 2002, 74, 1069–1074 CrossRef CAS PubMed.
- M. Green, F. M. Liu, L. Cohen, P. Kollensperger and T. Cass, Faraday Discuss., 2006, 132, 269–280 RSC.
- Y. Lu, Q. Huang, G. Meng, L. Wu and J. Zhang, Analyst, 2014, 139, 3083–3087 RSC.
- N. E. Marotta, K. R. Beavers and L. A. Bottomley, Anal. Chem., 2013, 85, 1440–1446 CrossRef CAS PubMed.
- C. M. Muntean, N. Leopold, A. Halmagyi and S. Valimareanu, J. Raman Spectrosc., 2011, 42, 844–850 CrossRef CAS.
- H. T. Ngo, H.-N. Wang, A. M. Fales, B. P. Nicholson, C. W. Woods and T. Vo-Dinh, Analyst, 2014, 139, 5655–5659 RSC.
- E. Papadopoulou and S. E. J. Bell, Chem. Commun., 2011, 47, 10966–10968 RSC.
- D. van Lierop, I. A. Larmour, K. Faulds and D. Graham, Anal. Chem., 2013, 85, 1408–1414 CrossRef CAS PubMed.
- A. Barhoumi and N. J. Halas, J. Am. Chem. Soc., 2010, 132, 12792–12793 CrossRef CAS PubMed.
- A. Barhoumi, D. Zhang, F. Tam and N. J. Halas, J. Am. Chem. Soc., 2008, 130, 5523–5529 CrossRef CAS PubMed.
- C. Otto, F. F. M. Demul, A. Huizinga and J. Greve, J. Phys. Chem., 1988, 92, 1239–1244 CrossRef CAS.
- A. Dallmann, L. Dehmel, T. Peters, C. Muegge, C. Griesinger, J. Tuma and N. P. Ernsting, Angew. Chem., Int. Ed., 2010, 49, 5989–5992 CrossRef CAS PubMed.
- J. M. Jean and K. B. Hall, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 37–41 CrossRef CAS.
- L. C. Sowers, G. V. Fazakerley, R. Eritja, B. E. Kaplan and M. F. Goodman, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 5434–5438 CrossRef CAS.
- S. König, L. Schwenkbier, S. Pollok, M. Riedel, S. Wagner, J. Popp, K. Weber and S. Werres, Plant Pathol., 2015, 64, 1176–1189 CrossRef.
- H. Schneidewind, T. Schueler, K. K. Strelau, K. Weber, D. Cialla, M. Diegel, R. Mattheis, A. Berger, R. Moeller and J. Popp, Beilstein J. Nanotechnol., 2012, 3, 404–414 CrossRef PubMed.
- T. Schueler, A. Steinbrueck, G. Festag, R. Moeller and W. Fritzsche, J. Nanopart. Res., 2009, 11, 939–946 CrossRef CAS.
- T. D. Miles, F. N. Martin and M. D. Coffey, Phytopathology, 2015, 105, 265–278 CrossRef CAS PubMed.
- N. H. Jang, Bull. Korean Chem. Soc., 2002, 23, 1790–1800 CrossRef CAS.
- L. Schwenkbier, S. Pollok, S. Koenig, M. Urban, S. Werres, D. Cialla-May, K. Weber and J. Popp, Anal. Methods, 2015, 7, 211–217 RSC.
- J. A. Tomlinson, M. J. Dickinson and N. Boonham, Phytopathology, 2010, 100, 143–149 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5an01156f |
| ‡ Both authors contributed equally to the paper. |
| This journal is © The Royal Society of Chemistry 2015 |