Open Access Article
Open Access ArticleFluorinated beta-sheet breaker peptides
Joana A.
Loureiro
a,
Rosa
Crespo
b,
Hans
Börner
c,
Pedro M.
Martins
ab,
Fernando A.
Rocha
a,
Manuel
Coelho
a,
M. Carmo
Pereira
*a and
Sandra
Rocha
*d
aLEPABE, Department of Chemical Engineering, Faculty of Engineering, University of Porto, Porto, Portugal. E-mail: mcsp@fe.up.pt
bICBAS – Instituto de Ciências Biomédicas Abel Salazar, University of Porto, Porto, Portugal
cDepartment of Chemistry, Laboratory for Organic Synthesis of Functional Systems, Humboldt-Universität zu Berlin, Germany
dDepartment of Chemical and Biological Engineering, Physical Chemistry, Chalmers University of Technology, Gothenburg, Sweden. E-mail: sandra.rocha@chalmers.se
First published on 14th March 2014
Abstract
The aggregation of amyloid-β peptide (Aβ) has been linked to the formation of neuritic plaques, which are pathological hallmarks of Alzheimer's disease. We synthesized peptides containing fluorinated amino acids and studied their effect on the Aβ aggregation. The peptides were based on the sequence LVFFD, in which valine was substituted by either 4,4,4-trifluorovaline or 4-fluoroproline, or the phenylalanine at position 3 was replaced by 3,4,5-trifluorophenylalanine. Our results demonstrate that fluorination of the hydrophobic residue valine or phenylalanine is effective in preventing the Aβ aggregation. This study opens up the possibility of using new sequences based on fluorinated amino acids to inhibit the amyloid-fibril formation.
Introduction
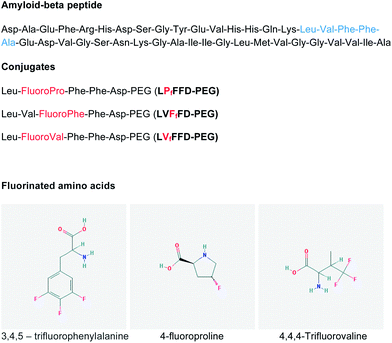
Amyloid β-peptide (Aβ) is about 4 kDa and it accumulates extracellularly in the brain as amyloid fibrils, which form the dense aggregates – neuritic plaques – that pathologically characterize Alzheimer's disease (AD).1 The aggregation process of Aβ is described as a nucleation process, with the formation of unstable intermediates that act as seeds. At least two intermediates have been identified during amyloid fibril formation: intermediates that are soluble and of low molecular weight, composed of dimeric or oligomeric peptide molecules, and the protofibrils that are short and flexible fibrils.2Increasing evidence suggests that soluble Aβ oligomers might be critical for the onset of AD.3–7 In the view of this hypothesis, a possible strategy to treat the disease is to inhibit the early stages of Aβ aggregation.8 The two hydrophobic cores of Aβ, the residues 17–21 and 30–42, have been associated with the peptide aggregation.9,10 Soto et al. designed a short peptide, based on the 17–21 hydrophobic core of Aβ (LVFFA), which binds to the full length Aβ and prevents its aggregation.11 This inhibitor has a similar degree of hydrophobicity to that of the 17–21 core but it has a very low propensity to adopt a β-sheet conformation due to the presence of a proline residue. Valine, which is considered a key residue for the β-sheet formation, was replaced by proline and alanine was substituted by aspartic acid to improve the peptide solubility.11–13 The sequence LPFFD, referred to as iAβ5, binds to Aβ and blocks the Aβ–Aβ molecule interactions, inhibiting the formation of amyloid fibrils. Certain fluorinated compounds have also been proposed as inhibitors of the Aβ aggregation.14–16 Fluorinated solvents such as hexafluoroisopropanol induce an α-helix structure in Aβ peptide. A similar effect is observed when Aβ interacts with poly(tetrafluoroethylene) surfaces or fluorinated nanoparticles.17–19 Since the hydrogenated analogues of the nanoparticles were not able to prevent the amyloid fibril formation, it has been postulated that fluorine atoms play an important role in the inhibition of Aβ aggregation. We have thus synthesized peptides containing fluorinated amino acids and studied their influence on Aβ oligomerization. The fluorinated peptides are also based on the hydrophobic central residues of Aβ(1–42) 17–21 (LVFFA) and on the iAβ5 beta-sheet breaker peptide. The aim is to study the impact of fluorine atoms on the beta-sheet breaker property of short peptide sequences and to understand the role of fluorine atoms on the Aβ(1–42) aggregation. The fluorinated sequences were covalently linked to polyethylene glycol (PEG) to increase their solubility (Fig. 1). We previously showed that coupling iAβ5 to PEG does not compromise its affinity to Aβ or the iAβ5 beta-sheet breaker activity.20 PEG has relatively low toxicity and is known to reduce proteolytic degradation.21 The use of non-natural residues such as fluorinated amino acids is also described to efficiently improve the stability of peptides or proteins against enzymatic degradation.22 The conjugates of fluorinated peptide–PEG were characterized by matrix-assisted laser desorption/ionization mass spectrometry. The kinetics of Aβ(1–42) aggregation, in the presence of the conjugates, were evaluated by the thioflavin T assay and the data were fitted using the crystallization-like model recently described by Crespo et al.23 The ultrastructure characterization of the Aβ aggregates was performed by transmission electron microscopy.
Material and methods
Synthesis of conjugates of fluorinated peptides and polyethylene glycol
The conjugates were prepared by the solid-phase supported strategy. Direct synthesis of the conjugate was performed applying a resin comprising a cleavable PEG spacer. Automated stepwise amino acid attachment was applied, following standard Fmoc-protocols. TentaGel PEG Attached Peptide resin (loading: 0.24 mmol g−1; Mn = 3200, PDI = 1.06 [GPC (THF, calibrated against linear PEG3200 standards, PSS, Germany)]) was purchased from Rapp, Polymere GmbH. Fmoc amino acid derivatives (Fmoc–phenylalanine OH, Fmoc–leucine OH, Fmoc–aspartic acid OH, Fmoc–valine OH, Fmoc–4,4,4-trifluoro-D,L-valine, Fmoc–3,4,5-trifluoro-L-phenylalanine and Fmoc–trans-4-fluoro-L-proline) were used as purchased from AnaSpec. The sequences of the fluorinated peptides are shown in Fig. 1. After the synthesis, the conjugates were dissolved in distilled water with 1% guanidinium hydrochloride and the pH was adjusted to 7 with 1 M sodium hydroxide. The samples were dialyzed against ultrapure water using a regenerate cellulose membrane (MWCO 1000 Da) for 4 days followed by lyophilization.Stock solutions of amyloid-β peptide
Aβ(1–42) (amyloid-β peptide 1–42, purity > 95.22%, MW: 4514.14, Selleck Chemicals) was dissolved in HFIP (1,1,1,3,3,3-hexafluoro-2-propanol, ≥99.8%, Sigma-Aldrich) at a concentration of 1.0 mg mL−1. HFIP was evaporated with nitrogen flow and the peptide film was dissolved in DMSO (dimethyl sulfoxide for molecular biology, ≥99.9%, FW: 78.13, Sigma-Aldrich) at a concentration of 9.0 mg mL−1.Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF)
MALDI-TOF-MS measurements were performed using a Voyager-DE STR BioSpectrometry Workstation MALDI-TOF mass spectrometer (Perceptive Biosystems, Inc., Framingham, MA, USA) at an acceleration voltage of 20 kV. The fluorinated peptide–PEG conjugates were dissolved in 0.1% TFA in acetonitrile–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) at a concentration of 0.1 mg mL−1. One microliter of the analyte solution was mixed with l μL of an alpha-cyano-4-hydroxycinnamic acid matrix solution consisting of 10 mg of the matrix dissolved in 1 mL of 0.1% TFA in acetonitrile–water (1
1, v/v) at a concentration of 0.1 mg mL−1. One microliter of the analyte solution was mixed with l μL of an alpha-cyano-4-hydroxycinnamic acid matrix solution consisting of 10 mg of the matrix dissolved in 1 mL of 0.1% TFA in acetonitrile–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). From the resulting mixture, 1 μL was applied to the sample plate. Samples were air-dried at room temperature (25 °C). Each spectrum is a mean of 250 laser shots.
1, v/v). From the resulting mixture, 1 μL was applied to the sample plate. Samples were air-dried at room temperature (25 °C). Each spectrum is a mean of 250 laser shots.
Thioflavin T (ThT) binding assay
For kinetic studies, Aβ(1–42) peptide (12.5 μM) was incubated at 37 °C in 96 well plates (Nunclon Delta Surface) with the fluorinated peptide–PEG conjugates (250 μM) in the presence of ThT (0.7 mg mL−1) in a PBS buffer. The ThT solution was filtered using a 0.2 μm syringe filter before adding to the Aβ samples. The fibrils conjugated with ThT have the excitation maximum at 450 nm and enhanced emission at 482 nm.24 The fluorescence intensity was measured every 30 minutes during 24 hours using a Biotek Synergy 2 fluorescence spectrometer with the excitation filter at 420/50 nm and the emission filter at 485/20 nm. Student's t-test statistical analysis was used to determine statistical significance between Aβ(1–42) incubated alone and Aβ(1–42) incubated with conjugates (n ≥ 3).Theoretical crystallization-like model
The crystallization-like model (CLM) is a generic two-parameter model that describes protein aggregation kinetics by a sequence of nucleation and growth steps. The CLM is represented by the following equation, in which α represents the normalized fraction of amyloid protein converted into fibrils.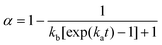 | (1) |
Transmission electron microscopy (TEM)
The Aβ(1–42) peptide (100 μM) was incubated at 37 °C with each of the fluorinated peptide–PEG conjugates (2 mM) in a PBS buffer (10 mM, pH: 7.4), for 48 hours. An aliquot of each sample (5 μL) was placed on carbon–formvar coated 200–400 mesh spacing grids and let to adsorb for five minutes. The negative staining was performed with a 2% filtered aqueous solution of uranyl acetate for 45 seconds. The grids were visualized using a Jeol JEM 1400 electron microscope at 80 kV.Results
Fluorinated peptide–PEG conjugates
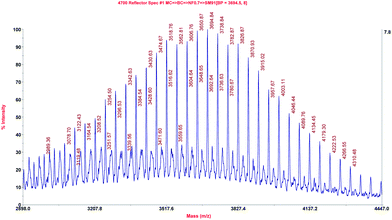
The fluorinated sequences conjugated to PEG were analysed by MALDI-TOF-MS. The spectrum of LPfFFD–PEG is depicted as representative of the results acquired for the conjugates and shows the typical distribution of the polymer PEG with the characteristic repeat unit of 44.1 ± 0.5 Da, which could be assigned to the ethylene oxide (EO) monomer of the PEG (Fig. 2). The experimentally found signal at m/z 3694.8, for example, can be assigned to [M + K] = 3692.3 Da by assuming an M[EO] of 44.05 Da, n = 68 units, a mass of the peptide M[peptide] = 655.8 Da, a mass of the polymer end groups 1H = 1 Da and the mass of a potassium counter ion M[counter ion] = 39.1 Da.Impact of the peptide–PEG conjugates on Aβ(1–42) fibrillization
The aggregation of Aβ(1–42) incubated at 37 °C with LPfFFD–PEG, LVFfFD–PEG and LVfFFD–PEG at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
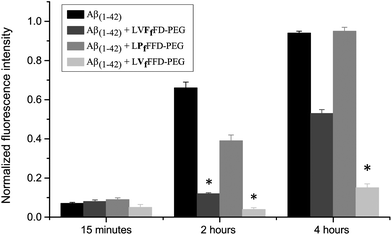
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 was evaluated by the ThT binding assay (Fig. 3 and 4). At 15 minutes incubation time, the ThT fluorescence intensity is low for all samples, which indicates low content of amyloid fibrils. After two hours, a large increase in the fluorescence intensity is observed for Aβ(1–42) alone and for the Aβ(1–42):LPfFFD–PEG samples, reaching the maximum at 4 hour incubation time (the fluorescence intensities were normalized to that of Aβ(1–42) incubated with ThT at 37 °C for 24 hours). The fluorescence of ThT in the presence of Aβ(1–42) and LVFfFD–PEG showed only a moderated increase (53%) after 4 hours, whereas that of the sample Aβ(1–42):LVfFFD–PEG remained low (15%). The conjugates in the absence of Aβ(1–42) do not induce any change in the fluorescence signal of ThT.
20 was evaluated by the ThT binding assay (Fig. 3 and 4). At 15 minutes incubation time, the ThT fluorescence intensity is low for all samples, which indicates low content of amyloid fibrils. After two hours, a large increase in the fluorescence intensity is observed for Aβ(1–42) alone and for the Aβ(1–42):LPfFFD–PEG samples, reaching the maximum at 4 hour incubation time (the fluorescence intensities were normalized to that of Aβ(1–42) incubated with ThT at 37 °C for 24 hours). The fluorescence of ThT in the presence of Aβ(1–42) and LVFfFD–PEG showed only a moderated increase (53%) after 4 hours, whereas that of the sample Aβ(1–42):LVfFFD–PEG remained low (15%). The conjugates in the absence of Aβ(1–42) do not induce any change in the fluorescence signal of ThT.
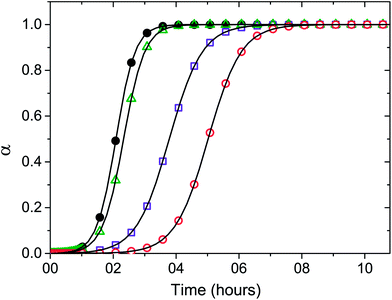 | ||
Fig. 4 Numerical fit (solid lines) of eqn (1) to the kinetic results of the Aβ(1–42) peptide incubated at 37 °C in PBS buffer in the absence (●) and in the presence of LPfFFD–PEG (Δ), LVFfFD–PEG (□), and LVfFFD–PEG (○) conjugates, monitored by ThT fluorescence. The Aβ(1–42) concentration was kept constant at 12.5 μM and the molar ratio of Aβ(1–42)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) conjugates was 1 conjugates was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. The aggregation rate (v50) was obtained by fitting a sigmoidal function to each kinetic trace according to eqn (1). 20. The aggregation rate (v50) was obtained by fitting a sigmoidal function to each kinetic trace according to eqn (1). | ||
The kinetic data were fitted according to the recently proposed CLM model (Fig. 4). The time at half fibril conversion (t50) and the aggregation rate (v50) were obtained by fitting a sigmoidal function to each kinetic trace according to the CLM (Table 1).23 In the case of sigmoidal-type aggregation kinetics such as those represented in Fig. 4, v50 is solely dictated by the growth rate constant ka. The parameter v50 did not change significantly in the presence of the conjugates. A marked increase in the value of t50 is, however, evident for the LVFfFD–PEG and LVfFFD–PEG, which can be explained by the CLM as the result of either nucleation-prevention or solubility-change effects. The latter hypothesis is discarded by the fact that the maximum fluorescence signal did not show significant variation in the presence of conjugates. Therefore, we ascribe the increased induction times to the inhibition of the nucleation step as a possible consequence of the stabilization of the Aβ molecule by the conjugates. The higher value of the induction time t50 is obtained for the conjugate containing the fluorinated valine (LVfFFD–PEG), suggesting a stronger inhibitory effect in this case. Inhibitors of the amyloid fibril formation may act upon the initial assembly of macromolecules forming the stable nuclei (nucleation step) or block the subsequent addition of new growth units (growth step).23 A third way of action involves the alteration of the thermodynamic solubility of the polypeptide. This alternative is more unlikely to result in vivo since cell and tissue media are in general buffered against non-specific interactions. Compared to the effect of a well-known and well accepted beta-sheet breaker peptide, iAβ5 (LPFFD), on Aβ(1–42) aggregation, a similar effect was obtained by the LVfFFD–PEG conjugate (3.1 hours) (Table 1). At the same molar ratio and with the same Aβ(1–42) batch, the reduction of the aggregation time of Aβ(1–42) is similar for iAβ5 and LVfFFD–PEG. The conjugate containing fluorinated phenylalanine (LVFfFD–PEG) induced an inhibitory effect of 1.5 hours and the LPfFFD–PEG did not inhibit the Aβ(1–42) aggregation.
| t 50 (hours) | Delay in aggregation (hours) | |
|---|---|---|
| Aβ(1–42) alone | 2.1 | — |
| Aβ(1–42) and LPfFFD–PEG | 2.4 | 0.2 |
| Aβ(1–42) and LVFfFD–PEG | 3.6 | 1.5 |
| Aβ(1–42) and LVfFFD–PEG | 5.2 | 3.1 |
| Aβ(1–42) and iAβ5 | 5.4 | 3.3 |
The Aβ(1–42)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) conjugate molar ratio of 1
conjugate molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 did not significantly change the aggregation parameters of Aβ, which indicates that there is a threshold concentration for the Aβ–conjugate interactions.
5 did not significantly change the aggregation parameters of Aβ, which indicates that there is a threshold concentration for the Aβ–conjugate interactions.
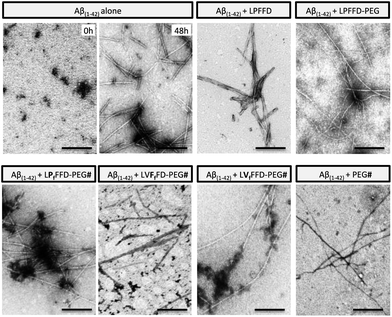
TEM analysis showed that Aβ(1–42) incubation at 37 °C for 48 hours resulted in amyloid-like, unbranched fibrils (Fig. 5). Aβ(1–42) also formed fibrils in the presence of the conjugates (or iAβ5), although the samples containing LVFfFD–PEG and LVfFFD–PEG show also small aggregates. This analysis demonstrates that the bioconjugates and the beta-sheet breaker iAβ5 are only delaying the Aβ(1–42) fibrillogenesis.
Discussion
Aβ(1–42) is an amphipathic peptide, which self-associates into oligomers that are toxic to cells.25 The peptide aggregation eventually leads to the formation of amyloid fibrils, which are known to bind to Congo red and thioflavin T dyes. The oligomerization and aggregation of Aβ(1–42) are difficult to control and although many attempts have been made to find a way to inhibit these processes, the understanding of how different factors can contribute to their initiation is still puzzling. The study on the self-assembly and structural transformation of other amyloid-fibril-forming peptides will provide important insights into the understanding of the fibril formation process and to find ways to prevent it.26–28We relied on the knowledge that fluorine atoms seem to play a role in the prevention of Aβ(1–42) aggregation and developed peptides with fluorinated amino acids to screen for fibrillization inhibitors. The ThT assay evidenced that two of the fluorinated peptides (LVFfFD–PEG and LVfFFD–PEG) are able to delay the aggregation of the Aβ(1–42). Hydrophobic interactions and H-bonding among side-chain groups are the two major driving forces that control protein aggregation.29 A large amount of evidence indicates that there are three distinct regions within the Aβ(1–42) sequence that might be involved in its aggregation: the central hydrophobic cluster corresponding to the 17–21 region, the 23–28 residues and the hydrophobic C-terminus.30–32 Considering that fluorine atoms increase the hydrophobicity of the sequences, it is expected that they would interact with the hydrophobic regions of Aβ(1–42) to be segregated away from the water. This interaction might contribute to the prevention of Aβ molecules to interact with each other, thus inhibiting their aggregation. The PEG molecules linked to the fluorinated peptides can also contribute to the interactions of the conjugates with Aβ(1–42). We have previously shown that the pegylation of a beta-sheet breaker peptide did not significantly change its binding to Aβ(1–42).20 PEG is described to bind to proteins such as albumin and lysozyme even in solution with an ionic strength similar to the physiological conditions.33 In the case of the fluorinated peptide–PEG conjugates, their effect on Aβ(1–42) cannot be simply explained by PEG and Aβ interactions since the three conjugates show different outcomes.
The fluorinated amino acids of the conjugates may play an important role in delaying the Aβ(1–42) aggregation by interacting with the hydrophobic residues of the peptide. There is also a geometric preference for the interactions between the C–F bond and the side-chain amides of glutamine (E) 15 and asparagine (N) 27 residues.34 Therefore, the fluorine atoms might be able to interact with the two stretches of Aβ hydrophobic residues responsible for their oligomerization (17–21 and 30–42 residues) and/or with residues in their vicinity by exploiting the same interactions as in the Aβ(1–42) assembly. A conformational tightened state is produced by the interplay of ionic and hydrophobic groups of both peptide and the conjugates.
Ferrão-Gonzales et al. showed that (1-anilino-8-naphthalene sulfonate)-derived molecules inhibit Aβ aggregation due to their dual nature provided by hydrophobic regions and charged groups.35 Vieira et al. demonstrated the effectiveness of different fluorinated alcohols in the β-to-α refolding process of Aβ(1–40).14 The observed effect was interpreted as a result of alteration of the hydration shell of the peptides and hydrophobic effects of fluorine groups. Montserret et al. have shown that although the folding of amphipathic α-helical peptides in hydrophilic SDS micellar solutions is mostly driven by electrostatic interactions, when hydrophobic peptides are considered, SDS–peptide hydrophobic interactions might be sufficiently strong to induce α-helical-rich structures.36 The effect of fluorinated complexes made of polyampholyte and the sodium salt of perfluorododecanoic acid on Aβ was attributed to the hydrophobic and acidic characters of the perfluorododecanoic acid.18,19 We propose that LVFfFD–PEG and LVfFFD–PEG have the necessary hydrophobicity and spatial geometry to allow the interaction with Aβ(1–42). LPfFFD–PEG, with one fluorine atom in the proline residue, was not able to prevent Aβ–Aβ interactions. Proline residues are generally solvent-exposed in proteins and they are considered binding motifs in protein–protein interactions.37 However it is possible that the presence of the fluorine atom in the proline residue of the LPfFFD–PEG prevents the exposure of the conjugate amino acids to Aβ. Comparing the LPfFFD–PEG with the non-fluorinated analogue, one can assume that the fluorine atom increases the steric hindrance of the conjugate reducing the access to the proline residue. On the other hand the hydrophobic interactions between LPfFFD–PEG and Aβ are probably not strong enough when compared to the Aβ–Aβ interactions. Apart from the apparently reduced interactions between Aβ and LPfFFD–PEG, the position of fluorine atoms at the extremity of the conjugate and the fluorination of hydrophobic residues seem to favour the inhibitory effect of the sequences on Aβ amyloid fibril formation.
Conclusions
The effects of fluorinated alcohols, molecules or particles on Aβ fibril formation are typically very different from those of their hydrogenated analogues. In general, it is possible to modify hydrogenated molecules or particles with fluorine atoms in such a way that they would be able to induce an α-helix structure on Aβ, a conformation which is less prone to aggregation. However, fluorination of molecules does not always lead to the inhibition of amyloid fibril formation but it can instead promote it. Therefore, it is important to understand the interactions of fluorinated molecules with Aβ peptide. We propose the use of beta-sheet breaker peptides as model molecules to study the effect of fluorination on the Aβ aggregation. Therefore, sequences that play an important role in Aβ amyloid fibril formation were modified with fluorine atoms. Our work shows that fluorination of hydrophobic amino acids such as valine or phenylalanine of sequences that interfere with amyloid fibril formation induces a significant delay in the Aβ aggregation process. This effect is likely a result of the interaction between the fluorinated amino acids and the hydrophobic residues of Aβ, which is strong enough to prevent the contact between Aβ molecules and thus it prevents the peptide aggregation. The fluorination of other amino acids or the modification of the fluorinated sequences with hydrophilic residues at different positions will contribute to better understand the interactions between Aβ and fluorinated peptides.Acknowledgements
This work was financed by FCT research project PTDC/QUI-BIQ/102827/2008.Notes and references
- A. E. Roher, J. D. Lowenson, S. Clarke, C. Wolkow, R. Wang, R. J. Cotter, I. M. Reardon, H. A. Zurcher-Neely, R. L. Heinrikson and M. J. Ball, et al. , J. Biol. Chem., 1993, 268, 3072–3083 CAS.
- C. Haass and D. J. Selkoe, Nat. Rev. Mol. Cell Biol., 2007, 8, 101–112 CrossRef CAS PubMed.
- W. L. Klein, G. A. Krafft and C. E. Finch, Trends Neurosci., 2001, 24, 219–224 CrossRef CAS.
- Y. Gong, L. Chang, K. L. Viola, P. N. Lacor, M. P. Lambert, C. E. Finch, G. A. Krafft and W. L. Klein, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 10417–10422 CrossRef CAS PubMed.
- R. Kayed, E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman and C. G. Glabe, Science, 2003, 300, 486–489 CrossRef CAS PubMed.
- G. M. Shankar, B. L. Bloodgood, M. Townsend, D. M. Walsh, D. J. Selkoe and B. L. Sabatini, J. Neurosci., 2007, 27, 2866–2875 CrossRef CAS PubMed.
- G. M. Shankar, S. Li, T. H. Mehta, A. Garcia-Munoz, N. E. Shepardson, I. Smith, F. M. Brett, M. A. Farrell, M. J. Rowan, C. A. Lemere, C. M. Regan, D. M. Walsh, B. L. Sabatini and D. J. Selkoe, Nat. Med., 2008, 14, 837–842 CrossRef CAS PubMed.
- J. Hardy and D. J. Selkoe, Science, 2002, 297, 353–356 CrossRef CAS PubMed.
- C. Hilbich, B. Kisters-Woike, J. Reed, C. L. Masters and K. Beyreuther, J. Mol. Biol., 1992, 228, 460–473 CrossRef CAS.
- J. T. Jarrett, E. P. Berger and P. T. Lansbury, Jr, Biochemistry, 1993, 32, 4693–4697 CrossRef CAS.
- C. Soto, M. S. Kindy, M. Baumann and B. Frangione, Biochem. Biophys. Res. Commun., 1996, 226, 672–680 CrossRef CAS PubMed.
- P. Y. Chou and G. D. Fasman, Annu. Rev. Biochem., 1978, 47, 251–276 CrossRef CAS PubMed.
- S. J. Wood, R. Wetzel, J. D. Martin and M. R. Hurle, Biochemistry, 1995, 34, 724–730 CrossRef CAS.
- E. P. Vieira, H. Hermel and H. Mohwald, Biochim. Biophys. Acta, 2003, 1645, 6–14 CrossRef CAS.
- S. L. Adamski-Werner, S. K. Palaninathan, J. C. Sacchettini and J. W. Kelly, J. Med. Chem., 2004, 47, 355–374 CrossRef CAS PubMed.
- M. Torok, M. Abid, S. C. Mhadgut and B. Torok, Biochemistry, 2006, 45, 5377–5383 CrossRef PubMed.
- C. E. Giacomelli and W. Norde, Macromol. Biosci., 2005, 5, 401–407 CrossRef CAS PubMed.
- S. Rocha, A. F. Thuneman, M. D. Pereira, M. Coelho, H. Mohwald and G. Brezesinski, Biophys. Chem., 2008, 137, 35–42 CrossRef CAS PubMed.
- A. M. Saraiva, I. Cardoso, M. C. Pereira, M. A. Coelho, M. J. Saraiva, H. Mohwald and G. Brezesinski, ChemBioChem, 2010, 11, 1905–1913 CrossRef CAS PubMed.
- S. Rocha, I. Cardoso, H. Borner, M. C. Pereira, M. J. Saraiva and M. Coelho, Biochem. Biophys. Res. Commun., 2009, 380, 397–401 CrossRef CAS PubMed.
- F. M. Veronese, Biomaterials, 2001, 22, 405–417 CrossRef CAS.
- B. C. Buer and E. N. G. Marsh, Protein Sci., 2012, 21, 453–462 CrossRef CAS PubMed.
- R. Crespo, F. A. Rocha, A. M. Damas and P. M. Martins, J. Biol. Chem., 2012, 287, 30585–30594 CrossRef CAS PubMed.
- H. LeVine, 3rd, Protein Sci., 1993, 2, 404–410 CrossRef PubMed.
- B. A. Chromy, R. J. Nowak, M. P. Lambert, K. L. Viola, L. Chang, P. T. Velasco, B. W. Jones, S. J. Fernandez, P. N. Lacor, P. Horowitz, C. E. Finch, G. A. Krafft and W. L. Klein, Biochemistry, 2003, 42, 12749–12760 CrossRef CAS PubMed.
- M. Reches and E. Gazit, Science, 2003, 300, 625–627 CrossRef CAS PubMed.
- X. H. Yan, P. L. Zhu and J. B. Li, Chem. Soc. Rev., 2010, 39, 1877–1890 RSC.
- X. H. Yan, Y. Su, J. B. Li, J. Fruh and H. Mohwald, Angew. Chem., Int. Ed., 2011, 50, 11186–11191 CrossRef CAS PubMed.
- A. T. Petkova, Y. Ishii, J. J. Balbach, O. N. Antzutkin, R. D. Leapman, F. Delaglio and R. Tycko, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16742–16747 CrossRef CAS PubMed.
- O. N. Antzutkin, J. J. Balbach and R. Tycko, Biophys. J., 2003, 84, 3326–3335 CrossRef CAS.
- M. Torok, S. Milton, R. Kayed, P. Wu, T. McIntire, C. G. Glabe and R. Langen, J. Biol. Chem., 2002, 277, 40810–40815 CrossRef PubMed.
- C. J. Pike, A. J. Walencewicz-Wasserman, J. Kosmoski, D. H. Cribbs, C. G. Glabe and C. W. Cotman, J. Neurochem., 1995, 64, 253–265 CrossRef CAS.
- J. Wu, Z. Wang, W. F. Lin and S. F. Chen, Acta Biomater., 2013, 9, 6414–6420 CrossRef CAS PubMed.
- K. Muller, C. Faeh and F. Diederich, Science, 2007, 317, 1881–1886 CrossRef PubMed.
- A. D. Ferrao-Gonzales, B. K. Robbs, V. H. Moreau, A. Ferreira, L. Juliano, A. P. Valente, F. C. L. Almeida, J. L. Silva and D. Foguel, J. Biol. Chem., 2005, 280, 34747–34754 CrossRef CAS PubMed.
- R. Montserret, M. J. McLeish, A. Bockmann, C. Geourjon and F. Penin, Biochemistry, 2000, 39, 8362–8373 CrossRef CAS PubMed.
- B. K. Kay, M. P. Williamson and P. Sudol, FASEB J., 2000, 14, 231–241 CAS.
| This journal is © The Royal Society of Chemistry 2014 |