Droplet microfluidics in (bio)chemical analysis
Evgenia Yu
Basova
*a and
Frantisek
Foret
*ab
aMasaryk University, CEITEC, Central European Institute Technology, 60200 Veveri 97, Brno, Czech Republic. E-mail: evgenia-basova@rambler.ru; foret@iach.cz
bInstitute of Analytical Chemistry of the Academy of Sciences of the Czech Republic, v. v. i., 60200 Veveri 97, Brno, Czech Republic
First published on 11th September 2014
Abstract
Droplet microfluidics may soon change the paradigm of performing chemical analyses and related instrumentation. It can improve not only the analysis scale, possibility for sensitivity improvement, and reduced consumption of chemical and biological reagents, but also the speed of performing a variety of unit operations. At present, microfluidic platforms can reproducibly generate monodisperse droplet populations at kHz or higher rates with droplet sizes suitable for high-throughput experiments, single-cell detection or even single molecule analysis. In addition to being used as microreactors with volume in the micro- to femtoliter range, droplet based systems have also been used to directly synthesize particles and encapsulate biological entities for biomedicine and biotechnology applications. This minireview summarizes various droplet microfluidics operations and applications for (bio)chemical assays described in the literature during the past few years.
Introduction
The past decade has seen increased interest in chemical reactions and the development of tools for fluid flow control at the microscale. This multidisciplinary field involves fundamental concepts from different areas including physics and electrical engineering to biology. Emulsions are also heavily used in personal care products, foods, and topical drug delivery; however, these applications do not require uniformity of droplet sizes. More demanding applications are now exponentially increasing and include, e.g., microreactors for catalysis and chemical synthesis, point-of-care diagnostics, drug delivery, cell/molecule compartmentalization and diagnostic testing.1–4 Most of the common laboratory unit operations can be performed quickly using minimum volumes of reagents and samples (volume reduction from milliliters to femtoliters). In addition short diffusion distances and higher surface to volume ratios can often reduce reaction times from hours to seconds or less.While the origins of the droplet based microfluidics can be formally traced back to segmented flow analysis and flow injection analysis concepts developed over 40 years ago, the current technology utilizing droplets in the micrometer to nanometer diameter range relates more to the development of microfabrication techniques, micro-total-analysis and the lab-on-chip concepts.5–7 Droplet-based microfluidics is one subcategory of microfluidics, where drops serve as transport and reaction vessels. The principle of droplet formation is based on using two immiscible liquids where the dispersed (droplet forming) phase is injected into the stream of a continuous phase, surrounding the droplets. Fast reaction times in such small compartments are induced by the high surface area to volume ratios, efficient heat and mass transfer and short diffusion distances. Most of the references deal with free floating droplets, which, unlike liquid plugs confined by the channel geometry, require using stabilizing surfactants.
In this review, we summarize recent advances in droplet formation and manipulation technology and chemical analysis applications with the stress on new developments in the past 4 years. For more comprehensive review of the field prior to 2010 we recommend the readers to refer to the excellent article by Huck et al.8
Principles of droplet formation
Emulsions (suspensions of droplets created using two immiscible fluids9) are widely used in a number of industrial areas including machining, civil engineering and mining. In turn, the formation of stable droplets is the key component of droplet microfluidics. Regardless whether their final use is in microreactors or in particle/cell-based applications10–13 the droplets must meet the desired properties, including the size, shape and monodispersity of droplets14 and resistance to coalescence during the use and storage.15–17 Droplets should also provide a biocompatible environment for experiments with living cells or higher organisms.18 The use of surfactants is essential for droplet formation and stability of the emulsion – resistance to coalescence.19 Their choice depends on the nature of the continuous phase influencing biocompatibility, viscosity defining the droplet formation through shear stress and exchange of gases.20 The most common continuous phases include hydrocarbon and fluorocarbon oils. In order to stabilize hydrocarbon oils (mineral oil, decanol, hexadecane, etc.), commercially available surfactants such as Abil EM (cetyl dimethicone copolyol) and Span 80 (sorbitane mono-oleate) are used.21–24 Perfluorinated hydrocarbons (perfluorodecalin, perfluorotributylamine FC-43, FC-70, etc.) are often preferred over mineral oils, due to their immiscibility with organic and aqueous solvents, their high oxygen transport capability, physical and chemical stability. Fluorocarbon oils are typically denser than the dispersed phase.25 Droplet emulsions in the fluorinated continuous phase can be stabilized by perfluoropolyether (PEPEs) based surfactants such as Krytox, (a commercial lubricant from DuPont), consisting of a PEPE tail and a carboxylic acid head group.26 This compound has been characterized in detail with regard to biocompatibility and long-term storage.26 In recent publications, several surfactants featuring polyethylene glycol (PEG), carboxy-PEPE, poly-L-lysine (pLL) or dimorpholino phosphate (DMP) head groups have also been used.26 Detailed information regarding the components of some of the described emulsions is listed in Table 1.| Type of emulsion | Continuous phase | Surfactants | Geometry |
|---|---|---|---|
| Fluorinated oils | |||
| W/O | 1H,1H,2H,2H-Perfluorodecyltrichlorosilane, Fluorinert FC 3283; FC 40 | PEPE–PEG; PFPE–PEG–PFPE triblock copolymers; PFPE–PEG–gold diblock surfactants (home-made); EA surfactant | T-junction; flow-focusing27–30 |
| Fluorinated HFE-7500 | EA surfactant; Krytox–Jeffamine–Krytox A–B–A triblock copolymer | Flow focusing31,32 | |
| Fluorinert FC 40 | PFPE–PEG block copolymer | Flow-focusing33 | |
| Fluorinert FC 70, FC 40 | PEPE–PEG | T-junction; flow-focusing25 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Hydrocarbon oils | |||
| W/O | Mineral | Span 80; Tween 80 | Main channel to the flow rate in the external access channel34 |
| Hexadecane | Span 80 | Single T, K-junction35,36 | |
| O/W | Soybean; hexadecane, silicon | PEG, sodium dodecyl sulphate (SDS) | Flow-focusing37,38 |
| Mineral; silicon | Pluronic F-68 | T-junction39 | |
| n-Hexadecane | SDS | T-junction40 | |
| G/L | Nitrogen | Dispersed phase – deionized water (90% (v/v) glycerol; 2 wt% SDS) | Flow-focusing41 |
| L/G | Propane, butane, air | 2 wt% Tween-20 | T-junction42 |
Droplet formation – microfluidic dynamics
Parameters describing microfluidic systems can be described by the balance of inertial force, viscous drag forces, buoyant forces and interfacial tension forces. There are a few of them useful for description of droplet formation in a microfluidic device – the Reynolds number (Re), Weber number (We), Bond number (Bo), capillary number (Ca) and flow rate ratio. Each one describes a particular aspect of fluid flow within a device. They can be used together to predict if a particular microfluidic system will allow droplet formation to occur and if the device can be used for a specific application.43Reynolds number
Re is a dimensionless number that characterizes the flow of fluid through a channel. It is the ratio between inertial forces and viscous forces which quantifies the relative importance of these two under given flow conditions. For flow in a pipe or around a spherical particle Re = ρVL/μ, where ρ is the fluid density, V is the mean velocity, L is the characteristic length and μ is the dynamic viscosity. Re can describe different flow regimes within similar fluids such as laminar and turbulent flows. Generally, in microfluidic systems operating with liquids, a value of Re ≪ 2300, indicates a laminar flow under practically all experimental conditions. A turbulent flow at Re > 2300 may be observed in microfluidic systems operating with gases.11Weber number
The Weber number describes the ratio of inertial forces to surface tension We = ρV2L/γ, where ρ is the fluid density, V is the characteristic velocity of the flow, R is the characteristic size (e.g., the droplet diameter) and γ is the surface tension. If We is larger than the critical value, Wecrit = 1.1, the droplets are strongly distorted by the inertia forces and breakup may occur.44 In the case of a very low We, inertia is negligible; for a moderate We, inertia deforms the droplet without disrupting it.Bond number
The Bond number determines the relationship between gravitational surface forces and surface tension forces Bo = ΔρgL2/γ, where Δρ is the difference in fluid densities (kg m−3), g is the gravitational acceleration (m s−2), L is the characteristic length scale, and γ is the surface tension between two fluids (N m−1). For Bo < 1, the gravity effects can be ignored.45Capillary number
One of the most important parameters for droplet formation is the capillary number used to compare the viscous stress with respect to interfacial tension phenomena at the liquid–liquid interfaces.46 The capillary number is defined as Ca = μV/γ, where μ is the fluid viscosity, V is the velocity, and γ is the surface tension. At low values of Ca (<1) it appears that the surface tension forces are dominant over the viscous force and droplets are spherical. At high values of Ca (≫1) the viscous force plays an important role, leading to deformation of the droplets by the flow, characterized by asymmetric shapes.47In summary, Re describes the relative importance of inertial forces to viscous forces; Ca represents the effect of viscous forces to interfacial tension forces and the We number compares the inertial effects to surface tension. Due to the fact that microfluidic systems are scaled in micrometer length, Re is less than 1. Thus the Ca and We values play a more important role and interfacial effects become dominant. It should be noted that the geometry of microfluidic channels often play a key role in droplet formation. A number of microfluidic configurations will be introduced in the next section.
Droplet generation
There are many methods for making emulsions; however, most have been developed for large batches in industrial use, leading to polydispersity. A variety of methods for droplet formation have been investigated and can be categorized into two groups, i.e., passive and active approaches. In both cases, the key parameters are the geometry of the microchannel, the flow rate ratio, and the capillary number to achieve droplet formation.48 Chen et al.,49 define several methods for droplet formation including:• hydrodynamics (T, Y-junctions, flow focusing, co-flowing)
• pneumatic pressure (the gas pressure is used as the shear force and diving force to form droplets)
• optically driven (the principle using optical force to manipulate particles in microfluidic chips was successfully applied for droplet generation)
• electrically driven techniques: dielectrophoresis (DEP) and (EWOD) electrowetting on dielectric.49,50
In this section, we will focus on the most important microfluidic configuration for droplet generation including T-junctions, flow focusing and co-flowing (hydrodynamic method).
T-junction
The most common microfluidic geometry used for the generation of droplets is the T-junction. In this configuration, the inlet channel, containing the dispersed phase, intersects the main channel, containing the continuous phase. The channels are typically arranged orthogonally and droplets form at the channel intersection. This geometry can also be used for the formation of bubbles and/or double emulsions. Depending on the capillary number three main regimes in the T-junction microfluidics can be distinguished – dripping, jetting, and squeezing. At high Ca, small droplets are formed, provided that the large viscous drag of the continuous phase shears the dispersed phase from the inlet of the channel. At low Ca, droplets larger than the microchannel could be obtained by blocking the microchannel downstream, as shown in Fig. 1A.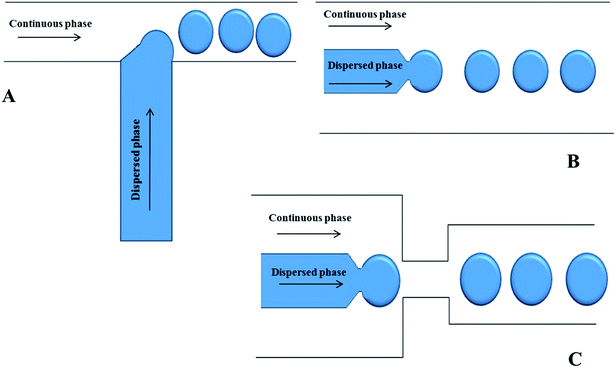 | ||
| Fig. 1 Schematics of microfluidic devices for making emulsions using (A) standard T-junction flow focusing junction; (B) co-flowing junction; (C) flow focusing junction. | ||
Co-flowing devices
There are two basic types of co-flow systems. Dripping and jetting are highly dependent on the flow rates of liquids. At low flow rates, dripping is the predominant regime and is the product of absolute instability as the forces acting on the stream of water occur at a specific frequency and location in the stream. At higher flow rates, jetting is predominant and is the result of convective instabilities. If monodisperse droplet formation is required, a system affected by absolute instabilities would be desired. In a microfluidic setup, this is achieved through careful control of the dynamics affecting the droplet formation. Droplet formation in co-flow systems highly depends on fluid velocities, viscosities, surface tension and the densities of the liquids. The geometry of the droplet formation region in the microfluidic device is important as well. Co-flow in microfluidics occurs as the dispersed phase is forced through a small capillary centered inside a larger diameter channel with the continuous phase flowing parallel to the dispersed phase as shown in Fig. 1B. As the continuous phase surrounds the dispersed phase, the viscous shear force becomes stronger as the diameter of the dispersed phase increases. Eventually, the drag and interfacial tension forces become equal at the tip of the capillary and a droplet is formed and pinched off from the dispersed phase. By establishing an absolute instability in the system and optimizing all the variables, droplets can be formed at uniform frequencies and sizes.Flow focusing devices
In this droplet formation arrangement, symmetric channels, supplying the continuous phase, surround the channel delivering the dispersed phase. As the dispersed phase flows into the open channel, the continuous phase forces the dispersed phase through a small orifice leading into a larger channel filled with the continuous phase. Droplets are formed as both phases are forced through the orifice. The continuous phase provides pressure to drive droplets through as well as the shear forces to separate the phases into different droplets. In flow focusing techniques, droplet size, velocity, and frequency can be tuned through the flow rates, controlling the inlet pressures, varying the phase viscosities, and the orifice size Fig. 1C.Droplet manipulation
Division of droplets
Droplet splitting or division into two or more “daughter droplets” is an important operation required for droplet microfluidic manipulations. The operation principle depends on the chip geometry and fluids. It can be achieved using a T-shaped channel when a droplet flows to the bifurcation section where it is stretched by the extension flow.51 If the droplet is large, it will be thinned and split into two or more droplets as it passes through the channels. It was demonstrated that asymmetric splitting can be achieved by changing the length of the branch channels. Fabricating a microfluidic tree network consisting of four Y-junctions Zhao et al. described the flow focusing junction approach for droplet splitting.47 The droplets were divided along their length in the flow and the number/size of the daughter droplets could be regulated by changing the flow rate of the splitting liquid. Other studies have shown that microfluidic devices allow nanoscale volume sampling from oil-segmented fluid plugs by passive splitting. The system allowed thousands of plugs to be asymmetrically split with no performance deterioration over time. The resulting plugs might be collected into separate channels for storage or other manipulation. In another approach fluid plug splitting is based on fluidic resistance.52 Kennedy et al. have developed a microfluidic device in which nanoliter oil segmented fluid plugs were pumped into a loop and asymmetrically split according to the flow resistance. The device with microfabricated posts inside the recombination region downstream of the split kept the stream of drops from coming into contact with each other and coalescence. It allowed to sample from plugs by asymmetrical splitting and collect the split aliquots for further use.53Sorting
Droplet sorting is important for selection of specific droplets from a larger population for the next step of analysis. There are many ways of influencing the movement of droplets in either a serial or parallel fashion. The forces necessary for influencing the droplet movement are most often based on fluid flow, acoustic waves or coulombic attraction/repulsion. Dielectrophoresis is an example of an electrically driven technique, which can be operated in both serial and parallel modes. The process is based on a force exerted on a particle with different polarizability relative to the surrounding medium in an electric field gradient. Dielectrophoresis is a rapidly growing field as recently documented by special issues of scientific journals.54,55 An example of a microfluidic device for high-speed sorting of water droplets using DEP has been described using microelectrodes prepared underneath polydimethylsiloxane (PDMS) channels of a microfluidic device. A sorting rate of over 1.6 kHz and a force of more than 10 nN has been reported.56 A similar approach has also been described for manipulation of water droplets using a microelectrode array and AC voltages across the electrodes with 25 V magnitude at frequencies ranging from 50 Hz to 1 MHz.57 In most cases the electrodes are prepared in the form of thin metal layers with a predefined geometry. In an alternative approach a uniform photosensitive semiconductor layer has been suggested for formation of “virtual” electrodes. In this case the photosensitive layer becomes electrically conductive only upon illumination. Thus the electrode geometry can be easily changed by projecting different light patterns on the semiconducting layer.58 Recently, the optically induced dielectrophoresis (ODEP) platform was investigated by Hung et al. using a new voltage transformation ratio simplifying the control of the photoconductive effect and the resulting ODEP force.59 The results for droplet manipulation showed that the numerical simulation conforms to the experimental data with a coefficient of variation of less than 6.2% (n = 3).An important application of the rapid serial sorting is the cell characterization/separation in flow cytometry (fluorescence-activated cell sorting, FACS). Ultrasensitive fluorescence monitoring is used for each cell and the speed of analysis is limited by mainly the serial detection process. While the speed of analysis can be increased by increasing the flow rate, at excessive flow velocities, the shear forces can damage or kill cells.60 Related to this limitation, Bai et al. proposed using the FACS microfluidic platform for droplet generation and protective cell encapsulation.61 Wu et al. presented integrated microfluidic droplet generation with fluorescence-activated droplet sorting (FADS) for single cell encapsulation, using hydrodynamically gated injection using a stopped flow technique.62–64 This method allowed encapsulation of cyan fluorescence protein transfected HeLa cells, with the encapsulation and recovery rates of 94.1 and 85%, respectively.
The application of acoustic forces (surface acoustic waves – SAW) is mostly related to the continuous flow particle sorting.65,66 Recently Li et al. demonstrated sorting of water in oil droplets into multiple outputs using standing surface acoustic waves (SSAW). The device integrated the microfluidic multichannel system with interdigital transducers (IDTs) to achieve droplet generation and sorting. The position of the pressure nodes could be changed by regulating the frequency of the SSAW excitation power with droplets sorted to multiple outlets at rates up to 222 droplets per s.67
Fusion
Chemical and biological analyses using reactions in droplets can be used for a number of applications, including chemical synthesis, microfractionation in droplets, monitoring of reaction kinetics, formation of particles, etc.68,69 Controlling coalescence (fusion) of droplets containing samples or reagents is a critical step for such applications. In practice, it is necessary to bring the droplets into a physical contact and overcome the stabilizing effects of surfactants.11 It should be noted that fusion of droplets depends on the viscosity ratio of the internal and external fluids as well as the presence of surfactants at the interface. There are several passive and active methods for controlling droplet fusion. Passive methods provide transfer of droplets into close proximity by the microchannel geometry to slow down the downstream drop until the upstream drop reaches it. Lower viscosities of droplets enable quick draining of the film between the immiscible fluids that facilitate coalescence of the droplets. Conversely, higher viscosity leads to reduction of the mobility of the interface and reduced chance for coalescence of droplets.70Detection methods for droplet microfluidics
Fluorescence
Fluorescence measurements using fluorescence microscopy with a camera to image the analyzed droplets belong to the most common detection modes. Marcoux et al. developed a microfluidic device for measurement of enzyme activities of single-cells.20 The technique is based on stochastic confinement of cells into picolitre sized microreactor droplets. Fluorogenic reporter molecule (4-methylumbelliferyl-β-D-glucuronide) served for monitoring of the enzyme activity during the time-lapse fluorescence imaging of droplets. Moreover, this approach leads to fast accumulation of fluorescent tracer and reduction of analysis time. Enzyme activity detection was also studied by Juul et al.71 They fabricated and integrated rolling-circle-enhanced-enzyme-activity for single-molecule-detection with a custom-designed microfluidic device allowing multiplexed detection of enzymatic events at aberrant cells in a population. Gu et al. developed a multifunctional droplet-based microfluidic platform named DropLab.72 The present device provides complex picolitre-scale droplet manipulation and single-cell enzyme activity detection using a fluorescent microscope. One of the simple microfluidic systems demonstrated laminar, diffusive mixing flow combined with parallel droplet formation in a T-shaped channel.73,74 Enzyme reactions have also been studied by Siuti et al. in a nanoporous container (19 pL) allowing to carry out reactions under continuous flow.75 It has been shown that a 10 nm pore size allows capturing of proteins and diffusive exchange of small molecules. Ismagilov et al. described a SlipChip design based on multiplexed reactions for screening of different reagents in nanoliter volumes.76 Quantitative measurements of fluorescence intensities and high-resolution imaging were performed in these glass SlipChips. Hatch and co-authors successfully demonstrated a single microfluidic device for rapid droplet generation, thermocycling, and wide-field fluorescence imaging of 1-million droplets with a micro-lens digital camera.21 This system was applied for DNA detection under digital PCR. The PCR amplification resulted in approximately 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies of the compartmentalized gene in each droplet. In the next step the droplets fused with droplets delivering a cell-free coupled transcription–translation (IVTT) system and the reagents for a fluorogenic assay. Genes were selectively recovered from droplets using fluorescence-activated electrocoalescence. This system can sort at 2000 droplets per s by selecting mixtures of lacZ and lacZmut genes.77
000 copies of the compartmentalized gene in each droplet. In the next step the droplets fused with droplets delivering a cell-free coupled transcription–translation (IVTT) system and the reagents for a fluorogenic assay. Genes were selectively recovered from droplets using fluorescence-activated electrocoalescence. This system can sort at 2000 droplets per s by selecting mixtures of lacZ and lacZmut genes.77
Fluorescence imaging can also provide spatial distribution of the molecules in droplets in a quantitative manner. This approach was used to characterize the development of the shape, size and the fluorescence intensity change inside the cells.78 Fluorescence imaging was also used in conjunction with dielectrophoresis and optical tweezers79 for detection of the viability of cells in individual droplets.80 The presented platform included single cell encapsulation in picolitre droplets, incubation for several hours and imaging using automated microscopy to determine cell viability (Fig. 2).81,82
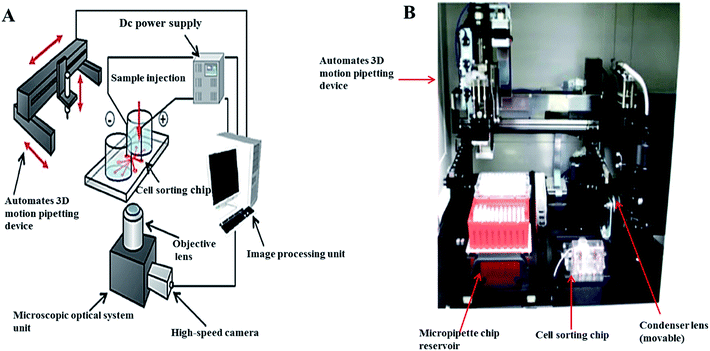 | ||
| Fig. 2 Automated on-chip imaging flow cytometry system with a disposable contamination-free plastic cultivation chip: (A) schematic diagram, (B) photograph. From ref. 82; with permission. | ||
The fluorescence lifetime imaging microscopy can also provide information about the internal environment of the observed droplets.83,84 Bennet et al. have shown the capability of optical tweezers for manipulation of objects with temperature-sensitive fluorescence lifetime imaging microscopy. Droplets with encapsulated Rhodamine B were used in the experiment. In addition to fluorescence the encapsulated fluorophore dye provided the refractive index differential required for optical trapping.79
Mass spectrometry
Mass spectrometry (MS) brings another dimension to droplet assays.85 In the simplest case (single quadrupole or linear time-of-flight based instruments) it can provide information about the mass to charge ratio of the analytes.86,87 More complex instruments, combining two or more mass analyzers, can often provide substantial structural information via the analysis of molecular fragments. Current instruments featuring electrospray ionization (ESI) or matrix assisted laser desorption/ionization (MALDI) offer very good sensitivity in the low attomole range.88 The mass spectrometer is usually the most complex part of the analytical system and the connected microfluidics act as the sample preparation tool.89–92 Since the sample volume consumed during the electrospray/mass spectrometry experiment is in the micro- to nanoliter range, the droplet/MS analysis represents a viable approach for limited sample quantities.93,94 Kennedy et al. successfully demonstrated the ESI-MC coupling to nanoliter scale flow reaction for label-free screening of reactions. The system was used for analysis of the cathepsin B inhibitor with ultralow reagent consumption and high-throughput.95 Yang et al. developed a segmented flow protocol to combine nano-liquid chromatography separation with MC in proteomics research. The protocol is based on segmenting of the liquid chromatography (LC) effluent and analysing the segments off-line by mass spectrometry. Tryptic digests of a T. tengcongensis sample were analyzed using the segmented flow interface between nano-LC and MS.96 Fang et al. developed a multifunctional microfluidic droplet-array system capable of performing multi-step sample pretreatment, introduction and ESI-MS detection using a capillary as a sampling probe and an electrospray emitter to interface an oil-covered two dimensional (2D) droplet array to the ESI-MS instrument.97 Girault et al. demonstrated a new approach for coupling droplet based microfluidics with MS using capacitive charge transfer across the boundary of immiscible liquids to monitor single-phase and biphasic reactions. An advantage of the presented system is dilution-free droplet ionization without the additional oil removal step, surfactant or sheath-flow.98 Kuester et al. described a novel interface between droplet microfluidics and matrix-assisted laser desorption/ionization (MALDI) mass spectrometry for label-free analysis of up to 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individual aqueous droplets on a microarray plate.92 The system enabled the synchronization between stage movement and the output of droplets from the capillary as well as the prevention of cross-contamination. The authors studied the efficiency of the system by monitoring the formation of an octapeptide (angiotensin II) from a decapeptide (angiotensin I) during an enzymatic reaction within several hours.92 Baker et al. successfully built a system consisting of a transfer capillary, a standard ESI needle, and a tapered gas nozzle addressing some of the challenges of integrated droplet microfluidics with MS detection. A transfer capillary was used to couple a microfluidic device to an ESI needle. The operation principle was based on applying a pulse of N2 to the nozzle; a pressure differential induced at the outlet of the ESI needle pushed the droplets from the device, passing the ESI needle and entering the flow of N2 to the MS inlet. Ionization potential and N2 pressure were 3206 V and 80 psi, respectively. Open and closed devices were demonstrated from 630 nL to 2.5 μL aqueous droplets mixed with droplets of caffeine and theophylline internal standard transferred from the device to the ESI/MS. The ratio of the caffeine/theophylline peak areas has been detected in the range of 5–250 μg mL−1 (12.5–625 ng per droplet) with a limit of detection (for caffeine) of 1 μg mL−1 (2.5 ng per droplet). The ability to rapidly mix droplets makes devices a promising platform for relative quantitation by MS.89 Smith et al. developed ESI-MS to measure femtomole amounts of proteins with molecular weights of 1 to >100 kDa in an individual droplet.99 The droplets were generated at a frequency of 171 Hz in a flow-focusing geometry microfluidic design using fluorocarbon oil which was found suitable as the continuous phase. The produced droplets (107 μm or 650 pL) containing α-chymotrypsinogen A (50 μM) were collected off-chip to be stored overnight at 4 °C and re-injected into emitter devices before ESI/MS analysis (Fig. 3). The mixture of droplets occurring at >150 per minute provided high quality mass spectra of surfactant stabilized droplets.
000 individual aqueous droplets on a microarray plate.92 The system enabled the synchronization between stage movement and the output of droplets from the capillary as well as the prevention of cross-contamination. The authors studied the efficiency of the system by monitoring the formation of an octapeptide (angiotensin II) from a decapeptide (angiotensin I) during an enzymatic reaction within several hours.92 Baker et al. successfully built a system consisting of a transfer capillary, a standard ESI needle, and a tapered gas nozzle addressing some of the challenges of integrated droplet microfluidics with MS detection. A transfer capillary was used to couple a microfluidic device to an ESI needle. The operation principle was based on applying a pulse of N2 to the nozzle; a pressure differential induced at the outlet of the ESI needle pushed the droplets from the device, passing the ESI needle and entering the flow of N2 to the MS inlet. Ionization potential and N2 pressure were 3206 V and 80 psi, respectively. Open and closed devices were demonstrated from 630 nL to 2.5 μL aqueous droplets mixed with droplets of caffeine and theophylline internal standard transferred from the device to the ESI/MS. The ratio of the caffeine/theophylline peak areas has been detected in the range of 5–250 μg mL−1 (12.5–625 ng per droplet) with a limit of detection (for caffeine) of 1 μg mL−1 (2.5 ng per droplet). The ability to rapidly mix droplets makes devices a promising platform for relative quantitation by MS.89 Smith et al. developed ESI-MS to measure femtomole amounts of proteins with molecular weights of 1 to >100 kDa in an individual droplet.99 The droplets were generated at a frequency of 171 Hz in a flow-focusing geometry microfluidic design using fluorocarbon oil which was found suitable as the continuous phase. The produced droplets (107 μm or 650 pL) containing α-chymotrypsinogen A (50 μM) were collected off-chip to be stored overnight at 4 °C and re-injected into emitter devices before ESI/MS analysis (Fig. 3). The mixture of droplets occurring at >150 per minute provided high quality mass spectra of surfactant stabilized droplets.
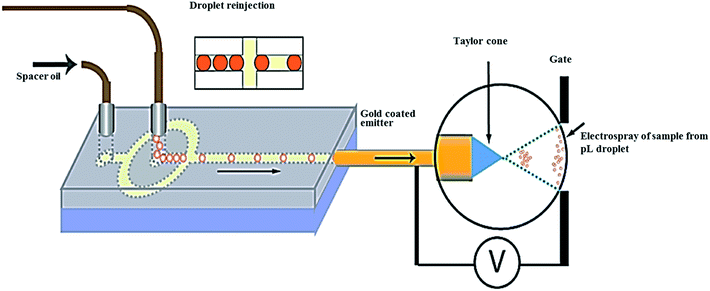 | ||
| Fig. 3 Picoliter droplets containing α-chymotrypsinogen A (50 μM) were generated at 171 Hz in a flow-focusing droplet generator, after storage the droplets at 4 °C, they were re-injected into the emitter device for mass spectrometric analysis. From ref. 99; with permission. | ||
Kirby et al. have broadened the application of the integrated system with in-line MS analysis and developed “microfluidic origami” microchemical reactions.91,100,101 This device included a microfluidic platform and a nanoelectrospray ionization (nanoESI) emitter formed on a single flexible polyimide film substrate. This device was used to qualitatively monitor the reaction progress of the Morita–Baylis–Hillman reaction in real-time. It was possible to carry out the microchemical synthesis through applying an in-line interface connecting the microfluidics and MS.
Raman spectrometry
Raman spectra can provide information about the analyte structure, in the form of a spectral fingerprint as well as can be combined with other detection methods. While the traditional Raman spectrometry is inherently insensitive, recent developments in surface enhanced Raman spectrometry (SERS) make this technique also suitable for microfluidic systems.102 Combining droplet techniques with SERS can significantly broaden the detection information in droplet systems.103 Cecchini and co-authors developed an ultra-high throughput SERS system for droplet reaction detection with submillisecond time resolution as a powerful tool. Such a technique may allow the study of millisecond enzyme dynamics and high throughput screening of protein evolution in a label-free fashion.104 In the subsequent studies, SERS was applied to high sensitive detection of Rhodamine 6G (R6G) by enhanced Au nanoparticle-coated chitosan microbeads, and paraquat in water with a limit of detection of 2 × 10−9 M.105,106 Qu et al. demonstrated the SERS detection approach by fabricating a novel Au/Ag bimetallic bell shaped droplet microfluidic SERS sensor on cellulose paper that could be integrated with portable analysis of substituted aromatic substances. In another paper, Walter et al. developed a SERS based microfluidic lab-on a-chip for bacteria identification using Raman spectroscopy fingerprinting. An advantage of this system is reducing the spectral recording time to 1 s with generating a database of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 spectra for a model system Escherichia coli including nine different strains. The validation accuracy of the support vector machine was up to 92.6% according to chemometric analysis. This new method has also demonstrated the potential for bacteria classification with high reliability.107 Currently, Li et al. reported a technique called “stamping surface-enhanced Raman spectroscopy” (S-SERS) for label-free, molecular sensing and imaging. This approach involves a technique that takes advantage of a polydimethylsiloxane (PDMS) thin film used as a carrier for target molecules, which were deposited on the PDMS form dried and stamped onto nanoporous gold disks with high-density plasmonic hot spots as the Raman signal enhancer. This design has enabled SERS measurement of “sandwiched” target molecules (R6D and urea) for concentrations ranging from 10 nM to 100 μM.108
200 spectra for a model system Escherichia coli including nine different strains. The validation accuracy of the support vector machine was up to 92.6% according to chemometric analysis. This new method has also demonstrated the potential for bacteria classification with high reliability.107 Currently, Li et al. reported a technique called “stamping surface-enhanced Raman spectroscopy” (S-SERS) for label-free, molecular sensing and imaging. This approach involves a technique that takes advantage of a polydimethylsiloxane (PDMS) thin film used as a carrier for target molecules, which were deposited on the PDMS form dried and stamped onto nanoporous gold disks with high-density plasmonic hot spots as the Raman signal enhancer. This design has enabled SERS measurement of “sandwiched” target molecules (R6D and urea) for concentrations ranging from 10 nM to 100 μM.108
Electrical measurements
One of the main challenges in the development of droplet microfluidic devices is related to the measurement of droplet size, frequency, conductivity, velocity and composition.109,110 While the electrochemical detection is less universal than mass spectrometry, it provides a scalable, low power, cost-effective alternative to microscope/fast-camera systems commonly used for monitoring droplets, measuring droplet size and velocity. Moiseeva et al. proposed a system to generate droplet trains that could be detected by measuring electrical impedance at thin-film electrodes.111 The developed system allowed simple low-cost measurements. Cahill et al. developed a method for impedance measurement of the conductivity of aqueous droplets in a segmented-flow system.112 A thin-walled glass capillary with electrodes in contact the outer surface was used to provide the contactless measurement of conductivity of the liquid within the capillary. The developed system provided a low-cost and robust method well suited for in-line measurement of impedance spectroscopy in a segmented flow.Electrophoresis
Recent years have seen the development of new methods for sampling and electrophoretic analysis of aqueous plugs segmented in an oil stream. The applications include sample manipulation for high-throughput screening, analysis of products of in-plug reactions, or microdialysis sampling.113–115 Chiu et al. explored coupled microfluidic devices with capillary electrophoresis separation by following laser-induced fluorescence of single femtoliter-volume droplets. The droplets were generated using a T-channel connected to microinjectors and moved towards the region with immiscible boundary isolating the CE separation channel. The device also incorporated patterned surface with differential wetting for selective deposition of a hydrophilic polymer in required regions of the chip.116 De Mello et al. described a simple platform incorporating droplet-based microfluidic module for the precise injection of pL to nL volume of sample to the capillary zone electrophoresis and capillary gel electrophoresis.117Biomolecule detection
Compartmentalization
The compartmentalization of reactions in a picoliter-sized drop is effectively used for a wide range of experiments where individual droplets contain key reagents for further experiments. The droplet technology avoids eliminating the dispersion of reactants and decreases the consumption of reagents.8 With lower concentration of samples, as well as the enclosed space of a droplet, compartmentalization allows control of the microenvironment for individual cell reactions. Massive parallel handling of millions of independent reactions enables the analysis of vast populations of single-cells to detect rare events. These technologies were employed by Tawfik and Griffith et al., where the fulfillment co-compartmentalization of single genes saved the genotype–phenotype linkage using aqueous droplets in oil emulsion.118 This system also allowed the study of in vitro transcription/translation solution for protein expression and enzymatic reactions. Fluorescence-based sorting of the droplets allowed timing of the initiation of an enzymatic reaction. A similar approach was demonstrated by Agresti et al., who carried out co-compartmentalization of yeast cells expressing horseradish peroxidase (HRP) on the cell surface parallel to a non-fluorescent substrate. This system has demonstrated a 10-fold increase of the catalytic efficiency of HRP and has also shown the possibility of using yeast cell surface display in conjunction with microfluids for enzyme engineering.119Microfluidic devices for PCR have been investigated by several groups. Geng et al. described the compartmentalization of genomic DNA from single-cells along with primer-functionalized microbeads in agarose-in-oil droplets.120 Ellefson et al. presented the phenotype compartmentalization partnered replication into water in oil droplets, where synthetic circuits were linked to the production of Taq DNA polymerase. More efficiently driven Taq DNA polymerase production was enriched by exponential amplification during a subsequent emulsion PCR step.121,122 Another system was demonstrated by Zhao et al. who described a microfluidic SlipChip device to compartmentalize target analyte bands in situ into separate microdroplets collected and moved for further analysis.123 The SlipChip generated droplets in a parallel fashion by slipping layers of a planar chip to different lateral positions. The system incorporated a microfluidic isoelectric focusing (IEF) separation followed by in situ compartmentalization as shown schematically in Fig. 4A–C.
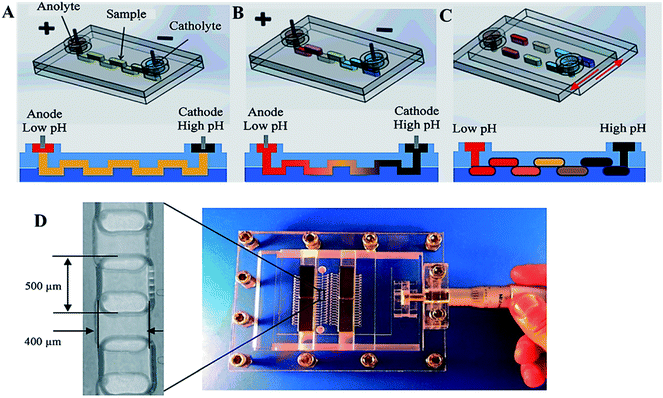 | ||
| Fig. 4 Scheme of the IEF separation and in situ compartmentalization in a SlipChip. (A) Sample loading to a continuous “zig-zag” channel. (B) pH gradient is established and IEF is performed after application of an electric field. (C) In situ compartmentalisation after IEF separation. (D) The microdevice and platform made of PMMA, contained a “zig-zag” channel with dimensions of 400 μm × 250 μm × 5 cm. From ref. 123; with permission. | ||
The device operation included the following steps: at first, IEF separation was performed in a microfluidic zig-zag channel composed of a sequence of wells formed in the two halves of the SlipChip (Fig. 4A) and the analyte was focused along the channel by application of an electric field (Fig. 4B). Slipping the chip disconnected the wells, leaving the analyte in isolated compartments or single droplets in each of the wells (Fig. 4C). The device was tested using standard pI markers as well with chip-based gel electrophoresis of a mixture containing 5 standard proteins (trypsin inhibitor, β-lactoglobulin A, carbonic anhydrase isozyme II, myoglobin, and lectin). It should be noted that this method is capable of avoiding remixing, is scalable and can be hyphenated with other analytical methods.
Single-cell detection in droplets
With the advancements in the field of cell biology and medicine, there is a considerable need for a device allowing multiparameter measurement and manipulation of single-cells.124 Microfabrication technologies are in many aspects well suited to bring such new tools for single-cell analyses avoiding problems related to common sample handling variability and contamination when the raw material from single-cells is diluted to microlitre volumes. With the development of microfluidic devices some of the problems can be minimized as far as devices comprise an interlinked series of channels, reaction microvalves, sample read-out devices, all of which are physically integrated on a microfabricated solid substrate.125,126In this section we focus on recent reports of single-cell analysis using fluorescently tagged probes including antibodies,127 nucleic acid,128 gene expression at the single-cell level, enzyme kinetic reaction and bioimaging.129 Detection of individual cells is performed by fluorescence microscopy imaging of cells inside microfluidic devices. Rane et al.130 have developed a microfluidic chip for the pathogenic cell detection and quantification by screening the fluorescent droplets using confocal fluorescence spectroscopy (CFS). The experimental setup included a custom metal plate to act as an interface between the microfluidic chip and the CFS setup. A Peltier heater under the ‘incubation zone’ on the microfluidic chip was included on the metal plate to maintain a fixed incubation temperature throughout the experiment. The setup allowed us to carry out sample digitization, cell lysis, probe–target hybridization and subsequent fluorescent detection with dual laser excitation (488 nm and 633 nm) as well as simultaneous dual color detection (520 nm and 670 nm). A peptide nucleic acid fluorescence resonance energy transfer probe (PNA beacon) was used to detect 16S rRNA in E. coli cells. The same device could perform the “sample-to-answer” pathogen detection of single-cells.130 A number of analytical approaches have utilized the flow cytometry for both quantitative and qualitative multi-parametric analyses of single cell in genetics, diagnosis, and cancer research.131,132 Some of the limitations common to conventional flow cytometry have been addressed by microfluidics-based miniature flow cytometry devices.133 In particular, fluorogenic assays by measuring enzymes markers can now be used for single cell studies.134,135
Recent years have witnessed the development of microfluidic approaches for the monodisperse droplet formation and manipulation in association with fluorescence detection.8,136 Arayanarakool et al. developed a micro- and nanochannel network for the femtoliter aqueous droplet generation. The tiny droplets provide an ideal environment for single-enzyme activity assay where enzyme and substrate solutions were confined into femtoliter volumes helping in achieving the high product concentration without increasing background noise. Researchers have demonstrated single enzyme molecule encapsulation on the chip and measurement of the kinetic activity of single β-glucosidase molecules.137 Most recently, Guan et al. have proposed a microfluidic method enabling single molecule counting and digital protein detection in picoliter droplets via enzyme catalyzed signal amplification. The developed chip allowed us to carry out an injection of two separate flows such as single β-galactosidase molecules and the fluorogenic substrate fluorescein di-β-D-galactopyranoside from different inlets. At a frequency of 6.6 kHz droplets were formed and approximately 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets were collected as a monolayer on the chip for imaging.138
000 droplets were collected as a monolayer on the chip for imaging.138
Methods combining droplet-based microfluidics and enzyme-linked immunosorbent assay (ELISA) are becoming quite popular due to low limits of detection.139 An integrated microfluidic chip with bead-based immunoassay enables measuring the activity of a single copy of enzyme and quantify an amount of biomarkers. The recently described prototype139 allows determination of the prostate-specific antigen (PSA) in buffer at a concentration of 1.2 pg mL; an improvement of nearly 2 orders of magnitude over the standard ELISA – Fig. 5.
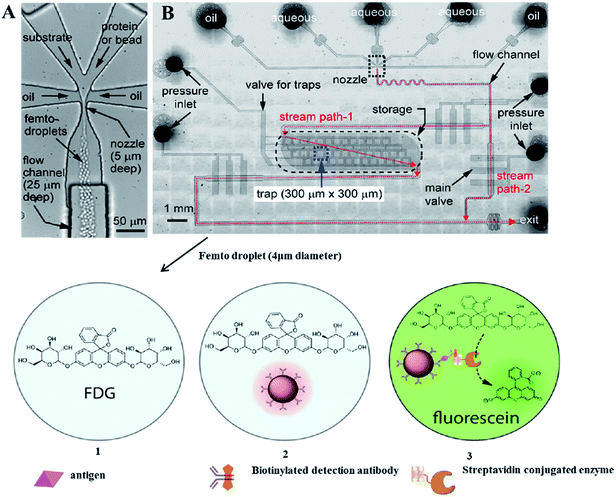 | ||
| Fig. 5 The microfluidic device used for femto droplet generation and manipulation. (A) Femto droplet formation with a typical volume of 32 fL at a frequency of around 3.5 × 105 Hz. (B) The multilayered PDMS device, where the upper layer consists of the nozzle (10 μm wide, 5 μm deep), flow channels (100 μm wide, 25 μm deep), and storage compartments (2 mm wide, 7 mm long 5 μm deep), with a capacity of ∼2 × 10−5 femto droplets. The bottom layer houses the monolithic valves used to control droplet flow and isolate the traps. If this valve is opened, droplets flow out of the device by stream path 2 due to the lower flow resistance encountered. After the on-chip incubation, three populations of femtodroplets are observed: (1) droplets containing no bead, (2) those containing a bead without immunocomplexes, and (3) those containing a bead with an immunocomplex exhibiting a positive fluorescence signal due to the enzymatic activity of a single β-galactosidase reporter. From ref. 139; with permission. | ||
Microfluidic droplet PCR
Polymerase chain reaction is an undoubtedly widely used process in different areas of DNA analysis ranging from evolutionary biology, environmental science, criminology, to microbial detection, clinical diagnostics and scientific research. While PCR is most commonly performed in the homogeneous phase, the use of droplets as reaction vessels has been previously developed for pathogen detection.140 One of the most promising approaches is the digital PCR, allowing the combination of the limiting dilution and partitioning of the diluted sample into hundreds or thousands of reaction chambers with position statistics. Thus, the measurement of the absolute number of DNA molecules can be easily achieved.141 Since its development, digital PCR has been utilized in a wide range of biological research studies including detection of bacteria, copy number variation and single cell genomics.135,141–146 An integrated microfluidic device with real-time PCR (RT-PCR) analysis has been demonstrated for RNA transcripts detection,147,148 reverse transcription PCR combined with fluorescence microscopy,149,150 monoclonal template amplification for bead-based gene sequencing,120,151 single RNA fragment detection152,153 and widely applied multiplex PCR.27,120,154,155 The recent work of Geng et al.120 used microfluidic droplets for single-molecule sequencing. The somatically acquired carcinogenic translocations were detected at low concentrations (<10−6) in biological samples by using a bead-based hemi-nested digital PCR (dPCR). Authors have isolated and quantified various clonal forms with 10−7 limit of detection. In addition, the transfer replicas of short tandem repeat (STR) targets from a single cell genomic DNA into the co-encapsulated microbeads have been investigated. STR products, bound on the primer-functionalized microbeads co-encapsulated in the droplets, were amplified by a secondary PCR reaction and detected by capillary electrophoresis fragment size analysis of target molecules. The microbead-based 9-plex PCR was optimized in bulk solution and in droplets using standard DNA solutions. A single cell STR typing method based on droplet microfluidics is shown in Fig. 6.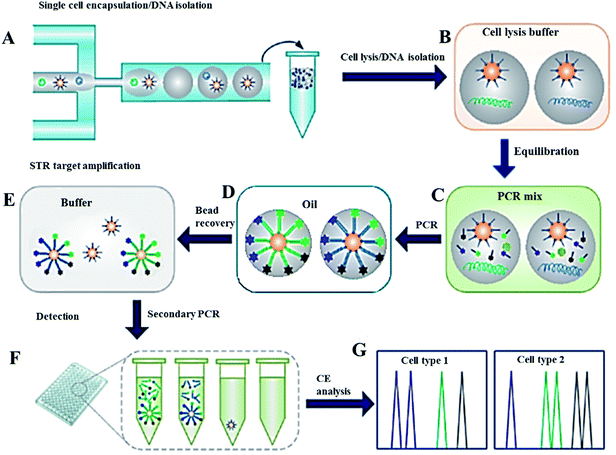 | ||
| Fig. 6 Analytical procedure for single cell forensic STR typing. (A) Microfluidic droplet generator for encapsulation of cells together with primer-functionalized microbeads within agarose microdroplets. (B) Incubation of the gelled droplets in the cell lysis buffer to release genomic DNA into the gel matrix. (C) Diffusion of the PCR components into the gel droplets by equilibrating in the PCR mix. (D) After re-dispersion of the droplets in oil, emulsion PCR is performed with a thermal cycler. (E) Recovery of the beads by melting the agarose, after the first STR amplification. (F) Dilution of the secondary PCR as a starting point from single beads in standard PCR plates. (G) Capillary electrophoresis system for fragment size analysis. From ref. 120; with permission. | ||
Compartmentalization of the sample prior to PCR amplification provides the basis for absolute quantification of target molecules.156 By dividing the diluted sample into a large number of small-volume reaction compartments, single copies of the nucleic acid template can be confined in isolated compartments and PCR amplified followed by a “yes or no” readout. The number of target molecules is then evaluated by performing a statistical analysis of the “positive” and “negative” signals. Digital PCR can be performed in a variety of formats, including well plates, microdroplets,157 pneumatically controlled microchips,158,159 spinning discs,160,161 the SlipChip76,162–164 and LabDisk platform.165 A recent example of single DNA molecule detection using an integrated self-priming compartmentalization on-chip digital PCR device was described by Zhu et al.166 In this design, the self-compartmentalization of the sample was based on surface tension differences in the chip fabricated in PDMS. The chip contained 5120 microchambers (5 nL) detecting an average of 480 to 4804 template molecules with the optimal number of positive chambers between 400 and 1250 as evaluated by the Poisson distribution.166 In another study, Hindson et al. investigated a high-throughput droplet digital PCR (ddPCR) system allowing processing of ∼2 million PCR reactions in a 96-well plate workflow.146 This approach allowed accurate measurement of germline copy number variations, sensitive detection of mutant DNA in a 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold excess of a wild type DNA and quantitation of circulating fetal and maternal DNA in cell-free plasma. The linearity of the response to DNA concentration and accuracy over the dynamic range of the 20
000-fold excess of a wild type DNA and quantitation of circulating fetal and maternal DNA in cell-free plasma. The linearity of the response to DNA concentration and accuracy over the dynamic range of the 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplet assay corresponded to more than 4 orders of magnitude of target DNA copy number per ddPCR. Copy number concentration and ratio with expanded uncertainties were evaluated using high density ddPCR and a lower density microfluidic chamber based digital PCR platform.167 A relative expanded uncertainty under 5% was obtained using ddPCR; a value much lower than currently typical for quantification of specific DNA target sequences.
000 droplet assay corresponded to more than 4 orders of magnitude of target DNA copy number per ddPCR. Copy number concentration and ratio with expanded uncertainties were evaluated using high density ddPCR and a lower density microfluidic chamber based digital PCR platform.167 A relative expanded uncertainty under 5% was obtained using ddPCR; a value much lower than currently typical for quantification of specific DNA target sequences.
Conclusions
This short review captures a snapshot of the field at this critical stage covering the recent advances in droplet microfluidics for (bio)chemical analysis with the focus on papers published in the past four years. A brief overview of the principle of droplet formation and manipulation, and detection methods in droplets including spectroscopy, fluorescence microscopy and electrical measurements is also provided. It is the belief of the authors that the rapid development of this field in the past decade will continue in the foreseeable future bringing solutions to many of the current bioanalytical challenges.Acknowledgements
Financial support from the Grant Agency of the Czech Republic (P206/12/G014) and the institutional support RVO: 68081715 is acknowledged. Part of the work was realized in CEITEC – Central European Institute of Technology with research infrastructure supported by the project CZ.1.05/1.1.00/02.0068 financed by European Regional Development Fund.References
- Z. Zhu, G. Jenkins, W. Zhang, M. Zhang, Z. Guan and C. J. Yang, Single-molecule emulsion PCR in microfluidic droplets, Anal. Bioanal. Chem., 2012, 403(8), 2127–2143 CrossRef CAS PubMed
.
- A. Golberg, M. L. Yarmush and T. Konry, Picoliter droplet microfluidic immunosorbent platform for point-of-care diagnostics of tetanus, Microchim. Acta, 2013, 180(9–10), 855–860 CrossRef CAS
.
- T. C. Scanlon, S. M. Dostal and K. E. Griswold, A High-Throughput Screen for Antibiotic Drug Discovery, Biotechnol. Bioeng., 2014, 111(2), 232–243 CrossRef CAS PubMed
.
- S. Neethirajan, I. Kobayashi, M. Nakajima, D. Wu, S. Nandagopal and F. Lin, Microfluidics for food, agriculture and biosystems industries, Lab Chip, 2011, 11(9), 1574–1586 RSC
.
- R. D. Begg, Dynamics of Continuous Segmented Flow Analysis, Anal. Chem., 1971, 43, 854–857 CrossRef CAS
.
- J. Ruzicka and E. Hansen, Flow Injection Analyses: Part 1. A new Concept of Fast Continuous-Flow Analysis, Anal. Chim. Acta, 1975, 78, 145–157 CrossRef CAS
.
- A. Manz, N. Graber and H. M. Widmer, Miniaturized Total Chemical-Analysis Systems – A Novel Concept For Chemical Sensing, Sens. Actuators, B, 1990, 1, 244–248 CrossRef CAS
.
- A. B. Theberge, F. Courtois, Y. Schaerli, M. Fischlechner, C. Abell and F. Hollfelder,
et al. Microdroplets in Microfluidics: An Evolving Platform for Discoveries in Chemistry and Biology, Angew. Chem., Int. Ed., 2010, 49(34), 5846–5868 CrossRef CAS PubMed
.
- S. S. Datta, D. D. Gerrard, T. S. Rhodes, T. G. Mason and D. A. Weitz, Rheology of attractive emulsions, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2011, 84(4), 041404 CrossRef
.
- M. Takinoue and S. Takeuchi, Droplet microfluidics for the study of artificial cells, Anal. Bioanal. Chem., 2011, 400(6), 1705–1716 CrossRef CAS PubMed
.
- C. N. Baroud, F. Gallaire and R. Dangla, Dynamics of microfluidic droplets, Lab Chip, 2010, 10(16), 2032–2045 RSC
.
- M. Windbergs and D. A. Weitz, Drug Dissolution Chip (DDC): A Microfluidic Approach for Drug Release, Small, 2011, 7(21), 3011–3015 CrossRef CAS PubMed
.
- S. Seiffert, M. B. Romanowsky and D. A. Weitz, Janus Microgels Produced from Functional Precursor Polymers, Langmuir, 2010, 26(18), 14842–14847 CrossRef CAS PubMed
.
- L. L. A. Adams, T. E. Kodger, S. H. Kim, H. C. Shum, T. Franke and D. A. Weitz, Single step emulsification for the generation of multi-component double emulsions, Soft Matter, 2012, 8(41), 10719–10724 RSC
.
- G. T. Vladisavljevic, I. Kobayashi and M. Nakajima, Production of uniform droplets using membrane, microchannel and microfluidic emulsification devices, Microfluid. Nanofluid., 2012, 13(1), 151–178 CrossRef CAS
.
- H. Hasinovic and S. E. Friberg, One-Step Inversion Process to a Janus Emulsion with Two Mutually Insoluble Oils, Langmuir, 2011, 27(11), 6584–6588 CrossRef CAS PubMed
.
- L. Mazutis and A. D. Griffiths, Selective droplet coalescence using microfluidic systems, Lab Chip, 2012, 12(10), 1800–1806 RSC
.
- D. J. Holt, R. J. Payne, W. Y. Chow and C. Abell, Fluorosurfactants for microdroplets: Interfacial tension analysis, J. Colloid Interface Sci., 2010, 350(1), 205–211 CrossRef CAS PubMed
.
- X. H. Ma, T. Y. Song, Z. Lan and T. Bai, Transient characteristics of initial droplet size distribution and effect of pressure on evolution of transient condensation on low thermal conductivity surface, Int. J. Therm. Sci., 2010, 49(9), 1517–1526 CrossRef CAS PubMed
.
- P. R. Marcoux, M. Dupoy, R. Mathey, A. Novelli-Rousseau, V. Heran and S. Morales,
et al. Micro-confinement of bacteria into w/o emulsion droplets for rapid detection and enumeration, Colloids Surf., A, 2011, 377(1–3), 54–62 CrossRef CAS PubMed
.
- A. C. Hatch, J. S. Fisher, A. R. Tovar, A. T. Hsieh, R. Lin and S. L. Pentoney,
et al. 1-Million droplet array with wide-field fluorescence imaging for digital PCR, Lab Chip, 2011, 11(22), 3838–3845 RSC
.
- A. M. Huebner, Ch. Abell and W. T. S. Huck,
et al. Monitoring a Reaction at Submillisecond Resolution in Picoliter Volumes, Anal. Chem., 2011, 83, 1462–1468 CrossRef CAS PubMed
.
- B. M. Paegel and G. F. Joyce, Microfluidic Compartmentalized Directed Evolution, Chem. Biol., 2010, 17(7), 717–724 CrossRef CAS PubMed
.
- N. Bremond, H. Domejean and J. Bibette, Propagation of Drop Coalescence in a Two-Dimensional Emulsion: A Route towards Phase Inversion, Phys. Rev. Lett., 2011, 106(21), 214502 CrossRef
.
- X. Z. Niu and A. J. deMello, Building droplet-based microfluidic systems for biological analysis, Biochem. Soc. Trans., 2012, 40, 615–623 CrossRef CAS PubMed
.
- J. C. Baret, Surfactants in droplet-based microfluidics, Lab Chip, 2012, 12, 422–433 RSC
.
- Q. Zhong, S. Bhattacharya, S. Kotsopoulos, J. Olson, V. Taly and A. D. Griffiths,
et al. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR, Lab Chip, 2011, 11(13), 2167–2174 RSC
.
- Z. N. Cao, F. Y. Chen, N. Bao, H. C. He, P. S. Xu and S. Jana,
et al. Droplet sorting based on the number of encapsulated particles using a solenoid valve, Lab Chip, 2013, 13(1), 171–178 RSC
.
- H. Zec, T. D. Rane and T. H. Wang, Microfluidic platform for on-demand generation of spatially indexed combinatorial droplets, Lab Chip, 2012, 12(17), 3055–3062 RSC
.
- I. Platzman, J. W. Janiesch and J. P. Spatz, Synthesis of Nanostructured and Biofunctionalized Water-in-Oil Droplets as Tools for Homing T Cells, J. Am. Chem. Soc., 2013, 135(9), 3339–3342 CrossRef CAS PubMed
.
- G. Woronoff, A. El Harrak, E. Mayot, O. Schicke, O. J. Miller and P. Soumillion,
et al. New Generation of Amino Coumarin Methyl Sulfonate-Based Fluorogenic Substrates for Amidase Assays in Droplet-Based Microfluidic Applications, Anal. Chem., 2011, 83(8), 2852–2857 CrossRef CAS PubMed
.
- Y. J. Ma, J. Thiele, L. Abdelmohsen, J. G. Xu and W. T. S. Huck, Biocompatible macro-initiators controlling radical retention in microfluidic on-chip photopolymerization of water-in-oil emulsions, Chem. Commun., 2014, 50(1), 112–114 RSC
.
- J. A. Stapleton and J. R. Swartz, Development of an In Vitro Compartmentalization Screen for High-Throughput Directed Evolution of FeFe Hydrogenases, PLoS One, 2010, 5(12), 15275 Search PubMed
.
- E. Cohen, N. Elfassy, G. Koplovitz, S. Yochelis, S. Shusterman and D. P. Kumah,
et al. Surface X-Ray Diffraction Results on the III–V Droplet Heteroepitaxy Growth Process for Quantum Dots: Recent Understanding and Open Questions, Sensors, 2011, 11(11), 10624–10637 CrossRef CAS PubMed
.
- J. Wegrzyn, A. Samborski, L. Reissig, P. M. Korczyk, S. Blonski and P. Garstecki, Microfluidic architectures for efficient generation of chemistry gradations in droplets, Microfluid. Nanofluid., 2013, 14(1–2), 235–245 CrossRef CAS
.
- Y. Elani, A. J. deMello, X. Z. Niu and O. Ces, Novel technologies for the formation of 2D and 3D droplet interface bilayer networks, Lab Chip, 2012, 12(18), 3514–3520 RSC
.
- K. van Dijke, I. Kobayashi, K. Schroen, K. Uemura, M. Nakajima and R. Boom, Effect of viscosities of dispersed and continuous phases in microchannel oil-in-water emulsification, Microfluid. Nanofluid., 2010, 9(1), 77–85 CrossRef CAS
.
- P. Jankowski, D. Ogonczyk, L. Derzsi, W. Lisowski and P. Garstecki, Hydrophilic polycarbonate chips for generation of oil-in-water (O/W) and water-in-oil-in-water (W/O/W) emulsions, Microfluid. Nanofluid., 2013, 14(5), 767–774 CrossRef CAS
.
- T. Krebs, C. Schroen and R. M. Boom, Coalescence kinetics of oil-in-water emulsions studied with microfluidics, Fuel, 2013, 106, 327–334 CrossRef CAS PubMed
.
- A. A. Maan, R. Boom and K. Schroen, Preparation of monodispersed oil-in-water emulsions through semi-metal microfluidic EDGE systems, Microfluid. Nanofluid., 2013, 14(5), 775–784 CrossRef CAS
.
- J. D. Wan and H. A. Stone, Coated Gas Bubbles for the Continuous Synthesis of Hollow Inorganic Particles, Langmuir, 2012, 28(1), 37–41 CrossRef CAS PubMed
.
- W. Choi, M. Hashimoto, A. K. Ellerbee, X. Chen, K. J. M. Bishop and P. Garstecki,
et al. Bubbles navigating through networks of microchannels, Lab Chip, 2011, 11(23), 3970–3978 RSC
.
- C. X. Zhao, Multiphase flow microfluidics for the production of single or multiple emulsions for drug delivery, Adv. Drug Delivery Rev., 2013, 65(11–12), 1420–1446 CrossRef CAS PubMed
.
- E. Movahednejad, F. Ommi and S. M. Hosseinalipour, Prediction of Droplet Size and Velocity Distribution in Droplet Formation Region of Liquid Spray, Entropy, 2010, 12(6), 1484–1498 CrossRef CAS PubMed
.
- H. Gu, M. H. G. Duits and F. Mugele, Droplets Formation and Merging in Two-Phase Flow Microfluidics, Int. J. Mol. Sci., 2011, 12(4), 2572–2597 CrossRef CAS PubMed
.
- B. M. Jose and T. Cubaud, Droplet arrangement and coalescence in diverging/converging microchannels, Microfluid. Nanofluid., 2012, 12(5), 687–696 CrossRef CAS
.
- C. X. Zhao and A. P. J. Middelberg, Two-phase microfluidic flows, Chem. Eng. Sci., 2011, 66(7), 1394–1411 CrossRef CAS PubMed
.
- Z. L. Xiao, M. L. Niu and B. Zhang, Droplet microfluidics based microseparation systems, J. Sep. Sci., 2012, 35(10–11), 1284–1293 CrossRef CAS PubMed
.
- J. S. Chen and J. H. Jiang, Droplet Microfluidic Technology: Microdroplets Formation and Manipulation, Chin. J. Anal. Chem., 2012, 40(8), 1293–1300 CrossRef CAS
.
- S. Park, Y. Zhang, S. Lin, T. H. Wang and S. Yang, Advances in microfluidic PCR for point-of-care infectious disease diagnostics, Biotechnol. Adv., 2011, 29(6), 830–839 CrossRef CAS PubMed
.
- G. T. Vladisavljevic, N. Khalid, M. A. Neves, T. Kuroiwa, M. Nakajima and K. Uemura,
et al. Industrial lab-on-a-chip: Design, applications and scale-up for drug discovery and delivery, Adv. Drug Delivery Rev., 2013, 65(11–12), 1626–1663 CrossRef CAS PubMed
.
- D. N. Adamson, D. Mustafi, J. X. J. Zhang, B. Zheng and R. F. Ismagilov, Production of arrays of chemically distinct nanolitre plugs via repeated splitting in microfluidic devices, Lab Chip, 2006, 6, 1178–1186 RSC
.
- J. Nie and R. T. Kennedy, Sampling from Nanoliter Plugs via Asymmetrical Splitting of Segmented Flow, Anal. Chem., 2010, 82, 7852–7856 CrossRef CAS PubMed
.
- B. H. Lapizco-Encinas and F. Foret, Dielectrophoresis I, Electrophoresis, 2011, 32(17), 2231 CrossRef CAS PubMed
.
- B. H. Lapizco-Encinas and F. Foret, Dielectrophoresis II, Electrophoresis, 2011, 32(17), 2401 CAS
.
- Y. Bai, E. Weibull, H. N. Joensson and H. Andersson-Svahn, Interfacing picoliter droplet microfluidics with addressable microliter compartments using fluorescence activated cell sorting, Sens. Actuators, B, 2014, 194, 249–254 CrossRef CAS PubMed
.
- E. Basova, J. Drs, J. Zemanek, Z. Hurak and F. Foret, Integrated microfluidic device for droplet manipulation, Chem. Listy, 2013, 107, 291–293 Search PubMed
.
- S. Zhou, W. Qiu, W. Hu, Y. Liu, F. Bai and D. Zhu, Photoconduction and application of oxotitanium phthalocyanine dual-layered thin films, Thin Solid Films, 2000, 375, 263–266 CrossRef CAS
.
- S.-H. Hung, S.-C. Huang and G.-B. Lee, Numerical Simulation of Optically-Induced Dielectrophoresis Using a Voltage-Transformation-Ratio Model, Sensors, 2013, 13(2), 1965–1983 CrossRef CAS PubMed
.
- S. M. Desmarais, H. P. Haagsman and A. E. Barron, Microfabricated devices for biomolecule encapsulation, Electrophoresis, 2012, 33(17), 2639–2649 CrossRef CAS PubMed
.
- Y. Bai, E. Weibull, H. N. Joensson and H. Andersson-Svahn, Interfacing picoliter droplet microfluidics with addressable microliter compartments using fluorescence activated cell sorting, Sens. Actuators, B, 2014, 194, 249–254 CrossRef CAS PubMed
.
- L. Wu, P. Chen, Y. Dong, X. Feng and B.-F. Liu, Encapsulation of single cells on a microfluidic device integrating droplet generation with fluorescence-activated droplet sorting, Biomed. Microdevices, 2013, 15(3), 553–560 CrossRef CAS PubMed
.
- J. Sun, P. Chen, X. Feng, W. Du and B.-F. Liu, Development of a microfluidic cell-based biosensor integrating a millisecond chemical pulse generator, Biosens. Bioelectron., 2011, 26(8), 3413–3419 CrossRef CAS PubMed
.
- T.-H. Wu, Y. Chen, S.-Y. Park, J. Hong, T. Teslaa and J. F. Zhong,
et al. Pulsed laser triggered high speed microfluidic fluorescence activated cell sorter, Lab Chip, 2012, 12(7), 1378–1383 RSC
.
- T. Laurell, F. Petersson and A. Nilsson, Chip integrated strategies for acoustic separation and manipulation of cells and particles, Chem. Soc. Rev., 2007, 36, 492–506 RSC
.
- O. Jakobsson, C. Grenvall, M. Nordin, M. Evander and T. Laurell, Acoustic actuated fluorescence activated sorting of microparticles, Lab Chip, 2014, 14, 1943–1950 RSC
.
- S. X. Li, X. Y. Ding, F. Guo, Y. C. Chen, M. I. Lapsley and S. C. S. Lin,
et al. An On-Chip, Multichannel Droplet Sorter Using Standing Surface Acoustic Waves, Anal. Chem., 2013, 85(11), 5468–5474 CrossRef CAS PubMed
.
- A. K. Price and A. B. MacConnell, Paegel BM Microfluidic Bead Suspension Hopper, Anal. Chem., 2014, 86, 5039–5044 CrossRef CAS PubMed
.
- V. Trivedi, A. Doshi, G. K. Kurup, E. Ereifej and P. J. Vandevord,
et.al. A modular approach for the generation, storage, mixing, and detection of droplet libraries for high throughput screening, Lab Chip, 2010, 10, 2433–2442 RSC
.
- C. G. Yang, Z. R. Xu and J. H. Wang, Manipulation of droplets in microfluidic systems, TrAC, Trends Anal. Chem., 2010, 29(2), 141–157 CrossRef CAS PubMed
.
- S. Juul, Y. P. Ho, J. Koch, F. F. Andersen, M. Stougaard and K. W. Leong,
et al. Detection of Single Enzymatic Events in Rare or Single Cells Using Microfluidics, ACS Nano, 2011, 5(10), 8305–8310 CrossRef CAS PubMed
.
- S. Q. Gu, Y. X. Zhang, Y. Zhu, W. B. Du, B. Yao and Q. Fang, Multifunctional Picoliter Droplet Manipulation Platform and Its Application in Single Cell Analysis, Anal. Chem., 2011, 83(19), 7570–7576 CrossRef CAS PubMed
.
- M. P. N. Bui, C. A. Li, K. N. Han, J. Choo, E. K. Lee and G. H. Seong, Enzyme Kinetic Measurements Using a Droplet-Based Microfluidic System with a Concentration Gradient, Anal. Chem., 2011, 83(5), 1603–1608 CrossRef CAS PubMed
.
- D. Lombardi and P. S. Dittrich, Droplet microfluidics with magnetic beads: a new tool to investigate drug-protein interactions, Anal. Bioanal. Chem., 2011, 399(1), 347–352 CrossRef CAS PubMed
.
- P. Siuti, S. T. Retterer, C.-K. Choi and M. J. Doktycz, Enzyme Reactions in Nanoporous, Picoliter Volume Containers, Anal. Chem., 2012, 84(2), 1092–1097 CrossRef CAS PubMed
.
- F. Shen, W. B. Du, E. K. Davydova, M. A. Karymov, J. Pandey and R. F. Ismagilov, Nanoliter Multiplex PCR Arrays on a SlipChip, Anal. Chem., 2010, 82(11), 4606–4612 CrossRef CAS PubMed
.
- A. Fallah-Araghi, J. C. Baret, M. Ryckelynck and A. D. Griffiths, A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution, Lab Chip, 2012, 12(5), 882–891 RSC
.
- Z. C. Gong, H. Zhao, T. H. Zhang, F. Nie, P. Pathak and K. M. Cui,
et al. Drug effects analysis on cells using a high throughput microfluidic chip, Biomed. Microdevices, 2011, 13(1), 215–219 CrossRef CAS PubMed
.
- M. A. Bennet, P. R. Richardson, J. Arlt, A. McCarthy, G. S. Buller and A. C. Jones, Optically trapped microsensors for microfluidic temperature measurement by fluorescence lifetime imaging microscopy, Lab Chip, 2011, 11(22), 3821–3828 RSC
.
- M. A. Khorshidi, P. K. P. Rajeswari, C. Wahlby, H. N. Joensson and H. A. Svahn, Automated analysis of dynamic behavior of single cells in picoliter droplets, Lab Chip, 2014, 14(5), 931–937 RSC
.
- K. Yasuda, A. Hattori, H. Kim, H. Terazono, M. Hayashi and H. Takei,
et al. Non-destructive on-chip imaging flow cell-sorting system for on-chip cellomics, Microfluid. Nanofluid., 2013, 14(6), 907–931 CrossRef CAS
.
- X. C. I. Solvas, M. Srisa-Art, A. J. Demello and J. B. Edel, Mapping of Fluidic Mixing in Microdroplets with 1 mu s Time Resolution Using Fluorescence Lifetime Imaging, Anal. Chem., 2010, 82(9), 3950–3956 CrossRef PubMed
.
- X. Casadevall i Solvas, X. Niu, K. Leeper, S. Cho, S.-I. Chang and J. B. Edel,
et al. Fluorescence detection methods for microfluidic droplet platforms, J. Visualized Exp., 2011,(58), 3437 Search PubMed
.
- J. Pei, Q. Li and R. T. Kennedy, Rapid and Label-Free Screening of Enzyme Inhibitors Using Segmented Flow Electrospray Ionization Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2010, 21, 1107–1113 CrossRef CAS PubMed
.
- Q. Li, J. Pei, P. Song and R. T. Kennedy, Fraction collection from capillary liquid chromatography and off-line electrospray Ionization Mass Spectrometry using oil segmented flow, Anal. Chem., 2010, 82, 5260–5267 CrossRef CAS PubMed
.
- S. Sun, R. T. Slaney and R. T. Kennedy, Label free screening of enzyme Inhibitors at femtomole scale using segmented flow electrospray ionization mass spectrometry, Anal. Chem., 2012, 84, 5794–5800 CrossRef CAS PubMed
.
- P. Song, N. D. Hershey, O. S. Mabrouk, T. R. Slaney and R. T. Kennedy, Mass Spectrometry “Sensor” for in Vivo Acetylcholine Monitoring, Anal. Chem., 2012, 84, 4659–4664 CrossRef CAS PubMed
.
- I. M. Lazar, J. Grym and F. Foret, Microfabricated devices: a new sample introduction approach to mass spectrometry, Mass Spectrom. Rev., 2006, 25, 573–594 CrossRef CAS PubMed
.
- C. A. Baker and M. G. Roper, Online Coupling of Digital Microfluidic Devices with Mass Spectrometry Detection Using an Eductor with Electrospray Ionization, Anal. Chem., 2012, 84(6), 2955–2960 CrossRef CAS PubMed
.
- X.-L. Wang, Y. Zhu and Q. Fang, Coupling liquid chromatography/mass spectrometry detection with microfluidic droplet array for label-free enzyme inhibition assay, Analyst, 2014, 139(1), 191–197 RSC
.
- A. E. Kirby and A. R. Wheeler, Microfluidic origami: a new device format for in-line reaction monitoring by nanoelectrospray ionization mass spectrometry, Lab Chip, 2013, 13(13), 2533–2540 RSC
.
- S. K. Kuester, S. R. Fagerer, P. E. Verboket, K. Eyer, K. Jefimovs and R. Zenobi,
et al. Interfacing Droplet Microfluidics with Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry: Label-Free Content Analysis of Single Droplets, Anal. Chem., 2013, 85(3), 1285–1289 CrossRef PubMed
.
- Y. Zhu and Q. Fang, Integrated Droplet Analysis System with Electrospray Ionization-Mass Spectrometry Using a Hydrophilic Tongue-Based Droplet Extraction Interface, Anal. Chem., 2010, 82(19), 8361–8366 CrossRef CAS PubMed
.
- J. Ji, L. Nie, L. Qiao, Y. Li, L. Guo and B. Liu,
et al. Proteolysis in microfluidic droplets: an approach to interface protein separation and peptide mass spectrometry, Lab Chip, 2012, 12(15), 2625–2629 RSC
.
- S. Sun, R. T. Slaney and R. T. Kennedy, Label free screening of enzyme Inhibitors at femtomole scale using segmented flow electrospray ionization mass spectrometry, Anal. Chem., 2012, 84, 5794–5800 CrossRef CAS PubMed
.
- L. Nie, G. Xu, J. Ji and Q. Liu,
et.al. Peptide-tight ESI/MSn analysis with segment of liquid chromatography effluent, Anal. Methods, 2013, 5, 3371 RSC
.
- Y. Su, Y. Zhu and Q. Fang, A multifunctional microfluidic droplet-array chip for analysis by electrospray ionization mass spectrometry, Lab Chip, 2013, 13, 1876 RSC
.
- N. Gasilova, Y. Yu, L. Qiao and H. H. Girault, On-Chip Spyhole Mass Spectrometry for Droplet-Based Microfluidics, Angew. Chem., Int. Ed., 2014, 53, 4408–4412 CrossRef CAS PubMed
.
- C. A. Smith, X. Li, T. H. Mize, T. D. Sharpe, E. I. Graziani and C. Abell,
et al. Sensitive, High Throughput Detection of Proteins in Individual, Surfactant-Stabilized Picoliter Droplets Using Nanoelectrospray Ionization Mass Spectrometry, Anal. Chem., 2013, 85(8), 3812–3816 CrossRef CAS PubMed
.
- A. E. Kirby, M. J. Jebrail, H. Yang and A. R. Wheeler, Folded emitters for nanoelectrospray ionization mass spectrometry, Rapid Commun. Mass Spectrom., 2010, 24(23), 3425–3431 CrossRef CAS PubMed
.
- A. E. Kirby and A. R. Wheeler, Digital Microfluidics: An Emerging Sample Preparation Platform for Mass Spectrometry, Anal. Chem., 2013, 85(13), 6178–6184 CrossRef CAS PubMed
.
- J. Prikryl, K. Kleparnik and F. Foret, Photodeposited silver nanoparticles for on-column surface-enhanced Raman spectrometry detection in capillary electrophoresis, J. Chromatogr. A, 2012, 1226, 43–47 CrossRef CAS PubMed
.
- A. Maerz, T. Henkel, D. Cialla, M. Schmitt and J. Popp, Droplet formation via flow-through microdevices in Raman and surface enhanced Raman spectroscopy-concepts and applications, Lab Chip, 2011, 11(21), 3584–3592 RSC
.
- M. P. Cecchini, J. Hong, C. Lim, J. Choo, T. Albrecht and A. J. deMello,
et al. Ultrafast Surface Enhanced Resonance Raman Scattering Detection in Droplet-Based Microfluidic Systems, Anal. Chem., 2011, 83(8), 3076–3081 CrossRef CAS PubMed
.
- W. Wang, C. Yang, X. Q. Cui, Q. L. Bao and C. M. Li, Droplet microfluidic preparation of au nanoparticles-coated chitosan microbeads for flow-through surface-enhanced Raman scattering detection, Microfluid. Nanofluid., 2010, 9(6), 1175–1183 CrossRef CAS
.
- R. Gao, N. Choi, S.-I. Chang, S. H. Kang, J. M. Song and S. I. Cho,
et al. Highly sensitive trace analysis of paraquat using a surface-enhanced Raman scattering microdroplet sensor, Anal. Chim. Acta, 2010, 681(1–2), 87–91 CrossRef CAS PubMed
.
- A. Walter, A. Maerz, W. Schumacher, P. Roesch and J. Popp, Towards a fast, high specific and reliable discrimination of bacteria on strain level by means of SERS in a microfluidic device, Lab Chip, 2011, 11(6), 1013–1021 RSC
.
- M. Li, J. Lu, J. Qi, F. Zhao, J. Zeng, J. C. Yu and W. C. Shih, Stamping surface-enhanced Raman spectroscopy for label-free, multiplexed, molecular sensing and imaging, J. Biomed. Opt., 2014, 5(19), 050501–050503 CrossRef PubMed
.
- C. Elbuken, T. Glawdel, D. Chan and C. L. Ren, Detection of microdroplet size and speed using capacitive sensors, Sens. Actuators, A, 2011, 171(2), 55–62 CrossRef CAS PubMed
.
- Y. Zhu and Q. Fang, Analytical detection techniques for droplet microfluidics-A review, Anal. Chim. Acta, 2013, 787, 24–35 CrossRef CAS PubMed
.
- E. V. Moiseeva, A. A. Fletcher and C. K. Harnett, Thin-film electrode based droplet detection for microfluidic systems, Sens. Actuators, B, 2011, 155(1), 408–414 CrossRef CAS PubMed
.
- B. P. Cahill, R. Land, T. Nacke, M. Min and D. Beckmann, Contactless sensing of the conductivity of aqueous droplets in segmented flow, Sens. Actuators, B, 2011, 159(1), 286–293 CrossRef CAS PubMed
.
- D. T. Chiu, Interfacing droplet microfluidics with chemical separation for cellular analysis, Anal. Bioanal. Chem., 2010, 397, 3179–3183 CrossRef CAS PubMed
.
- G. T. Roman, M. Wang, K. N. Shultz, C. Jennings and R. T. Kennedy, Sampling and electrophoretic analysis of segmented flow streams using virtual walls in a microfluidic device, Anal. Chem., 2008, 80, 8231–8238 CrossRef CAS PubMed
.
- J. Nie and R. T. Kennedy, Capillary liquid chromatography fraction collection and postcolumn reaction using segmented flow microfluidics, Journal of Separation Science, 2013, 36, 3471–3477 CrossRef CAS PubMed
.
- J. S. Edgar, C. P. Pabbati, R. M. Lorenz, M. He, G. S. Fiorini and D. T. Chiu, Capillary Electrophoresis Separation in the presence of an immiscible boundary for droplet analysis, Anal. Chem., 2006, 78, 6948–6954 CrossRef CAS PubMed
.
- X. Niu, F. Pereira, J. B. Edel and A. J. Mello, Droplet-interfaced microchip and capillary electrophoretic separations, Anal. Chem., 2013, 85, 8654–8660 CrossRef CAS PubMed
.
- I. Levin and A. Aharoni, Evolution in Microfluidic Droplet, Chem. Biol., 2012, 19(8), 929–931 CrossRef CAS PubMed
.
- J. J. Agresti, E. Antipov, A. R. Abate, K. Ahn, A. C. Rowat and J. C. Baret,
et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(9), 4004–4009 CrossRef CAS PubMed
.
- T. Geng, R. Novak and R. A. Mathies, Single cell Forensic Short Tandem Repeat Typing within Microfluidic Droplets, Anal. Chem., 2014, 86(1), 703–712 CrossRef CAS PubMed
.
- J. W. Ellefson, A. J. Meyer, R. A. Hughes, J. R. Cannon, J. S. Brodbelt and A. D. Ellington, Directed evolution of genetic parts and circuits by compartmentalized partnered replication, Nat. Biotechnol., 2014, 32(1), 97–101 CrossRef CAS PubMed
.
- T. Schneider, J. Kreutz and D. T. Chiu, The Potential Impact of Droplet Microfluidics in Biology, Anal. Chem., 2013, 85(7), 3476–3482 CrossRef CAS PubMed
.
- Y. Zhao, F. Pereira, A. J. deMello, H. Morgan and X. Z. Niu, Droplet-based in situ compartmentalization of chemically separated components after isoelectric focusing in a SlipChip, Lab Chip, 2014, 14(3), 555–561 RSC
.
- K. Kleparnik and F. Foret, Recent advances in the development of single cell analysis-A review, Anal. Chim. Acta, 2013, 800, 12–21 CrossRef CAS PubMed
.
- D. G. Spiller, C. D. Wood, D. A. Rand and M. R. H. White, Measurement of single cell dynamics, Nature, 2010, 465(7299), 736–745 CrossRef CAS PubMed
.
- S. Lindstrom and H. Andersson-Svahn, Miniaturization of biological assays – Overview on microwell devices for single cell analyses, Biochim. Biophys. Acta Gen. Subj., 2011, 1810(3), 308–316 CrossRef PubMed
.
- T. Konry, M. Dominguez-Villar, C. Baecher-Allan, D. A. Hafler and M. L. Yarmush, Droplet-based microfluidic platforms for single T cell secretion analysis of IL-10 cytokine, Biosens. Bioelectron., 2011, 26(5), 2707–2710 CrossRef CAS PubMed
.
- M. Y. Wu, M. Piccini, C. Y. Koh, K. S. Lam and A. K. Singh, Single Cell MicroRNA Analysis Using Microfluidic Flow Cytometry, PLoS One, 2013, 8(1), 55044 Search PubMed
.
- C.-Y. Koh, A. K. Singh, K. S. Lam, M. Wu and M. Piccini, Single Cell MicroRNA Analysis Using Microfluidic Flow Cytometry, Figshare, 2013, 8(1), 55044 Search PubMed
.
- T. D. Rane, H. C. Zec, C. Puleo, A. P. Lee and T.-H. Wang, Droplet microfluidics for amplification-free genetic detection of single cells, Lab Chip, 2012, 12(18), 3341–3347 RSC
.
- M. Rosenauer and M. J. Vellekoop, Characterization of a microflow cytometer with an integrated three-dimensional optofluidic lens system, Biomicrofluidics, 2011, 5(3), 044101 Search PubMed
.
- N. Hashemi, J. S. Erickson, J. P. Golden and F. S. Ligler, Optofluidic characterization of marine algae using a microflow cytometer, Biomicrofluidics, 2011, 5(3), 034102 CrossRef PubMed
.
- X. Mao, A. A. Nawaz, S.-C. S. Lin, M. I. Lapsley, Y. Zhao and J. P. McCoy,
et al. An integrated, multiparametric flow cytometry chip using “microfluidic drifting” based three-dimensional hydrodynamic focusing, Biomicrofluidics, 2012, 6(2), 02410 CrossRef PubMed
.
- M. L. Kovarik and N. L. Allbritton, Measuring enzyme activity in single cells, Trends Biotechnol., 2011, 29(5), 222–230 CrossRef CAS PubMed
.
- M. Najah, A. D. Griffiths and M. Ryckelynck, Teaching Single cell Digital Analysis Using Droplet-Based Microfluidics, Anal. Chem., 2012, 84(3), 1202–1209 CrossRef CAS PubMed
.
- M. T. Guo, A. Rotem, J. A. Heyman and D. A. Weitz, Droplet microfluidics for high-throughput biological assays, Lab Chip, 2012, 12(12), 2146–2155 RSC
.
- R. Arayanarakool, L. Shui, S. W. M. Kengen, A. van den Berg and J. C. T. Eijkel, Single-enzyme analysis in a droplet-based micro- and nanofluidic system, Lab Chip, 2013, 13(10), 1955–1962 RSC
.
- Z. C. Guan, Y. Zou, M. X. Zhang, J. Q. Lv, H. L. Shen and P. Y. Yang,
et al. A highly parallel microfluidic droplet method enabling single-molecule counting for digital enzyme detection, Biomicrofluidics, 2014, 8(1) CrossRef CAS PubMed
.
- J.-u. Shim, R. T. Ranasinghe, C. A. Smith, S. M. Ibrahim, F. Hollfelder and W. T. S. Huck,
et al. Ultrarapid Generation of Femtoliter Microfluidic Droplets for Single-Molecule-Counting Immunoassays, ACS Nano, 2013, 7(7), 5955–5964 CrossRef CAS PubMed
.
- P. Neuzil, C. D. M. Campos, C. C. Wong, J. B. W. Soon, J. Reboud and A. Manz, From chip-in-a-lab to lab-on-a-chip: towards a single handheld electronic system for multiple application-specific lab-on-a-chip (ASLOC), Lab chip, 2014, 14, 2168–2176 RSC
.
- M. Baker, Digital PCR hits its stride, Nat. Methods, 2012, 9(6), 541–544 CrossRef CAS
.
- A. D. Tadmor, E. A. Ottesen, J. R. Leadbetter and R. Phillips, Probing Individual Environmental Bacteria for Viruses by Using Microfluidic Digital PCR, Science, 2011, 333(6038), 58–62 CrossRef CAS PubMed
.
- Z. F. Quan, N. Ye, S. J. Chen, S. J. Cao, M. He and Q. G. Yan, High-throughput digital PCR in a low-cost and practical format introduction: the application progress in copy number variation, low-level detection and absolute quantification, Rev. Med. Microbiol., 2013, 24(4), 89–93 CrossRef
.
- A. K. White, K. A. Heyries, C. Doolin, M. VanInsberghe and C. L. Hansen, High-Throughput Microfluidic Single cell Digital Polymerase Chain Reaction, Anal. Chem., 2013, 85(15), 7182–7190 CrossRef CAS PubMed
.
- V. Sanchez-Freire, A. D. Ebert, T. Kalisky, S. R. Quake and J. C. Wu, Microfluidic single cell real-time PCR for comparative analysis of gene expression patterns, Nat. Protoc., 2012, 7(5), 829–838 CrossRef CAS PubMed
.
- B. J. Hindson, K. D. Ness, D. A. Masquelier, P. Belgrader, N. J. Heredia and A. J. Makarewicz,
et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number, Anal. Chem., 2011, 83(22), 8604–8610 CrossRef CAS PubMed
.
- D. J. Eastburn, A. Sciambi and A. R. Abate, Picoinjection Enables Digital Detection of RNA with Droplet RT-PCR, PLoS One, 2013, 8(4), 62961 Search PubMed
.
- V. Sanchez-Freire, Microfluidic single cell real-time PCR for comparative analysis of gene expression patterns, Nat. Protoc., 2012, 7(5), 829–838 CrossRef CAS PubMed
.
- Y. Li, Fast identification of foodborne pathogenic viruses using continuous-flow reverse transcription-PCR with fluorescence detection, Microfluid. Nanofluid., 2011, 10(2), 367–380 CrossRef CAS
.
- F. Moltzahnl, Microfluidic-Based Multiplex qRT-PCR Identifies Diagnostic and Prognostic microRNA Signatures in the Sera of Prostate Cancer Patients, Cancer Res., 2010, 71(2), 550–560 CrossRef PubMed
.
- Y. Zeng, R. Novak, J. Shuga, M. T. Smith and R. A. Mathies, High-Performance Single Cell Genetic Analysis Using Microfluidic Emulsion Generator Arrays, Anal. Chem., 2010, 82(8), 3183–3190 CrossRef CAS PubMed
.
- D. C. Saunders, G. L. Holst, C. R. Phaneuf, N. Pak, M. Marchese and N. Sondej,
et al. Rapid, quantitative, reverse transcription PCR in a polymer microfluidic chip, Biosens. Bioelectron., 2013, 44, 222–228 CrossRef PubMed
.
- K. A. Hagan, C. R. Reedy, M. L. Uchimoto, D. Basu, D. A. Engel and J. P. Landers, An integrated, valveless system for microfluidic purification and reverse transcription-PCR amplification of RNA for detection of infectious agents, Lab Chip, 2011, 11(5), 957–961 RSC
.
- A. Didelot, S. K. Kotsopoulos, A. Lupo, D. Pekin, X. Y. Li and I. Atochin,
et al. Multiplex Picoliter-Droplet Digital PCR for Quantitative Assessment of DNA Integrity in Clinical Samples, Clin. Chem., 2013, 59(5), 815–823 CAS
.
- V. Taly, D. Pekin, L. Benhaim, S. K. Kotsopoulos, D. Le Corre and X. Y. Li,
et al. Multiplex Picodroplet Digital PCR to Detect KRAS Mutations in Circulating DNA from the Plasma of Colorectal Cancer Patients, Clin. Chem., 2013, 59(12), 1722–1731 CAS
.
- J. Shuga, Y. Zeng, R. Novak, Q. Lan, X. J. Tang and N. Rothman,
et al. Single molecule quantitation and sequencing of rare translocations using microfluidic nested digital PCR, Nucleic Acids Res., 2013, 41(16), 159 CrossRef PubMed
.
- X. F. Leng, W. H. Zhang, C. M. Wang, L. A. Cui and C. J. Yang, Agarose droplet microfluidics for highly parallel and efficient single molecule emulsion PCR, Lab Chip, 2010, 10(21), 2841–2843 RSC
.
- Z. R. Xu, X. Wang, X. F. Fan and J. H. Wang, An extrusion fluidic driving method for continuous-flow polymerase chain reaction on a microfluidic chip, Microchim. Acta, 2010, 168(1–2), 71–78 CrossRef CAS
.
- J. R. Scherer, P. Liu and R. A. Mathies, Design and operation of a portable scanner for high performance microchip capillary array electrophoresis, Rev. Sci. Instrum., 2010, 81(11), 113105 CrossRef PubMed
.
- S. O. Sundberg, C. T. Wittwer, C. Gao and B. K. Gale, Spinning Disk Platform for Microfluidic Digital Polymerase Chain Reaction, Anal. Chem., 2010, 82(4), 1546–1550 CrossRef CAS PubMed
.
- G. H. Wang, H. P. Ho, Q. L. Chen, A. K. L. Yang, H. C. Kwok and S. Y. Wu,
et al. A lab-in-a-droplet bioassay strategy for centrifugal microfluidics with density difference pumping, power to disc and bidirectional flow control, Lab Chip, 2013, 13(18), 3698–3706 RSC
.
- F. Shen, B. Sun, J. E. Kreutz, E. K. Davydova, W. B. Du and P. L. Reddy,
et al. Multiplexed Quantification of Nucleic Acids with Large Dynamic Range Using Multivolume Digital RT-PCR on a Rotational SlipChip Tested with HIV and Hepatitis C Viral Load, J. Am. Chem. Soc., 2011, 133(44), 17705–17712 CrossRef CAS PubMed
.
- F. Shen, E. K. Davydova, W. B. Du, J. E. Kreutz, O. Piepenburg and R. F. Ismagilov, Digital Isothermal Quantification of Nucleic Acids via Simultaneous Chemical Initiation of Recombinase Polymerase Amplification Reactions on SlipChip, Anal. Chem., 2011, 83(9), 3533–3540 CrossRef CAS PubMed
.
- F. Shen, W. B. Du, J. E. Kreutz, A. Fok and R. F. Ismagilov, Digital PCR on a SlipChip, Lab Chip, 2010, 10(20), 2666–2672 RSC
.
- O. Strohmeier, N. Marquart, D. Mark, G. Roth, R. Zengerle and F. von Stetten, Real-time PCR based detection of a panel of food-borne pathogens on a centrifugal microfluidic “LabDisk” with on-disk quality controls and standards for quantification, Anal. Methods, 2014, 6(7), 2038–2046 RSC
.
- Q. Y. Zhu, L. Qiu, B. W. Yu, Y. N. Xu, Y. B. Gao and T. T. Pan,
et al. Digital PCR on an integrated self-priming compartmentalization chip, Lab Chip, 2014, 14(6), 1176–1185 RSC
.
- L. B. Pinheiro, V. A. Coleman, C. M. Hindson, J. Herrmann, B. J. Hindson and S. Bhat,
et al. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification, Anal. Chem., 2012, 84(2), 1003–1011 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2015 |


