 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The potential of chiroptical and vibrational spectroscopy of blood plasma for the discrimination between colon cancer patients and the control group†
Michal
Tatarkovič
*a,
Michaela
Miškovičová
b,
Lucie
Šťovíčková
a,
Alla
Synytsya
a,
Luboš
Petruželka
b and
Vladimír
Setnička
a
aDepartment of Analytical Chemistry, University of Chemistry and Technology, Prague, Technická 5, 166 28 Prague 6, Czech Republic. E-mail: michal.tatarkovic@vscht.cz; Fax: +420 220 444 352; Tel: +420 220 443 762
bDepartment of Oncology, First Faculty of Medicine, Charles University and General University Hospital, U Nemocnice 2, 128 00 Prague 2, Czech Republic
First published on 17th February 2015
Abstract
Colorectal cancer is one of the most abundant causes of cancer deaths in the world. At an early stage, the established clinical procedures have low reliability and sensitivity. Therefore, we tested a novel approach based on chiroptical methods such as electronic circular dichroism (ECD) and Raman optical activity (ROA). These methods are suitable for detecting slight changes in the 3D structure of chiral biomolecules, some of which may be caused by pathological processes occurring during cancer growth. Fifty-five blood plasma samples were analyzed using the combination of ECD and ROA supplemented by conventional Raman and FT-IR spectroscopy. All obtained spectra were evaluated together by linear discriminant analysis. The accuracy of sample discrimination reached 100% and the subsequent leave-one-out cross-validation resulted in 93% sensitivity and 81% specificity. The achieved results indicate that chiroptical methods supplemented by Raman and FT-IR spectroscopy might be new supporting and minimally invasive tools in the clinical diagnosis of colon cancer.
Introduction
In 2012, about 694![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths worldwide were caused by colorectal cancer.1 Unfortunately, the number of deaths is growing every year.2 The current diagnostic approach involves screening by Fecal Occult Blood Test (FOBT) followed by colonoscopy. However, FOBT has very low reliability and sensitivity and is able to detect only 25% of cancer cases.3 In spite of a higher level of accuracy colonoscopy is sometimes refused by patients due to its invasiveness and discomfort. In addition, the investigation by colonoscopy might cause colon perforation.3
000 deaths worldwide were caused by colorectal cancer.1 Unfortunately, the number of deaths is growing every year.2 The current diagnostic approach involves screening by Fecal Occult Blood Test (FOBT) followed by colonoscopy. However, FOBT has very low reliability and sensitivity and is able to detect only 25% of cancer cases.3 In spite of a higher level of accuracy colonoscopy is sometimes refused by patients due to its invasiveness and discomfort. In addition, the investigation by colonoscopy might cause colon perforation.3
To avoid the relatively inaccurate colonoscopy or histopathology, new diagnostic methods based on molecular spectroscopy analysis of cancer tissues or biofluids have been investigated.3–7
In comparison with tissues, few spectroscopic studies deal with the blood plasma or serum8–10 of colon cancer patients although they are more easily accessible and their spectroscopic analysis has the potential to serve as a standard screening tool in clinical laboratory practice, since it measures the chemical variations of the structure and composition at the molecular level and is fast, minimally invasive and reagent-free. Focusing on the variations of molecular structure of plasmatic biomolecules, we selected chiroptical spectroscopy that is among the few methods inherently sensitive to the 3D structure of chiral biomolecules.11–13 We presume that chiroptical methods have the potential for the detection of slight conformational and stereochemical changes in the structure of biomolecules caused by pathological processes occurring during several diseases, such as cancer or others.14 To the best of our knowledge, we are the first group to have measured real biofluids by chiroptical spectroscopy.15–18 Our previous studies showed that the human blood plasma can be successfully measured by both ECD and ROA spectroscopy.16,17 Therefore in this study, we used ECD and ROA supplemented by conventional Raman and FT-IR spectroscopy as tools for the investigation of blood plasma samples of colon cancer patients and healthy controls.
Materials and methods
Blood plasma samples
Fifty-five samples of blood plasma including 28 samples from patients with colon cancer were selected for this study. The samples involved all clinical stages from incipience stage I to stage IV with disseminated carcinoma (including metastases in other organs), comprising different degrees of grading from well differentiated (Grade 1) to poorly differentiated (Grade 3). The predominant types of samples were stages II and III, i.e. without metastases or metastases just in lymphatic nodes, and Grade 1 and 2 carcinoma. The patients were between 37 and 86 years old with an average of 65 and a median of 66 years. The diagnosis of all patients was verified by histopathology. The control group consisted of 27 samples without any form of cancer. The age of the control group was comparable to the patient group. The individuals from the control group were between 25 and 82 years old with an average of 52 and a median of 55 years.The whole blood of all individuals was collected by venipuncture and anticoagulant-treated with K3EDTA (tripotassium salt of ethylenediaminetetraacetic acid, BD Vacutainer Systems, Plymouth, UK). For plasma separation, the blood was centrifuged at 1500g and 25 °C for 10 minutes16,17 at the Department of Oncology, First Faculty of Medicine, Charles University and General University Hospital in Prague. The obtained plasma fraction was immediately frozen and stored at −75 °C. Before each analysis, the frozen plasma samples were thawed at room temperature and filtered using a centrifuge filter with a 0.45 μm PVDF membrane (Grace, Chicago, IL, USA) at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 15 °C for 10 minutes.17
000g and 15 °C for 10 minutes.17
All optical cells were cleaned prior to and after the spectral measurements using “Starna CellClean” solution (Starna Scientific Ltd, Essex, UK), repeatedly rinsed with demineralized water and methanol, and finally dried.
The study was carried out according to the principles expressed in the Declaration of Helsinki; the study was approved by the Ethics Committee of the First Faculty of Medicine and the General University Hospital, Prague. A written informed consent was secured from all subjects.
Electronic circular dichroism
The ECD spectra were recorded on a J-815 spectrometer equipped with a CDF-426S/16 Peltier unit (Jasco, Japan) to control the sample temperature. The filtered plasma was diluted four-times with phosphate buffer (pH = 7.4, NaCl 137 mM, KCl 2.7 mM, KH2PO4 1.5 mM, Na2HPO4 8 mM) and measured in a 0.01 mm Suprasil quartz cuvette (Hellma, Germany) in the 280–185 nm spectral range. The following parameters were used: 2 nm bandwidth, 50 nm min−1 scanning speed, 2 s data integration time, Peltier unit set to 23 °C. The spectra represent the average of 5 accumulations.Raman optical activity and Raman spectroscopy
Raman optical activity/Raman spectra were recorded simultaneously on the ChiralRaman-2X spectrometer (BioTools, Inc., USA) equipped with a 532 nm laser system Opus 2W/MPC6000 (Laser Quantum, UK), and a homemade Peltier cell holder to control the sample temperature. The resolution of the ROA spectrometer is about 7 cm−1. The 4 × 4 mm quartz cell (BioTools, Inc., USA) was tempered to 15 °C. The filtered plasma (100 μl) was treated with 10 mg of NaI for the suppression of undesirable fluorescence background by the procedure described in our previous work.17 The reduction of the fluorescence background was supported by “bleaching” in a laser beam (280 mW) for 12 hours. Then, the laser power was set to 250 mW and the illumination period was set according to the optimal working range of the CCD detector (from 1 s to 2.5 s depending on individual samples). The total exposure time for the ROA spectra was kept at 24 hours. The real laser power on the sample was monitored by using an Optical power meter 1916-R with an 818-P sensor (Newport, USA). A residual distorted baseline in the raw ROA/Raman spectra was corrected by a procedure described in the literature.17,19Infrared spectroscopy
A Nicolet 6700 (Thermo Scientific, USA) infrared spectrometer with Fourier transformation (FT-IR) was used to measure the sample spectra in the spectral range of 4000–600 cm−1. Because of a strong absorption of water, an attenuated total reflection (ATR) technique with a ZnSe crystal was used. The measurements were performed collecting 512 scans with a resolution of 4 cm−1 for each sample. Before each measurement, a new background was measured and the ATR crystal was cleaned following the same process as with the optical cells. The obtained spectra were corrected by subtracting the spectrum of water as a background and the spectrum of water vapor.Finally, the spectra were smoothened by using a Gaussian filter with a segment size of 5 and aligned by linear baseline correction in the Unscrambler X (Camo, Norway) software.
Data processing for multivariate analysis
The Raman spectra were processed by a detrending function (3rd order polynomial) in the Unscrambler X software (Camo, Norway). The detrended Raman spectra and the spectra from other methods (ECD, FT-IR, ROA) were processed using principal component analysis (PCA); and according to loadings, significantly contributing bands were selected. These bands were used for the discrimination of the control group and the group of patients by linear discriminant analysis (LDA). The further optimization of band selection was based on correlation coefficients, standardized canonical discriminant coefficients and overall success in cross-validation.Results and discussion
Spectral analysis
The average ECD spectra of patients with colon cancer and healthy controls are presented in Fig. 1. The bands at 209 and 222 nm belong to n–π* and that at 192 nm to π–π* transitions of the peptide bond in proteins.20 In both cases, the spectral pattern is typical for mainly α-helical proteins, due to ∼60% content of albumin (34–47 g L−1 of albumin per 70–75 g L−1 of total proteins) in the blood plasma of healthy persons.21 The most obvious changes between the control and patient groups are partly in the intensity of bands and partly in the relative ratio of the bands at 209 and 222 nm. The decreased intensity of these bands in the patient group cannot be explained only by the potential variation of pathlengths during the ECD analyses using a demountable 10 μm cuvette. We performed additional experiments measuring the ECD spectra of one sample, which was prepared independently three times (individual sample preparation, cuvette filling, measurement, etc.). The results clearly demonstrate that the variability in ECD intensities is almost negligible within one sample (see Fig. S1 in ESI†). However, the decreased intensities of these bands in the patient spectra can be at least partly explained by the biochemical data of the samples obtained from Department of Oncology, First Faculty of Medicine, Charles University and General University Hospital in Prague. The average content of albumin for patients was 36.7 ± 8.4 g L−1 and total protein was 60.7 ± 8.5 g L−1, and for controls 48.8 ± 3.1 g L−1 and 73.4 ± 6.4 g L−1, respectively. The ratios of albumin to total protein in the blood plasma for patients and controls were 0.59 and 0.66, respectively. However, the differences in the ECD (as well as ROA) intensities are much larger, probably not only due to the difference in the protein concentration, but also due to a higher content of less ordered structures (probably random coil) in the patient group.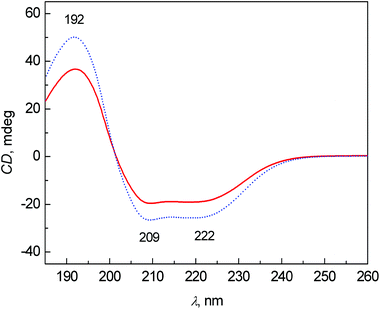 | ||
| Fig. 1 Average ECD spectra of human blood plasma samples: control group (dotted line) and colon cancer patients (solid line). | ||
Fig. 2 shows the average Raman spectra for patients and healthy controls. The most intense Raman bands at 1006, 1156 and 1517 cm−1 can be assigned to carotenoids.7,15,22–25 Although their concentration in blood plasma is low,26,27 the observed high intensities are caused by the resonance enhancement due to laser excitation at 532 nm.17,22,28 In comparison with the control group, a significant intensity decrease of carotenoid bands was observed for the patient group. This observation is in agreement with the study of cancer tissues by Raman spectroscopy.23,24,29 Other important bands at 1657 cm−1 (amide I), 1270 and 1285 cm−1 (amide III) are related to proteins and vibrations of their peptide bonds.11 In the amide III region, other significant changes between the control group and patients can be observed. The bands at 1270 and 1285 cm−1 were much more intense for the control group than for patients. In addition, the band at 1285 cm−1 was more intense in the control group than the band at 1270 cm−1, whereas the intensity was comparable in the patient group. Other bands associated with the proteins, 879 cm−1 and partly 1450 cm−1 (due to aliphatic side chains), overlap with phospholipids.15 Amino acids phenylalanine and tryptophan show specific bands at 1588, 1196 cm−1 and also a band at 1006 cm−1 (phenylalanine, overlapped with carotenoids). Lastly, the spectra show a band of carbohydrates at 960 cm−1. A more detailed interpretation of the particular spectral features in Raman/ROA was published previously in our work.15 Other differences between the patients and controls in the Raman spectra were especially visible after processing by multivariate analysis.
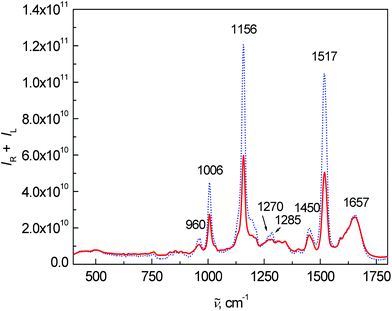 | ||
| Fig. 2 Average Raman spectra of human blood plasma samples: control group (dotted line) and colon cancer patients (solid line). | ||
Fig. 3 shows the average ROA spectra of samples from the control and patient groups. The overall spectral pattern is typical for predominant α-helical proteins represented by a negative–positive couplet at (−)1645 and (+)1674 cm−1 (amide I).11,30,31 The positive bands at 1295, 1311 and 1345 cm−1 (extended amide III region)11,31 are especially sensitive to the secondary structure of proteins.11 Changes in the intensity and ratio of these bands can be also observed in this case. The positive bands between 870 and 960 cm−1 confirmed the observation from ECD – the overall spectral pattern of blood plasma corresponds to mainly α-helical proteins and their content is lower in the patient group. A band at 1248 cm−1 and its spectral pattern relate mostly to a higher contribution of particular β-structure conformations, the detailed structure of this band can be found in the literature.11,32
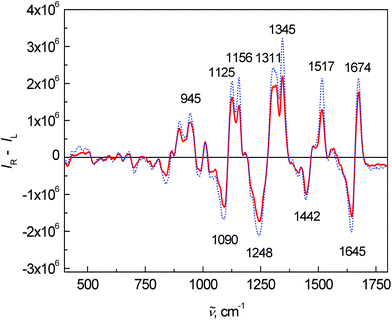 | ||
| Fig. 3 Average ROA spectra of human blood plasma samples: control group (dotted line) and colon cancer patients (solid line). | ||
Lastly, the average FT-IR spectra are presented in Fig. 4. Two dominant bands at 1649 and 1547 cm−1 belong to the vibrations of amide I and amide II, respectively. The most significant changes in the FT-IR spectra were observed in the intensity and ratio between the amide I and amide II bands. The band at 1448 cm−1 arose from the bending vibrations of CH3 and CH2 groups in the side chains of proteins mixed with phospholipids. The 1260–1390 cm−1 spectral region coheres with the symmetric stretching vibration of protein carboxyl groups.2 However in this fingerprint area, many bands overlap and a deeper interpretation is almost impossible with such a complex matrix as blood plasma.
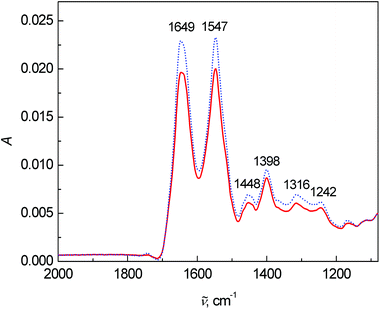 | ||
| Fig. 4 Average FT-IR spectra of human blood plasma samples: control group (dotted line) and colon cancer patients (solid line). | ||
For Raman, ROA and FT-IR spectroscopy, we also performed the same experiment as for ECD, i.e. three independent measurements of one sample (Fig. S2–S4 in ESI†). Raman and FT-IR spectroscopy showed small differences between repeated measurements. In the case of ROA, we observed relatively large changes in comparison with other used spectroscopies. However, the changes are within the naturally higher noise level of ROA spectra.
Statistical analysis
The spectra of each spectral method were processed by PCA to obtain significant bands for further evaluation by LDA (for the results of PCA see Fig. S5–S8 in ESI†). The LDA results for individual spectral methods (Fig. 5) are presented in squared Mahalanobis distance.33 Clearly, the control and patient groups were partly discriminated in each spectral method, but both groups overlapped. The overall classification ability of particular models reached 81–87%. In addition, each spectral model was also processed by leave-one-out cross-validation (LOOCV), see Table 1.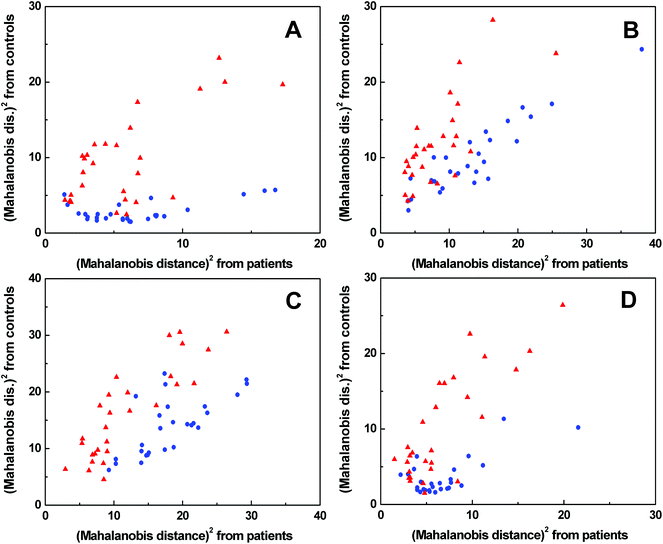 | ||
Fig. 5 Graphical results of LDA for individual spectral methods, ECD (A), Raman (B), ROA (C), FT-IR (D);  control group, control group,  patient group. patient group. | ||
| ECD spectroscopy | Raman spectroscopy | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| From/to | Cancer | Control | Total | %Correct | From/to | Cancer | Control | Total | %Correct |
| Cancer | 22 | 6 | 28 | 79% | Cancer | 20 | 8 | 28 | 71% |
| Control | 3 | 24 | 27 | 89% | Control | 7 | 20 | 27 | 74% |
| Total | 25 | 30 | 55 | 84% | Total | 27 | 28 | 55 | 73% |
| ROA spectroscopy | FT-IR spectroscopy | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| From/to | Cancer | Control | Total | %Correct | From/to | Cancer | Control | Total | %Correct |
| Cancer | 20 | 8 | 28 | 71% | Cancer | 21 | 7 | 28 | 75% |
| Control | 5 | 22 | 27 | 81% | Control | 4 | 23 | 27 | 85% |
| Total | 25 | 30 | 55 | 76% | Total | 25 | 30 | 55 | 80% |
According to LOOCV, ECD spectroscopy exhibited the highest sensitivity (79%) and specificity (89%), whereas the lowest sensitivity was observed in both ROA and Raman spectroscopy (71%). However, the lowest specificity (74%) was obtained from Raman spectroscopy, while in the case of ROA it reached 81%. The statistical evaluation of ECD data yielded the highest overall accuracy (84%) after LOOCV, whereas the lowest level (73%) was reached for Raman spectroscopy. Usually, the misclassified samples differed for each spectroscopic method. Each spectroscopy is sensitive to different properties of the studied biomolecules/samples. While ECD is sensitive to electronic transitions of chromophores and their chirality, ROA is based on the change of the polarizability of functional groups and their chirality. Raman spectroscopy deals with the change of the polarizability of functional groups and FT-IR with the change of the dipole moment of functional groups. Therefore, the obtained results were significantly improved by evaluating all spectral methods together, for compensating the inadequacies in the classification of certain samples by individual spectroscopic methods. After this combination, both the investigated groups were well separated (Fig. 6). The discrimination ability in this case achieved 100% overall accuracy. The quality of the combined statistical model was confirmed by LOOCV, where sensitivity and specificity reached 93% and 81%, respectively. Only 5 samples from the control group and 2 samples from the patient group were misclassified (Table 2). The overall accuracy for LOOCV was 87%.
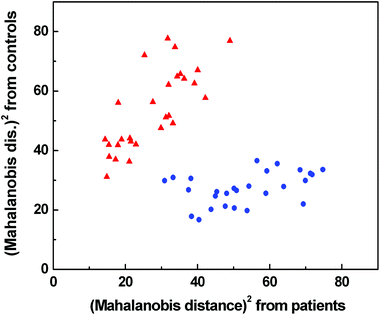 | ||
Fig. 6 Graphical result of LDA for the combination of Raman, IR, ROA and ECD spectroscopic data;  control group, control group,  patients group. patients group. | ||
| From/to | Cancer | Control | Total | %Correct |
|---|---|---|---|---|
| Cancer | 26 | 2 | 28 | 93% |
| Control | 5 | 22 | 27 | 81% |
| Total | 31 | 24 | 55 | 87% |
For LDA, we combined 27 bands: 3 bands from ECD (192, 209 and 222 nm), 8 bands from Raman (1270, 1285, 1341, 1357, 1391, 1450, 1517 and 1586 cm−1), 12 bands from ROA (833, 956, 1264, 1295, 1301, 1311, 1345, 1442, 1604, 1645, 1665 and 1674 cm−1) and 4 bands from FT-IR (1244, 1400, 1547 and 1639 cm−1). Most of these bands belong to proteins or other compounds (e.g. carotenoids, phospholipids) described previously in this article.
Conclusion
In this paper, we showed that chiroptical spectroscopy can be used in the combination with Raman and FT-IR spectroscopy for the discrimination of the control group from patients with colon cancer. The detailed results of LDA showed that each particular spectral method (ECD, ROA, Raman, FT-IR) has limited ability to discriminate the control group from patients, but the best results were achieved from their combination. In this case, the discrimination ability achieved 100%. The mathematical model was sequentially confirmed by leave-one-out cross-validation, where the sensitivity and specificity reached 93% and 81%, respectively. The overall accuracy for the leave-one-out cross-validation was 87%. The achieved results indicate that the chiroptical spectroscopy has the potential to become a new supporting method in the clinical diagnosis of colon cancer based on the analysis of blood plasma, which is minimally invasive for patients. Our future work will focus on the sample set extension, data validation and testing the interference with other diseases.Acknowledgements
The work was financially supported by the Ministry of Health of the Czech Republic (project no. NT13259-3).References
- B. W. Stewart and C. Wild, World Cancer Report 2014, World Health Organization, 2014 Search PubMed.
- J. A. A. C. Piva, J. L. R. Silva, L. Raniero, A. A. Martin, H. G. Bohr and K. J. Jalkanen, Theor. Chem. Acc., 2011, 130, 1261–1273 CrossRef CAS PubMed.
- G. Ekelund, J. Manjer and S. Zackrisson, Int. J. Colorectal Dis., 2010, 25, 1269–1275 CrossRef PubMed.
- L. Dong, X. Sun, Z. Chao, S. Zhang, J. Zheng, R. Gurung, J. Du, J. Shi, Y. Xu, Y. Zhang and J. Wu, Spectrochim. Acta, Part A, 2014, 122, 288–294 CrossRef CAS PubMed.
- L. Mavarani, D. Petersen, S. F. El-Mashtoly, A. Mosig, A. Tannapfel, C. Kotting and K. Gerwert, Analyst, 2013, 138, 4035–4039 RSC.
- C. Krafft, B. Dietzek and J. Popp, Analyst, 2009, 134, 1046–1057 RSC.
- N. Stone, C. Kendall, J. Smith, P. Crow and H. Barr, Faraday Discuss., 2004, 126, 141–157 RSC.
- D. Lin, S. Feng, J. Pan, Y. Chen, J. Lin, G. Chen, S. Xie, H. Zeng and R. Chen, Opt. Express, 2011, 19, 13565–13577 CrossRef CAS PubMed.
- M. Tatarkovič, S. Kykal, A. Synytsya, M. Miškovičová, L. Petruželka and V. Setnička, Eur. Biophys. J., 2013, 42, S107–S107 Search PubMed.
- D. Lin, J. Pan, H. Huang, G. Chen, S. Qiu, H. Shi, W. Chen, Y. Yu, S. Feng and R. Chen, Sci. Rep., 2014, 4 DOI:10.1038/srep04751.
- L. D. Barron, L. Hecht, E. W. Blanch and A. F. Bell, Prog. Biophys. Mol. Biol., 2000, 73, 1–49 CrossRef CAS.
- N. Berova, P. L. Polavarapu, K. Nakanishi and R. W. Woody, Comprehensive Chiroptical Spectroscopy, Wiley, New Jersey, 2012 Search PubMed.
- V. Parchaňský, J. Kapitán and P. Bouř, RSC Adv., 2014, 4, 57125–57136 RSC.
- D. P. Minde, Z. Anvarian, S. G. Rudiger and M. M. Maurice, Mol. Cancer, 2011, 10, 101 CrossRef CAS PubMed.
- A. Synytsya, M. Judexová, T. Hrubý, M. Tatarkovič, M. Miškovičová, L. Petruželka and V. Setnička, Anal. Bioanal. Chem., 2013, 405, 5441–5453 CrossRef CAS PubMed.
- M. Tatarkovič, Z. Fišar, J. Raboch, R. Jirák and V. Setnička, Chirality, 2012, 24, 951–955 CrossRef PubMed.
- M. Tatarkovič, A. Synytsya, L. Šťovíčková, B. Bunganič, M. Miškovičová, L. Petruželka and V. Setnička, Anal. Bioanal. Chem., 2015, 407, 1335–1342 CrossRef PubMed.
- L. Šťovíčková, M. Tatarkovič, H. Logerová, J. Vavřinec and V. Setnička, Analyst, 2015 10.1039/C4AN01874E.
- M. Člupek, P. Matějka and K. Volka, J. Raman Spectrosc., 2007, 38, 1174–1179 CrossRef.
- S. M. Kelly, T. J. Jess and N. C. Price, Biochim. Biophys. Acta, Proteins Proteomics, 2005, 1751, 119–139 CrossRef CAS PubMed.
- R. K. Murray, D. K. Granner, P. A. Mayes and V. W. Rodwell, Harper's Illustrated Biochemistry, McGraw-Hill, New York, 26th edn, 2003 Search PubMed.
- M. E. Darvin, I. Gersonde, H. Albrecht, W. Sterry and J. Lademann, Laser Phys. Lett., 2007, 4, 452–456 CrossRef CAS.
- T. R. Hata, T. A. Scholz, I. V. Ermakov, R. W. McClane, F. Khachik, W. Gellermann and L. K. Pershing, J. Invest. Dermatol., 2000, 115, 441–448 CrossRef CAS PubMed.
- D. C. B. Redd, Z. C. Feng, K. T. Yue and T. S. Gansler, Appl. Spectrosc., 1993, 47, 787–791 CrossRef CAS.
- F. Schulte, J. Mäder, L. W. Kroh, U. Panne and J. Kneipp, Anal. Chem., 2009, 81, 8426–8433 CrossRef CAS PubMed.
- N. J. Ollberding, G. Maskarinec, S. M. Conroy, Y. Morimoto, A. A. Franke, R. V. Cooney, L. R. Wilkens, L. Le Marchand, M. T. Goodman, B. Y. Hernandez, B. E. Henderson and L. N. Kolonel, Blood, 2012, 119, 5817–5823 CrossRef CAS PubMed.
- R. S. Parker, J. Nutr., 1989, 119, 101–104 CAS.
- L. Feltl, V. Pacáková, K. Štulík and K. Volka, Curr. Anal. Chem., 2005, 1, 93–102 CrossRef.
- A. Synytsya, M. Judexova, D. Hoskovec, M. Miskovicova and L. Petruzelka, J. Raman Spectrosc., 2014, 45, 903–911 CrossRef CAS.
- M. Urbanová, Chirality, 2009, 21, E215–E230 CrossRef PubMed.
- M. N. Kinalwa, E. W. Blanch and A. J. Doig, Anal. Chem., 2010, 82, 6347–6349 CrossRef CAS PubMed.
- T. Weymuth and M. Reiher, J. Phys. Chem. B, 2013, 117, 11943–11953 CrossRef CAS PubMed.
- R. De Maesschalck, D. Jouan-Rimbaud and D. L. Massart, Chemom. Intell. Lab. Syst., 2000, 50, 1–18 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4an01880j |
| This journal is © The Royal Society of Chemistry 2015 |
