Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Solution-based DNA-templating of sub-10 nm conductive copper nanowires†
Jonathan
Pate
a,
Felix
Zamora
b,
Scott M. D.
Watson
a,
Nicholas G.
Wright
c,
Benjamin R.
Horrocks
a and
Andrew
Houlton
*a
aChemical Nanoscience Laboratories, School of Chemistry, Newcastle University, Bedson Building, Newcastle upon Tyne, NE1 7RU, UK. E-mail: andrew.houlton@ncl.ac.uk
bDepartamento de Química Inorgánica Universidad Autónoma de Madrid, 28049 Madrid, Spain
cSchool of Electrical, Electronic and Computing Engineering, Newcastle University, Newcastle upon Tyne, NE1 7RU, UK
First published on 25th September 2014
Abstract
Templating the electroless reduction of metal ions on DNA is now an established route to the preparation of nanowires and can be particularly useful for the formation of nanowires in the desirable <10 nm size range. However, different preparation conditions produce nanowires of widely different morphologies and conductivities. We describe a method for the synthesis of Cu nanowires in which electroless metal deposition is carried out on DNA ‘template’ molecules in bulk solution. Though analogous to previous surface-based routes, importantly this now produces conductive material. AFM was used to evaluate the size and morphology of the resulting nanowires; a mean nanowire diameter of 7.1 nm (standard deviation = 4.7 nm) was determined from a statistical analysis of 100 nanowires and the Cu coatings were continuous and smooth. These findings represent a notable improvement in nanowire morphology in comparison to the previous surface-based routes. UV-vis spectroscopy, X-ray diffraction, and X-ray photoelectron spectroscopy were used to confirm formation of Cu(0) metal takes place during nanowire synthesis, and additional scanning probe microscopy techniques were employed to probe the electrical properties of the nanowires. The nanowires are less conductive [resistivity ∼ 2 Ω cm] than bulk Cu, but much more conductive than nanowires prepared by the analogous method on surface-bound DNA. Using an extension of our thermodynamic model for DNA-templating, we show that the templating process in bulk solution favours the formation of continuous nanowires compared to templating on surface-bound DNA.
Introduction
The development of new approaches for the fabrication of Cu nanowires offers much value to the semiconductor industry due to their potential application as interconnects in future nanoelectronic devices.1–3 Cu remains the material of choice for interconnect design due to its low electrical resistivity (1.7 × 10−6 Ω cm), high resistance to electromigration,4,5 and relatively low cost. The use of DNA as a template to promote low-dimensional material growth is well-established for a wide range of materials.6–34 The metallisation of DNA with Cu as a route to preparing 1D Cu nanostructures was first reported by Woolley et al.9,10 It was later shown by our group, however, that Cu nanostructures formed by this type of electroless deposition at surface-immobilised DNA were not electrically conducting.11 The absence of conductivity was proposed to be due to the highly granular nature of the Cu coatings, which consisted of linear arrangements of Cu nanoparticles packed along the DNA in a “beads-on-a-string” arrangement.Metal-seeding approaches have been combined with immobilised DNA; Kudo et al. used Pd seeding prior to electroless Cu deposition.12 However, no chemical or electrical characterisation was reported in this work. More recently, Woolley has also employed seeding in the fabrication of DNA/Cu structures, using Ag13 and Pd14 to promote electroless Cu deposition at DNA. Multiple seeding steps were applied in these instances, in order to provide good seed uniformity and density on the DNA. By using an electroless plating process to then form continuous Cu coatings upon the seeded DNA, structures with diameters in the region of 500 nm (Ag-seeded) and 40 nm (Pd-seeded) were demonstrated to be produced. In the case of the Pd-seeded structures, they were confirmed to be electrically conductive, though their resistivity was found to be orders of magnitude greater than that of bulk Cu.14 To the best of our knowledge, these are the only example of electrically conducting DNA-templated Cu nanostructures that have been reported to date.‡
Increases in the measured resistivity of metal wires are well-known when their dimensions are reduced to the nanoscale.16–19 Trends in the literature have previously been noted which highlight that the resistivities of metal nanowires tend to be larger as nanowires are reduced in diameter.35 In contrast to Ag where conductive DNA-templated nanowires have been prepared6,36 the formation of electrically conducting DNA-templated Cu nanowires of dimensions at the lower end of the nanoscale (i.e. sub-10 nm) offers significant challenges.
In the studies reported here, we explore a synthetic approach for the preparation of DNA-templated Cu nanostructures which is carried out in bulk solution. It is known from previous cases of DNA-templating reactions involving conducting polymers25 and inorganic semiconducting materials,26 that surface-immobilised DNA can yield a quite different morphology to solution-phase, with the former tending to show granular structure while the latter can be more regular and continuous. A reduction in the granular character of the Cu coatings formed around the DNA could be expected to result in reduced electron scattering, leading to improvements in conductivity. Here, we investigate the solution-based approach without hetero-metal seeding in order to allow more direct comparison with previous surface-based DNA/Cu nanostructures11 as well as offering a route to fabrication of 1D structures at the lower end of the nanometre scale range.
Results and discussion
Synthesis of DNA-templated Cu nanowires and AFM analysis
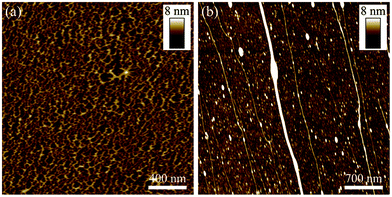
In contrast to previous templating at DNA pre-immobilised on a support surface (e.g. silicon wafer),9–11 the method used here involved mixing aqueous solutions of duplex DNA and Cu(NO3)2 in order to first ‘dope’ the DNA with Cu(II) cations. Divalent metal cations are generally known to be able to associate with the DNA duplex structure through the formation of interactions involving either binding to the phosphate groups in the DNA backbone, or donor sites in the bases.37–39 Formation of the final DNA-templated Cu structures was initiated by the addition of a solution of ascorbic acid, which acts to reduce the Cu(II) cations to Cu(0). During this step, as well as reduction of the dopant Cu(II) species bound to the DNA, further Cu(0) deposition can also take place due to the abundant Cu(II) cations in the solution which are available to undergo reduction at the surface of the DNA/Cu(0) structures as the templating reaction proceeds.In order to isolate the product material for AFM analysis, the reaction mixture was centrifuged to separate the DNA-templated Cu structures from non-templated Cu material also formed in the solution (see ESI, Fig. S1†). The structures were then isolated from the lower fraction of the solution and immobilised upon a Si wafer (modified with a trimethylsilane (TMS) monolayer) via molecular combing methods.25,40 Initial efforts to prepare the DNA-templated Cu structures showed that the Cu(II) concentration used in the templating solution had a notable effect upon the structural character of the product material. For example, templating solutions containing higher concentrations of Cu(II) (25–500 mM) were typically found to result in the DNA/Cu product material forming as dense networks, see Fig. 1(a). This network formation was attributed to association of the metal cations to the DNA causing effective charge neutralisation of the DNA's polyanionic phosphodiester backbone. Minimisation of the repulsive electrostatic forces between the DNA molecules as a result of this charge neutralisation allows the DNA to aggregate into the dense networks which were observed by AFM. In contrast, the use of much lower Cu(II) concentrations in the templating solutions (10–50 μM) appeared insufficient to promote such aggregation, and the resulting DNA/Cu product material was observed as more distinct 1D structures, see Fig. 1(b). Based on these initial studies, a set of reaction conditions were established which facilitated the production of distinct DNA-templated Cu nanowires with relatively uniform metallisation along the DNA. These were: λ-DNA (300 μg mL−1), Cu(NO3)2 (50 μM) and ascorbic acid (2 mM) mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v/v) ratio, and incubated at room temperature for 3 hours. An example of a DNA-templated Cu structure prepared under these conditions is shown in Fig. 2(a). Line profile measurements revealed the structure height to be ∼4.7 nm, confirming that metallisation of the DNA has taken place (the theoretical diameter of a duplex DNA molecule being ∼2 nm)§. The Cu coating around the DNA appears relatively smooth and continuous with no evident breaks. The morphology of the structures prepared in bulk solution here is, critically, different to the more granular structures observed when carrying out analogous templating reactions on surface-bound DNA.11 To have the possibility to act as true nanowires a continuous coverage along the structure length is required to provide the conductance pathway.
1 (v/v/v) ratio, and incubated at room temperature for 3 hours. An example of a DNA-templated Cu structure prepared under these conditions is shown in Fig. 2(a). Line profile measurements revealed the structure height to be ∼4.7 nm, confirming that metallisation of the DNA has taken place (the theoretical diameter of a duplex DNA molecule being ∼2 nm)§. The Cu coating around the DNA appears relatively smooth and continuous with no evident breaks. The morphology of the structures prepared in bulk solution here is, critically, different to the more granular structures observed when carrying out analogous templating reactions on surface-bound DNA.11 To have the possibility to act as true nanowires a continuous coverage along the structure length is required to provide the conductance pathway.
The observed difference between the current preparation method and previous ones9,10,12,41 can be rationalised by considering a thermodynamic model of the templating process which describes how the formation of materials such as metals, semiconductors and conjugated polymers on a long, thin template, like DNA, will produce smooth 1D structures under the appropriate conditions.41 This outcome results from the competition between the surface tension of the material γ and the line energy σ (representing its adhesion to the template). In the absence of this line energy, a cylindrical wire is unstable and will break up into spherical droplets; this is the well-known Savart–Plateau–Rayleigh instability. The effect of the line energy can be appreciated by considering states of the templated material characterised by a linear dimension, r, which may be the radius of a series of identical droplets or the thickness of a cylinder. The free energy of the system per unit volume of templated material has two terms; one proportional to r−1 which originates in the surface tension, and a second proportional to r−2 from the line energy (eqn (1); written for the case of spheres). When the volume of templated material per unit length (V) is small enough, the line energy dominates and the lowest free energy state is a smooth cylinder, as we have previously discussed in more detail elsewhere.41
 | (1) |
However, if we extend this theoretical description of the templating process to a scenario in which the template lies on a solid substrate, the situation changes because there are additional terms in the free energy to consider; the adhesion of the material to the substrate (∼r−1) and a line energy at the 3-phase boundary at the edge of the region where the templated material meets the substrate (∼r−2) (see ESI, Fig. S2†):
 | (2) |
Chemical characterisation of DNA-templated Cu nanowires
In order to verify that reduction of the Cu(II) proceeds in the templating solution, experiments were carried out in which the reaction was monitored by UV-vis spectroscopy. Formation of the desired Cu(0) was evident in the UV-vis spectrum by the presence of broad plasmon band in the 540–630 nm region, consistent with nanostructured Cu(0) material (see ESI, Fig. S3†).43–45However, it should be noted that whilst UV-vis studies provided evidence of the reduction of the Cu(II) species to Cu(0), this data does not offer information which verifies that the Cu(0) forms directly on the DNA. To explore this aspect of the Cu(0) formation, FTIR spectroscopy was carried out upon samples of ‘bare’ DNA and DNA/Cu material isolated from the templating solution (see ESI, Fig. S4 and Table S1†). Comparison of the spectra showed several small shifts in wavenumber, as well as changes in intensity, of IR bands associated with vibrations of the DNA backbone and bases. These changes in the spectrum are attributed to interactions between the DNA and Cu(0), indicating that Cu(0) forms directly on the DNA; observations of IR band changes have previously been reported for the preparation of other materials on DNA.23–25
X-ray diffraction data (XRD) data obtained from a powder sample of the DNA/Cu material¶ confirmed that the Cu(0) produced is of high purity. No additional peaks where evident in the diffraction pattern indicating the presence of impurities (e.g. CuO, Cu2O) (see ESI, Fig. S5†) even following a 24 hour period, where the sample was allowed to stand in air. We ascribe this to the relative lack of surface sensitivity of the technique and to indicate the amorphous nature of the oxidized surface (vide infra). Further chemical analysis of the DNA/Cu material was carried out using X-ray photoelectron spectroscopy (XPS) in order to verify if the Cu possesses any thin surface oxide layer which was beyond the detection limits of the XRD instrumentation. Fig. 3 shows a high resolution XP spectrum of the Cu2p region, acquired from a sample of the DNA/Cu material. The curve fitting shows that the experimental data can be fitted to a minimum of two doublets with binding energies of 932.8 eV and 934.9 eV (Cu2p3/2), respectively. The doublet at lower binding energy (1) is consistent with the presence of Cu(0),46–48 in agreement with the findings of the XRD data. It is noted here that it can often be difficult to distinguish between Cu(0) and Cu(I) species in Cu2p XP spectra due to their similar binding energies, meaning that one could also consider the possibility of the presence of Cu2O, along with Cu(0), at the metal surface. However, the O1s XP spectrum acquired from the sample shows little evidence of a component in the O1s envelope centred around the expected binding energy (∼530.4 eV) for oxygen species related to Cu2O,48,49 suggesting this oxide is not present (see ESI, Fig. S6†).
The doublet at higher binding energy (2) is consistent with the presence of Cu(II) species, as are the two characteristic satellite peaks seen in the 938–947 eV region of the spectrum.48,49 This can be attributed to Cu(II) species, originating from the Cu(NO3)2 starting material.||
Electrical characterisation of DNA-templated Cu nanowires
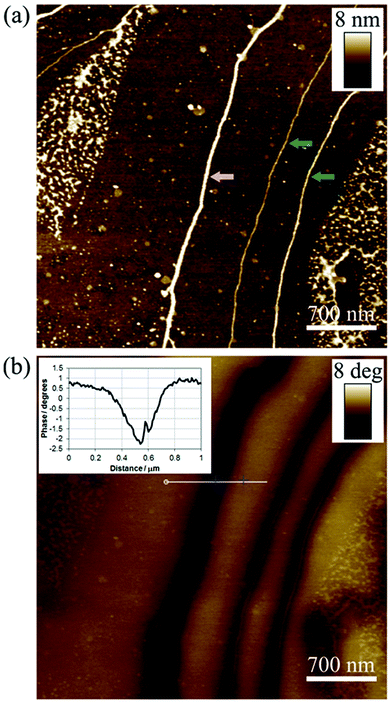
In order to determine the suitability of the resulting DNA/Cu structures to function as nanowires – i.e. act as 1D nanostructures along which an electrical current can be passed – it was important to evaluate their electrical properties. Given the non-conducting nature of highly granular Cu nanostructures prepared using surface-bound DNA templates,11 the more regular morphologies of the DNA/Cu structures prepared here proved encouraging in this regard. Initially, the electrical properties of individual DNA/Cu structures were probed in a qualitative fashion using scanning conductance microscopy (SCM).50–52Fig. 4(a) shows an AFM height image of three DNA-templated Cu structures immobilised upon a TMS-modified Si/200 nm SiO2 wafer. The main DNA/Cu structure (white arrow) seen in the image has a mean diameter of 5.5 nm, whilst the additional two structures (green arrows) have mean diameters of 2.8 nm and 4.2 nm, respectively. The corresponding SCM phase map (Fig. 4(b)), acquired at an applied direct current (dc) bias of −10 V, shows dark contrast corresponding to the positions of all three DNA/Cu structures on the wafer surface. Dark phase contrast associated with the structures was observed over the full range of bias potentials applied (−10 V to +10 V), with the magnitude of the phase shifts being proportional to V2 (see ESI, Fig. S9†). It has previously been demonstrated in other SCM experiments that the occurrence of negative phase shifts in this manner can only result if the object under analysis is electrically conducting,50,51 confirming that the DNA-templated Cu structures here are, to some extent, electrically conductive. It is of interest to note that even the smallest of the three DNA/Cu structures examined (2.8 nm diameter) is shown to be conductive. Hence it can be concluded that with this solution-based synthetic approach to nanowire synthesis, even relatively thin Cu coatings formed upon the DNA molecules, the morphology is such that a continuous electrical conducting pathway is available along the structure.In order to obtain quantitative information regarding the electrical properties of the DNA-templated Cu structures, conductive AFM (cAFM) was carried out, using our previously established procedure.32,33 Briefly, this involved depositing a dense network of the DNA/Cu nanowires onto a TMS-modified Si/200 nm SiO2 wafer; at the periphery of this dense deposit, small bundles of DNA/Cu nanowires could found protruding radially outwards, see Fig. 5(a). The dense nanowire deposit was used to create an electrical contact between the sample material and the metallic chuck. The DNA/Cu structures located at the edge of the dense nanowire deposits were then able to be probed by cAFM, with the Ga–In eutectic/nanowire network acting as one electrical contact, and the conductive AFM tip used as the second contact. The areas of the sample surface scanned during cAFM measurements were typically located ≤1 mm away from the eutectic contact (see ESI, Fig. S10†).
Fig. 5(b) shows a contact mode height image, acquired at the edge of a dense DNA/Cu nanowire deposit, in which several DNA/Cu structures can be seen extending across the surface of the Si/SiO2 wafer. The heights of these structures (typically ∼11–20 nm in this instance), as well as the presence of ‘branching’, indicates them to consist of multiple DNA/Cu nanowires bundled together into thicker rope-like structures. The use of the these larger ‘nanoropes’ has generally been found to be a necessary requirement for these cAFM experiments in order to achieve reliable imaging of the samples in contact mode operation.** A current map, simultaneously acquired alongside the height data at an applied bias of +8 V, is shown in Fig. 5(c). The conductive nature of the DNA/Cu structures is evident, with current signals of the order of tens of nanoamps seen to correlate with the positions of the DNA/Cu nanoropes (see Fig. 5(d)). In contrast, no current signal was detected across regions where the AFM tip was in contact with the dielectric SiO2 surface. A current–voltage (I–V) plot of an individual DNA/Cu nanorope was produced through recording a series of current maps, each at a different applied bias (see Fig. 6). An approximate value for the current passing through the nanorope was determined from each current map using line profile measurements carried out at the same point of the nanorope. The resulting plot clearly shows a linear relationship between the current and applied bias. A non-zero current value at zero bias was also noted to be indicated from plot trend line, possibly a consequence of stray capacitance in the circuit. From the slope of the regression line, a resistance of ∼107 MΩ was estimated for the nanorope. In order to calculate a resistivity value, the diameter was taken to be ∼20 nm, based upon AFM height measurements, and the length ∼1.5 μm, which represented the approximate distance between the edge of the nanowire network and the point at which the cAFM line profile measurements were made. Based upon these dimensions, a resistivity value of ∼2 Ω cm was determined. This is two orders of magnitude higher than that reported by Woolley for Pd-seeded DNA-templated Cu nanostructures (3.6 × 10−2 Ω cm),14 and six orders of magnitude higher than that of pure bulk Cu (1.7 × 10−6 Ω cm).
We attribute this high resistivity to excess grain boundaries and surface scattering of electrons in the nanowire which are known to inhibit electrical conductivity in thin Cu wires.53 The reduced size of the structures prepared here, in comparison to the previous larger (≥40 nm) Pd-seeded DNA/Cu structures, is expected to play a significant role in causing the increased resistivity, with the effects of grain boundaries and electron surface scattering becoming more significant with the downscaling in structure size.35
Based on these findings, the resistivities of the DNA-templated Cu nanowires prepared in the current studies, are in fact more comparable with that of a lightly doped specimen of silicon, rather than a true metal. Whilst a significant improvement is shown here compared to our previous results for DNA-templated Cu nanowires prepared using surface-bound DNA, which were absent of conductivity,11 there is still a considerable effect of resistance increasing mechanisms, such as grain boundaries, in these samples. These samples are therefore best described as granular metals.54,55
Conclusions
The solution-based synthesis of DNA-templated Cu nanostructures has been demonstrated for the preparation of 1D structures. These show improved metallisation of the DNA template in comparison to our previous, surface-based approach.11 This improvement in the metallisation of the DNA was observed in the form of more extensive metal deposition along the DNA molecules, and smoother morphologies associated with the final DNA/Cu structures. The distinction between the structures produced by the two different approaches to fabrication can be rationalised using a thermodynamic model to describe templated metal growth in the two different scenarios. The model highlights that, under appropriate conditions, the formation of smooth nanowires can be expected when the template reaction is carried out in bulk solution, whilst the additional influence of the substrate surface when carrying out the templating reaction via the surface-based route, will instead lead to formation of a series of metal particles decorating the length of the DNA.SPM studies confirmed the DNA/Cu structures to be electrically conductive, with improvements in the electrical properties, compared to our previous examples of DNA-templated Cu structures.11 This was largely attributed to differences in the morphologies of the metal coatings formed in the two different approaches to synthesis. However, it was also noted that the resistivity of the structures remained several orders of magnitude higher than that of bulk Cu. This suggests that whilst improvements in the morphologies of the Cu coatings have been demonstrated here, there remain notable issues, such as the presence of grain boundaries, causing significant increases in the resistivities of the DNA/Cu structures produced.
Experimental
Materials
All chemicals, unless otherwise stated, were purchased from Sigma-Aldrich Company Ltd., of Analar grade or equivalent, and used as received. Ascorbic acid (99+%) was purchased from Alfa Aesar. Lambda DNA (Bacteriophage lambda, λ-DNA, 500 μg mL−1, 10 mM tris-HCl, 1 mM EDTA, pH 8, Cat no. N3011S) was purchased from New England Biolabs (UK) Ltd. Calf thymus DNA (CT-DNA, highly polymerised, 6% sodium) was purchased from Sigma-Aldrich Company Ltd. Silicon wafers (100 mm diameter, 525 ± 50 μm thickness, polished on one side with reverse etched, phosphorus doped, 1–10 Ω cm resistance) and silicon wafers (100 mm diameter, 500 ± 25 μm, double side polished, arsenic doped, <0.005 Ω cm resistance, thermal oxide layer 2000 Å ± 10% thickness) were purchased from Compart Technology Ltd. NANOpure® deionised water (18 MΩ cm resistivity) was obtained from a NANOpure® DIamond™ Life Science ultrapure water system equipped with a DIamond™ RO Reverse Osmosis System (Barnstead International).Cleaning and preparation of silicon wafers
Silicon < p-100 > wafers were cut into ∼10 mm × 10 mm chips, and washed in serial fashion in acetone, propanol, and NANOpure® water. The wafers were subsequently washed in a hot surfactant solution (sodium dodecyl sulfate, 0.01 g per 100 mL water) for 20 min, before rinsing with copious amounts of NANOpure® water. This was followed by treating the wafers in ‘Piranha’ solution (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 H2SO4
1 H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2) for 45 min at ∼50 °C. (Caution! Piranha solution should be handled with extreme care; it is a strong oxidant and reacts violently with many organic materials. It also presents an explosion danger). Upon removal from the Piranha solution, the wafers were rinsed again with NANOpure® water and baked in an oven for 30 min.
H2O2) for 45 min at ∼50 °C. (Caution! Piranha solution should be handled with extreme care; it is a strong oxidant and reacts violently with many organic materials. It also presents an explosion danger). Upon removal from the Piranha solution, the wafers were rinsed again with NANOpure® water and baked in an oven for 30 min.
Chemical modification of the clean Si wafer surfaces with a self-assembled monolayer of trimethylsilane (TMS) was carried out by exposing the wafers to the vapour of chlorotrimethylsilane (Me3SiCl) for 8 min, at room temperature.
For SCM and cAFM experiments, Si wafers with 200 nm thick SiO2 capping layer were used as the substrate supports for the sample material. In the case of these wafers, the SiO2 layer was required to be stripped from one side. This was carried out by applying a solution of HF (aq. 10%) to one side of the wafer, which acts to etch the SiO2.
Preparation of Cu-DNA nanowires
The preparation of DNA-templated Cu nanostructures which provided the most uniform, and continuous metal coatings around the DNA templates was carried out as follows: a solution of Cu(NO3)2·3H2O (aq. 50 μM, 10 μL) was added to λ-DNA solution (aq. 300 μg mL−1, 10 μL), followed by addition a solution of ascorbic acid (aq. 2 mM, 10 μL). The templating reaction was left to proceed for 3 h, with gentle stirring on a mechanical roller. To separate the suspension of nanowires from non-templated Cu nanoparticles also formed, the sample was centrifuged at 8000 rpm for 2 min. The bottom fraction, containing the desired DNA/Cu nanowire material, was then collected for analysis.Alignment of Cu-DNA nanostructures upon substrates
DNA-templated Cu nanostructures were aligned on TMS-modified Si surfaces using a molecular combing/drop-drying method. A 5 μL volume of the templating solution containing the DNA/Cu nanowires product material was applied to the wafer surface by micropipette. The nanowires were then ‘combed’ across the surface by slowly withdrawing the solution from the wafer, and leaving a small residual amount of the solution behind. The sample was allowed to dry, leaving a small stain on the surface from near which individual nanostructures could be identified by AFM.X-ray photoelectron spectroscopy (XPS)
XPS analysis was carried out using a Thermo K-Alpha XPS system with a monochromated Al Kα X-ray source. An electron/ion gun was used to compensate for charge build-up on the sample during analysis, and all binding energies were referenced to the C1s hydrocarbon peak at 284.6 eV. Shirley background subtractions used for the Cu2p spectra, and peaks fitted using Gaussian–Lorentzian functions. Samples were prepared in powder form, in a similar manner as that described for XRD experiments, with samples drop-cast onto a clean Si wafer and dried in vacuo.Scanning probe microscopy
TappingMode™ AFM imaging was performed in air, using Multimode Nanoscope IIIa and Dimension Nanoscope V instruments (Veeco Instruments Inc., Metrology Group, Santa Barbara, CA), with TESP (n-doped Si cantilevers, resonant frequency = 230–410 kHz, spring force constant = 20–80 N m−1 Veeco Instruments Inc.), and Tap300Al-G (Si cantilevers, resonant frequency = 200–400 kHz, spring force constant = 20–75 N m−1, BudgetSensors) probes. Data acquisition was carried out using Nanoscope software version 5.12b36 (Multimode) and Nanoscope software version 7.00b19 (Dimension Nanoscope V) (Veeco Instruments Inc., Digital Instruments). For both AFM systems, vibrational noise was reduced with an isolation table (Veeco Inc., Metrology Group).Scanning conductance microscopy was performed in air, on a Dimension Nanoscope V system, using MESP probes (n-doped Si cantilevers, metallic Co/Cr coating, resonant frequency = 50–100 kHz, spring force constant = 1–5 N m−1, Veeco Instruments Inc.). Data acquisition was carried out using Nanoscope version 7.00b19 software. SCM measurements were performed using a two-pass method, in which the first pass of the tip over a scan line gathers topographical data in standard TappingMode™, and the second pass of the tip records the SCM phase information. During the second pass, the tip travelled over the sample surface at a constant lift height (typically set between 40–100 nm), whilst an independently controlled dc bias was applied to the sample, with the tip grounded.
Conductive AFM (cAFM) experiments were performed on a Dimension Nanoscope V AFM, using MESP probes. Both topographical data (obtained in contact mode) and current maps were acquired simultaneously during cAFM operation. Samples were prepared by drop casting 5–40 μL of the DNA/Cu nanowire suspension onto a TMS-modified Si/200 nm SiO2 wafer, and leaving the droplet to evaporate at room temperature. The surface tension created by the receding meniscus is enough to align the wires on the substrate parallel to the direction of solvent withdrawal. This results in a dense deposit of nanomaterial on the wafer surface, with aligned wires extending from the periphery of this dense body of material. A drop of Ga–In eutectic was applied between the dense nanomaterial deposit and the AFM metallic chuck to provide an electrical contact. The metallic AFM tip was used to act as the second electrical contact to the sample. The AFM tip was positioned over the surface region of interest for data acquisition, approximately ≤1 mm from the eutectic contact. A bias of between 1–10 V was applied to the sample in order to obtain sufficient current signals for analysis. All measurements were made at room temperature.
Acknowledgements
X-ray photoelectron spectra were obtained at the National EPSRC XPS User's Service (NEXUS) at Newcastle University, an EPSRC mid-range facility. This work was financially supported by Newcastle University, EU ITN NANOEMBRACE (Contract No. 316751) and Intel Ireland Ltd (with special thanks to Bernie D. Capraro, Research Programme Manager).Notes and references
- Y. Huang, X. Duan, Y. Cui, L. J. Lauhon, K.-H. Kim and C. M. Lieber, Science, 2001, 294, 1313–1317 CrossRef CAS PubMed
.
- Y. Huang, X. Duan, Q. Wei and C. M. Lieber, Science, 2001, 291, 630–633 CrossRef CAS PubMed
.
- C. M. Lieber, MRS Bull., 2003, 28, 486–491 CrossRef CAS
.
- M. Lane, R. H. Dauskardt, N. Krishna and I. Hashim, J. Mater. Res., 2000, 15, 203–211 CrossRef CAS
.
- S. Strehle, J. W. Bartha and K. Wetzig, Thin Solid Films, 2009, 517, 3320–3325 CrossRef CAS PubMed
.
- E. Braun, Y. Eichen, U. Sivan and G. Ben-Yoseph, Nature, 1998, 391, 775–778 CrossRef CAS PubMed
.
- A. Houlton and S. M. D. Watson, Annu. Rep. Prog. Chem., Sect. A: Inorg. Chem., 2011, 107, 21–42 RSC
.
- A. Houlton, A. R. Pike, M. Angel Galindo and B. R. Horrocks, Chem. Commun., 2009, 1797–1806 RSC
.
- C. F. Monson and A. T. Woolley, Nano Lett., 2003, 3, 359–363 CrossRef CAS
.
- R. M. Stoltenberg and A. T. Woolley, Biomed. Microdevices, 2004, 6, 105–111 CrossRef CAS
.
- S. M. D. Watson, N. G. Wright, B. R. Horrocks and A. Houlton, Langmuir, 2010, 26, 2068–2075 CrossRef CAS PubMed
.
- H. Kudo and M. Fujihira, IEEE Trans. Nanotechnol., 2006, 5, 90–92 CrossRef
.
- J. Liu, B. Uprety, S. Gyawali, A. T. Woolley, N. V. Myung and J. N. Harb, Langmuir, 2013, 29, 11176–11184 CrossRef CAS PubMed
.
- Y. Geng, A. C. Pearson, E. P. Gates, B. Uprety, R. C. Davis, J. N. Harb and A. T. Woolley, Langmuir, 2013, 29, 3482–3490 CrossRef CAS PubMed
.
- S. M. D. Watson, H. D. A. Mohamed, B. R. Horrocks and A. Houlton, Nanoscale, 2013, 5349–5359 RSC
.
- M. Al Hinai, N. G. Wright, A. Horsfall, R. Hassanien, B. R. Horrocks and A. Houlton, IEEE Sens. J., 2011, 1–4 Search PubMed
.
- K. Keren, M. Krueger, R. Gilad, G. Ben-Yoseph, U. Sivan and E. Braun, Science, 2002, 297, 72–75 CrossRef CAS PubMed
.
- J. Richter, M. Mertig, W. Pompe, I. Mönch and H. K. Schackert, Appl. Phys. Rev., 2001, 78, 536–538 CAS
.
- S. H. Park, M. W. Prior, T. H. LaBean and G. Finkelstein, Appl. Phys. Lett., 2006, 89, 033901 CrossRef PubMed
.
- H. A. Becerril, R. M. Stoltenberg, C. F. Monson and A. T. Woolley, J. Mater. Chem., 2004, 14, 611–616 RSC
.
- H. A. Becerril, R. M. Stoltenberg, D. R. Wheeler, R. C. Davis, J. N. Harb and A. T. Woolley, J. Am. Chem. Soc., 2005, 127, 2828–2829 CrossRef CAS PubMed
.
- K. Nguyen, M. Monteverde, A. Filoramo, L. Goux-Capes, S. Lyonnais, P. Jegou, P. Viel, M. Goffman and J.-P. Bourgoin, Adv. Mater., 2008, 20, 1099–1104 CrossRef CAS
.
- H. D. A. Mohamed, S. M. D. Watson, B. R. Horrocks and A. Houlton, Nanoscale, 2012, 4, 5936 RSC
.
- R. Hassanien, S. A. F. Al-Said, L. Šiller, R. Little, N. G. Wright, A. Houlton and B. R. Horrocks, Nanotechnology, 2012, 23 Search PubMed
.
- L. Dong, T. Hollis, S. Fishwick, B. A. Connolly, N. G. Wright, B. R. Horrocks and A. Houlton, Chem.–Eur. J., 2007, 13, 822–828 CrossRef CAS PubMed
.
- L. Dong, T. Hollis, B. A. Connolly, N. G. Wright, B. R. Horrocks and A. Houlton, Adv. Mater., 2007, 19, 1748–1751 CrossRef CAS
.
- W. U. Dittmer and F. C. Simmel, Appl. Phys. Lett., 2004, 85, 633–635 CrossRef CAS PubMed
.
- N. Li, F. Gao, L. Hou and D. Gao, J. Phys. Chem. C, 2010, 114, 16114–16121 CAS
.
- D. S. Hopkins, D. Pekker, P. M. Goldbart and A. Bezryadin, Science, 2005, 308, 1762–1765 CrossRef CAS PubMed
.
- Y. Ma, J. Zhang, G. Zhang and H. He, J. Am. Chem. Soc., 2004, 126, 7097–7101 CrossRef CAS PubMed
.
- S. A. F. Al-Said, R. Hassanien, J. Hannant, M. A. Galindo, S. Pruneanu, A. R. Pike, A. Houlton and B. R. Horrocks, Electrochem. Commun., 2009, 11, 550–553 CrossRef PubMed
.
- J. Hannant, J. H. Hedley, J. Pate, A. Walli, S. A. Farha Al-Said, M. A. Galindo, B. A. Connolly, B. R. Horrocks, A. Houlton and A. R. Pike, Chem. Commun., 2010, 46, 5870–5872 RSC
.
- R. Hassanien, M. Al-Hinai, S. A. Farha Al-Said, R. Little, L. Siller, N. G. Wright, A. Houlton and B. R. Horrocks, ACS Nano, 2010, 4, 2149–2159 CrossRef CAS PubMed
.
- S. M. D. Watson, J. H. Hedley, M. A. Galindo, S. A. F. Al-Said, N. G. Wright, B. A. Connolly, B. R. Horrocks and A. Houlton, Chem.–Eur. J., 2012, 18, 12008–12019 CrossRef CAS PubMed
.
- K. Critchley, B. P. Khanal, M. Ł. Górzny, L. Vigderman, S. D. Evans, E. R. Zubarev and N. A. Kotov, Adv. Mater., 2010, 22, 2338–2342 CrossRef CAS PubMed
.
- S. H. Park, H. Yan, J. H. Reif, T. H. LaBean and G. Finkelstein, Nanotechnology, 2004, 15, S525–S527 CrossRef CAS
.
- G. L. Eichhorn, Nature, 1962, 194, 474–475 CrossRef CAS
.
- Metal Ions in Biological Systems: Interactions of Metal Ions with Nucleotides, Nucleic Acids, and Their Constituents, vol. 32, ed. A. Sigel and H. Sigel, CRC Press, New York, 1996 Search PubMed.
- G. L. Eichhorn and Y. A. Shin, J. Am. Chem. Soc., 1968, 90, 7323–7328 CrossRef CAS
.
- J. Li, C. Bai, C. Wang, C. Zhu, Z. Lin, Q. Li and E. Cao, Nucleic Acids Res., 1998, 26, 4785–4786 CrossRef CAS PubMed
.
- S. M. D. Watson, A. Houlton and B. R. Horrocks, Nanotechnology, 2012, 23 Search PubMed
.
- Y. Liu, J. Wang and X. Zhang, Sci. Rep., 2013, 3, 2008 Search PubMed
.
- H. H. Huang, F. Q. Yan, Y. M. Kek, C. H. Chew, G. Q. Xu, W. Ji, P. S. Oh and S. H. Tang, Langmuir, 1997, 13, 172–175 CrossRef CAS
.
- Z. Liu, Y. Yang, J. Liang, Z. Hu, S. Li, S. Peng and Y. Qian, J. Phys. Chem. B, 2003, 107, 12658–12661 CrossRef CAS
.
- D. Mott, J. Galkowski, L. Wang, J. Luo and C.-J. Zhong, Langmuir, 2007, 23, 5740–5745 CrossRef CAS PubMed
.
- Y. Wang, A. V. Biradar, G. Wang, K. K. Sharma, C. T. Duncan, S. Rangan and T. Asefa, Chem.–Eur. J., 2010, 16, 10735–10743 CrossRef CAS PubMed
.
- J. Hernandez, P. Wrschka and G. S. Oehrlein, J. Electrochem. Soc., 2001, 148, G389–G397 CrossRef CAS PubMed
.
- I. Platzman, R. Brener, H. Haick and R. Tannenbaum, J. Phys. Chem. C, 2008, 112, 1101–1108 CAS
.
- N. S. McIntyre and M. G. Cook, Anal. Chem., 1975, 47, 2208–2213 CrossRef CAS
.
- M. Bockrath, N. Markovic, A. Shepard, M. Tinkham, L. Gurevich, L. P. Kouwenhoven, M. W. Wu and L. L. Sohn, Nano Lett., 2002, 2, 187–190 CrossRef CAS
.
- C. Staii, A. T. Johnson Jr and N. J. Pinto, Nano Lett., 2004, 4, 859–862 CrossRef CAS
.
- S. Pruneanu, S. A. F. Al-Said, L. Dong, T. A. Hollis, M. A. Galindo, N. G. Wright, A. Houlton and B. R. Horrocks, Adv. Funct. Mater., 2008, 18, 2444–2454 CrossRef CAS
.
- A. Bid, A. Bora and A. K. Raychaudhuri, Phys. Rev. B: Condens. Matter Mater. Phys., 2006, 74, 035426 CrossRef
.
- A. L. Efros and B. I. Shklovskii, J. Phys. C: Solid State Phys., 1975, 8, L49 CrossRef CAS
.
- B. Abeles, P. Sheng, M. D. Coutts and Y. Arie, Adv. Phys., 1975, 24, 407 CrossRef CAS
.
- Z. Zhou, Y. Du and S. Dong, Anal. Chem., 2011, 83, 5122–5127 CrossRef CAS PubMed
.
- L. Zhang, J. Zhao, H. Zhang, J. Jiang and R. Yu, Biosens. Bioelectron., 2013, 44, 6–9 CrossRef CAS PubMed
.
- Y.-R. Liu, R. Hu, T. Liu, X.-B. Zhang, W. Tan, G.-L. Shen and R.-Q. Yu, Talanta, 2013, 107, 402–407 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available: AFM data of non-templated Cu material and DNA-templated Cu structures, cartoon scheme highlighting aspects of the thermodynamic model describing material growth on a surface-bound template, UV-vis spectra obtained from DNA/Cu(II) templating solutions, FTIR spectra and peak assignments for bare DNA and DNA/Cu material, powder XRD data for DNA/Cu material, XP spectrum of O1s and N1s core levels from DNA/Cu material, SCM phase data plot for DNA/Cu nanowire, and additional cAFM data for DNA/Cu nanowires. See DOI: 10.1039/c4tc01632g |
| ‡ As this work is concerned with the preparation of continuous conducting 1D structures we do not consider the various reports that seek to prepare copper cluster/particles on DNA as reported elsewehere.56–58 |
| § The theoretical diameter of a molecule of duplex DNA is 2 nm, though AFM is well-known to underestimate this size (likely due to tip-induced compression of the molecule), with values as low as <1.0 nm commonly measured. |
| ¶ Samples for XRD analysis required some changes to the sample preparation compared to samples produced for AFM analysis; specifically, the reagent concentrations in the templating solution were increased. This was necessary in order to obtain sufficient amounts of the product material, in powder form, from which meaningful XRD data could be acquired. As a result, certain distinctions between the DNA/Cu materials produced for AFM and XRD studies were noted. In particular, Scherrer analysis of the XRD data showed the powder sample to have an average crystallite size of ∼35 nm; this was notably larger than the mean DNA/Cu nanostructure diameter of ∼7 nm, determined for AFM samples. |
| || The binding energy of the fitted component relating to Cu(II) species in the Cu2p spectrum also falls in good agreement with the expected value for Cu(OH)2; this presumably appearing as a thin layer on the surface of the DNA/Cu material. However, due to the close overlap between expected binding energies of oxygen species in Cu(OH)2 and in DNA (∼531.0–531.8 eV), it cannot be readily verified from the O1s spectrum if Cu(OH)2 is present. |
| ** cAFM studies were limited to studying small bundles of DNA/Cu nanowires due to cAFM measurements being required to be carried out in contact mode in order to maximise the time that the tip is in electrical contact with the sample. The high forces exerted upon the sample by the tip when operating in contact mode frequently results in individual nanowires being ‘brushed’ around the Si/SiO2 surface during scanning. The larger DNA/Cu nanorope structures were less prone to displacement by the tip during scanning, though careful regulation of the applied force by the tip was still required. |
| This journal is © The Royal Society of Chemistry 2014 |