Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
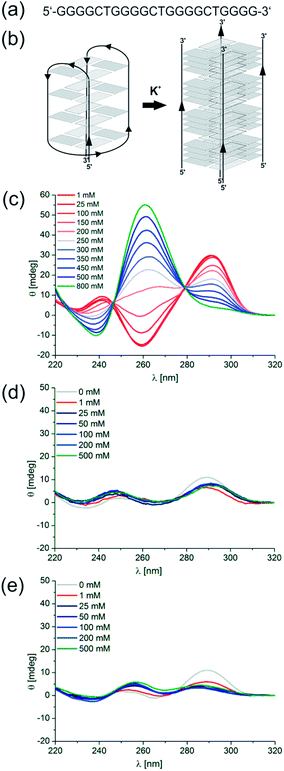
A bacterial DNA quadruplex with exceptional K+ selectivity and unique structural polymorphism†
Charlotte
Rehm
,
Isabelle T.
Holder
,
Andreas
Groß
,
Filip
Wojciechowski
,
Maximilian
Urban
,
Malte
Sinn
,
Malte
Drescher
and
Jörg S.
Hartig
*
Department of Chemistry and Konstanz Research School Chemical Biology (KoRS-CB), University of Konstanz, Universitätsstr. 10, 78457 Konstanz, Germany. E-mail: joerg.hartig@uni-konstanz.de
First published on 20th March 2014
Abstract
The G-rich sequence d[(G4CT)3G4] was recently identified as a potential quadruplex-forming sequence associated with loci involved in antigenic variation in the human pathogen Treponema pallidum. We found this motif to be enriched in eubacterial genomes. Employing a combination of CD spectroscopy, EPR spectroscopy, analytical ultracentrifugation, and EMSA, we demonstrate that d[(G4CT)3G4] displays unique features among the many G-quadruplex-forming sequences studied so far. To our knowledge d[(G4CT)3G4] shows a so far unprecedented selectivity for K+ with even high concentrations of Na+ unable to induce pronounced G-quadruplex formation. A remarkable continuous and complete transition from an anti-parallel, monomolecular structure into a tetrameric, parallel conformation is observed upon increasing K+-concentrations. Furthermore we investigate the effects of cation selectivity, quadruplex loop composition and length as well as G-tract length on quadruplex conversion.
Introduction
Guanines can interact via Hoogsteen basepairing to form a tetrameric square arrangement, a so-called tetrad. In G-rich oligonucleotides several tetrads can stack upon each other to form a G-quadruplex structure. A quadruplex is stabilized by metal cations bound in the central cavity, primarily by the monovalent cations Na+ and K+ that coordinate to the O6 carboxy oxygen of the guanines, yielding compact and stable structures.1,2 Potential G-quadruplex forming sequences are found in eukaryotic and prokaryotic genomes.3–5 Proof for abundant quadruplex formation in vivo is increasing. For example Balasubramanian and co-workers have recently shown the distribution of G-quadruplexes on eukaryotic chromosomes and their regulation during cell cycle progression using a G-quadruplex-specific antibody in different types of human cell lines.6 Putative cellular roles that have been assigned to quadruplex structures are the involvement in organization and protection of the telomeres, stalling of the replication fork machinery, promotion of homologous recombination and regulating transcription.7 In particular, quadruplex-forming sequences have been characterized within promoter regions, where they serve as transcriptional regulators.8–10 When comparing the quadruplex folding abilities of a variety of DNA sequences and their respective RNA counterparts we found that all RNA sequences exclusively formed G-quadruplex structures with parallel strand orientations that were often more stable than the structures adopted by the homologous DNA sequences.11 In mammalian cells we were able to show that synthetic RNA G-quadruplexes inserted into the 5′UTR in front of a luciferase reporter gene provided predictable repression of gene expression by acting as translational suppressors.12,13 Furthermore a whole-transcriptome analysis conducted in HeLa S3 cells detected specific changes for quadruplex containing genes upon treatment of the cells with G-quadruplex-specific bisquinolinium compounds.14Although these and many other studies hint at diverse roles for quadruplexes in eukaryotic cells, relatively little is known about the potential functions of quadruplex-forming sequences in bacteria. In an artificial setup we were able to show that quadruplexes masking the ribosomal binding site within an mRNA lead to repression of gene expression in Escherichia coli and that the level of repression correlated with the thermodynamic stability of the quadruplex.15 Next to inhibition of translation initiation, suppression of translation elongation by quadruplex-forming sequences found in protein-coding sequences (ORFs) in E. coli has recently been demonstrated.16 In a follow-up study it was shown that ribosomal stalling by such an RNA quadruplex can cause a −1 ribosomal frameshift in cellulo.17 In a computational study Chowdhury and co-workers identified potential quadruplex forming sequences in promoter regions in bacteria and found them to be enriched in certain gene classes. Specifically, in Deinococcus radiodurans potential quadruplex sequences are located in regulatory regions of genes contributing to radioresistance. Upon treatment with a quadruplex-binding ligand attenuation of the radioresistance was observed.18 In a landmark study Seifert and co-workers identified a cis-acting quadruplex sequence that is necessary for pilin antigenic variation in Neisseria gonorrhoeae.19 Antigenic variation takes place via a non-homologous recombination event between a single expressed pilE locus and many silent donor loci. Mutation of the G-rich sequence upstream of pilE inhibited recombinational switching at the variable locus. Quadruplex formation is required for nicking the DNA, the break site is then further processed by the recombination machinery.19 Furthermore Seifert and co-workers identified a conserved promoter sequence adjacent to the pilE quadruplex motif; transcription of a cis-acting, non-coding small RNA from this promoter is essential for antigenic variation to commence.20
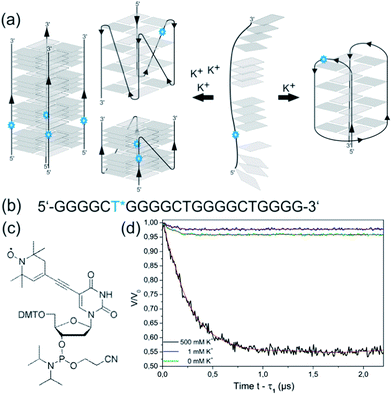
We recently reported that the sequence d[(G4CT)3G4] (Fig. 1a) studied in this report is potentially involved in the antigenic variation of the surface-exposed variable antigen TprK protein in the human pathogen Treponema pallidum.21 TprK heterogeneity is generated by nonreciprocal gene conversion between the TprK expression and donor sites. A series of G-rich sequences that were found to be associated with these sites were investigated and demonstrated folding into stable quadruplex structures. However, despite its elevated G-score22 d[(G4CT)3G4] with 100 mM KCl did not show the expected quadruplex circular dichroism (CD) spectrum in contrast to other potential quadruplex-folding sequences studied in the same context.21 This unexpected result motivated us to further characterize the quadruplex folding properties of d[(G4CT)3G4]. Here we report that the sequence displays several remarkable and to some extend unique properties. We find that the folding of the quadruplex proceeds with very high activation energies, a feature that caused it to be overlooked in the initial screening for quadruplex-forming sequences.21 Moreover, the sequence shows high selectivity for K+ as a G-quadruplex-stabilizing cation and most importantly displays a K+-concentration-dependent conformational polymorphism ranging from an exclusively anti-parallel conformation at low K+ to a parallel topology at high concentrations of K+. Several different DNA quadruplex conformations are known, largely dependent on the length and to a lesser extent the identity of the loop sequences as well as the number of stacked tetrads.23–25 In addition to intramolecular quadruplexes, where the DNA sequence folds back upon itself, intermolecular quadruplexes exist with two, four or more individual strands interacting. Furthermore, it is known that the structural polymorphism of some quadruplexes depends on the nature of the stabilizing cation. A well-studied example for the polymorphic nature of G-quadruplexes is the human telomeric sequence, hTel d[(TTAGGG)n]. Crystal structures of d[AG3(T2AG3)3]26 reveal a parallel topology when folded in the presence of K+, whereas an anti-parallel conformation in solution containing Na+ was detected for the same sequence by NMR.27 Furthermore, several co-existing structures have been postulated for this sequence in solution when stabilized by K+.28–32 Using electron paramagnetic resonance (EPR) spectroscopy we have been able to elucidate the polymorphic nature of d[(G3T2A)3G3] in K+ solution, in which a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of the parallel propeller and the anti-parallel basket structure was detected.33 We found the same distribution when the quadruplex sequence was injected into Xenopus oocytes for in cellulo measurements.34 However, when studying individual G-quadruplex units within the context of extended sequences composed of the human telomeric DNA repeats, we found that a (3 + 1) hybrid structure is formed.35
1 mixture of the parallel propeller and the anti-parallel basket structure was detected.33 We found the same distribution when the quadruplex sequence was injected into Xenopus oocytes for in cellulo measurements.34 However, when studying individual G-quadruplex units within the context of extended sequences composed of the human telomeric DNA repeats, we found that a (3 + 1) hybrid structure is formed.35
The Oxytricha telomeric sequence d[(G4T4)3G4] as well as the Tetrahymena-related telomeric sequence d[(G4T2)3G4] were also shown to adopt multiple conformations in solution depending on the stabilizing ion in electrophoretic assays and CD measurements. In the case of d[(G4T2)3G4], Na+ promoted the anti-parallel conformation, however, the parallel and tetrameric conformation was formed in the presence of Sr2+ or K+.36 Thomas and co-workers have demonstrated the structural polymorphism of Oxytricha telomeric DNA by Raman spectroscopy. At low concentrations of Na+ or K+ the sequence adopted an anti-parallel foldback quadruplex; with increasing alkali ion concentrations interquadruplex conversion took place yielding a parallel quadruplex.37,38 Part of the Oxytricha telomeric sequence d[G4T4G4] has been shown to fold into a dimeric anti-parallel quadruplex in solution with Na+ by Sugimoto and co-workers.39,40 Addition of divalent cations, particularly Ca2+, leads to oligomerization of the sequence and switches the conformation to the parallel topology.24 However, to the best of our knowledge there is no example known where varying K+ concentration alone results in a pronounced structural transition of quadruplex conformations.
Here we characterized the quadruplex-folding sequence d[(G4CT)3G4] (Fig. 1a). Initially identified associated to loci involved in antigenic variation in T. pallidum, we found this sequence motif highly over-represented in bacterial genomes. We report a remarkable selectivity for K+ as a stabilizing cation. Upon increasing the K+ concentration a very pronounced structural transition from the intramolecular, anti-parallel conformation to a parallel conformation is revealed. By utilizing a diverse range of methods including CD spectroscopy, EPR distance measurements, electrophoretic mobility shift (EMSA) assays, NMR, and analytical ultracentrifugation (AUC) we demonstrate that at high K+-concentrations the sequence forms a parallel-stranded tetrameric complex as compared to a monomeric, anti-parallel fold in low K+-concentrations. Fig. 1b shows a simplified scheme of quadruplex conversion showing the so-called basket structure as an example for an anti-parallel quadruplex structure, formation of e.g. a chair topology is equally possible. Participation of all guanosines of a G-tract in tetrad formation is assumed. In addition, we studied the occurrence of this remarkable structural transition and its dependence on loop composition, loop length, and G-tract length.
Results and discussion
K+ selectivity and structural transition
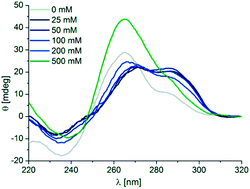
We employed CD spectroscopy as a convenient method to quickly assess the structural properties of quadruplexes in solution. A typical CD spectrum of an anti-parallel quadruplex shows a maximum at around 290 nm and a minimum at 265 nm, whereas a typical spectrum of a parallel quadruplex displays a maximum at about 265 nm and a minimum at 240 nm.41 Oligonucleotides were prepared as a 5 μM solution in 10 mM Tris–HCl pH 7.5 supplemented with KCl, NaCl, LiCl or MgCl2 as noted and denatured by heating to 98 °C for 5 min, followed by slow cooling to 20 °C overnight to induce quadruplex folding. Interestingly, with increasing concentrations of K+ a continuous structural transition of d[(G4CT)3G4] from the anti-parallel conformation observed at 0.5 mM KCl to the all parallel conformation at 800 mM is revealed by CD spectroscopy (Fig. 1c, see Fig. S1a† for more concentrations). The presence of isodichroic points at approximately 250 nm and 280 nm indicate a two state transition from the anti-parallel to the parallel conformation. Intriguingly, addition of the same concentrations of Na+ did not induce the formation of any pronounced G-quadruplex structure detectable by CD spectroscopy (Fig. 1d), although Na+ usually is a strong stabilizer of quadruplex structures. Likewise, quadruplexes did not fold in the presence of Li+ (Fig. 1e), which is more expected from experiences with other quadruplex sequences. Mg2+ partially stabilized the parallel conformation, but not to the same extent as K+, and it did not stabilize the anti-parallel conformation (Fig. S1b†). Although Na+ and Li+ alone did not show significant stabilization of a quadruplex structure, the parallel conformer was stabilized when NaCl or LiCl were employed in addition to KCl. d[(G4CT)3G4] folded mostly into the anti-parallel conformer at 100 mM K+, when increasing concentrations of NaCl or LiCl were added to the mixture structural transition to the parallel conformer could be observed (Fig. 2a and b). This suggests that K+ is required for initial stabilization of the quadruplex structure, but high ionic strength irrespective of cation nature is needed to induce the parallel fold. In addition we tested the influence of the counter ion of K+ on the observed topological switching. Identical spectra and structural transition as observed with KCl was also induced by KBr, KF, KI and was therefore not dependent on the anion (Fig. S2†).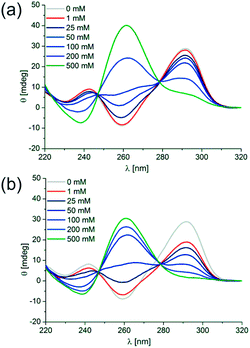 | ||
| Fig. 2 CD spectra of 5 μM d[(G4CT)3G4] in the presence of 100 mM K+ and 0 mM (gray), 1 mM (red), 25 mM to 200 mM (blue) and 500 mM (green) (a) NaCl or (b) LiCl. | ||
When d[(G4CT)3G4] was folded with equal amounts of the complementary strand present during denaturation and slow overnight cooling, the quadruplex structure rather than the duplex was formed at high K+ concentrations (Fig. 3). The C-rich strand alone did not fold into a specific structure upon incubation with K+ (Fig. S1c†). K+ is the major cation in the bacterial cell, Shabala and co-workers determined a cytosolic concentration of about 200 mM in E. coli.42 In fact higher K+ concentrations can be reached in bacterial cells e.g. under osmotic shock or salt stress conditions. Epstein and Schultz reported an increase in the intracellular K+ concentration from 150 to 550 mM in exponentially growing E. coli when the osmolality of the growth medium was increased from 0.1 to 1.2 osm by addition of glucose, NaCl or sucrose.43 Hence the K+-concentrations utilized in the described experiments are within a range that can be expected to be present in bacterial cells. In this respect the finding that a quadruplex sequence is folded even in the presence of a complementary strand makes at least the transient formation of such DNA structures in genomes not unlikely.
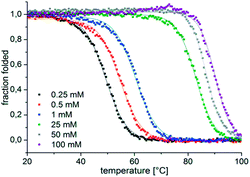
To assess the stability of the different quadruplex species we performed melting experiments. We determined melting temperatures T1/2 of 49.3 °C, 54.6 °C, 60.5 °C, 82.9 °C, 86.9 °C and 90.1 °C for the anti-parallel conformer at 0.25, 0.5, 1, 25, 50 and 100 mM K+, respectively, by monitoring ellipticity (Fig. 4). A T1/2 of the anti-parallel and parallel conformers at KCl concentrations higher than 250 mM could not be accurately determined as both structures were extremely thermostable and only started to denature at temperatures above 85 °C for 250–350 mM K+ and above 90 °C for 400–500 mM K+ (Fig. S3†). Similarly, we determined a T1/2 of 59.3 °C for the anti-parallel conformer at 1 mM KCl by UV thermal denaturation by measuring absorption at 295 nm, likewise the parallel conformer was stable above 90 °C (Fig. S4†). Although CD spectra did not show detectable quadruplex formation in the presence of NaCl, a T1/2 of 50.8 °C and 79.8 °C was determined for d[(G4CT)3G4] for an undefined structure formed with 1 mM and 500 mM NaCl, respectively (Fig. S4†). Formation of a mixture of structures with opposing CD signatures might be preventing characterization by CD spectroscopy. We therefore measured the 1H-NMR spectra of d[(G4CT)3G4] in the presence of 1 mM, 200 mM and 500 mM NaCl (Fig. S5a and b†). NMR spectra in the presence of 1 mM NaCl showed weak signals between 11 and 12 ppm for imino protons indicative of minor G-quadruplex formation, however the imino proton signal intensity differed profoundly from the distinct signals detected in the presence of KCl (Fig. S5c†) (for detailed characterization of NMR spectra with KCl see below, Fig. S11†). Notably no pronounced imino proton signals were recorded for d[(G4CT)3G4] in the presence of 500 mM NaCl, suggesting that the structure is not a well-defined quadruplex (Fig. S5d†). d[(G4CT)3G4] in the absence of monovalent cations formed a structure with a T1/2 of 32.7 °C and was minimally stabilized in the presence of 1 and 500 mM LiCl with T1/2 of 38.6 °C and 46.3 °C, respectively (Fig. S4†).
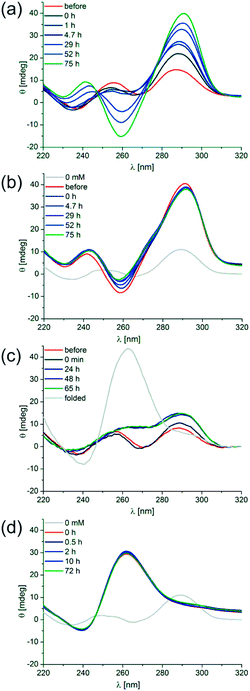
We were surprised to find the sequence d[(G4CT)3G4] displaying such interesting properties, even more so since we had overlooked the quadruplex-forming potential during the initial characterization of Treponema sequence motifs.21 In order to clarify the kinetic aspects of quadruplex formation, the sequence d[(G4CT)3G4] was denatured with 1 mM or 500 mM KCl present, then folding was carried out comparing different cooling rates. Denatured samples were either slowly cooled to room temperature over the course of several hours or immediately transferred to room temperature or put on ice. In the presence of 500 mM K+ the parallel conformation readily formed irrespective of quick or slow cooling rates, whereas at 1 mM K+ a smaller fraction of oligonucleotides formed the anti-parallel conformer upon quick cooling. Only slow cooling yielded the fully anti-parallel spectrum. Likewise, a very minor fraction of the anti-parallel conformer also formed under slow cooling conditions in the presence of 500 mM K+, the spectrum of the parallel conformer is more pronounced after quick cooling (Fig. S6†).
To determine the isothermal folding kinetics d[(G4CT)3G4] was denatured and slow-cooled without K+ present. KCl was then added to a final concentration of 1 mM at 20 °C and folding of the quadruplex was observed by CD spectroscopy. The anti-parallel conformer folded within 75 hours at 20 °C (Fig. 5a). It was then stable at 20 °C and could not even be converted to the parallel conformation after further addition of KCl to 500 mM, only minor changes in the ellipticity of the anti-parallel conformer were detectable after its incubation with 500 mM K+ for an additional 75 h (Fig. 5b). Addition of 500 mM K+ to d[(G4CT)3G4] folded without K+ present did not induce detectable quadruplex formation up to 65 hours, however after the same sample was denatured at 98 °C the parallel fold was readily adopted after cooling (Fig. 5c). Furthermore when d[(G4CT)3G4] was folded in a parallel conformation at 500 mM and diluted to 50 mM K+ at room temperature, the structure was stable for at least 72 hours at 20 °C and did not convert to the anti-parallel conformer (Fig. 5d), although the latter is more stable at this K+ concentration. Taken together the observed stabilities and folding kinetics suggest extraordinarily high energy barriers for folding and hence slow folding and structure conversion rates of this quadruplex at room temperature.
Similar effects have been observed by Mergny and co-workers, who studied the kinetics of tetramolecular quadruplexes containing a single G-tract.44 They found no dissociation for quadruplexes with G-tracts consisting of 4 or more guanosines in the presence of K+. Isothermal renaturation was highly dependent on the concentration of oligonucleotides used. At low temperature renaturation was found to be minimal for samples containing less than 20 μM oligonucleotides. For d[TG4T] at 10 μM half-association times of 110 days and 2.2 days in 110 mM Na+ and K+, respectively, were found at 4 °C. Generally, increasing the temperature had a deleterious effect on the rates since folding was found to take approximately 15 times longer at room temperature.
Taking the rather short biological timescales into account (e.g. a typical bacterial mRNA lifetime of only a few minutes) the finding of very high activation barriers for folding, unfolding, and structure switching might suggest that a formed quadruplex could be very persistent during genetic and other processes. However, in our opinion it cannot easily be concluded from the in vitro data whether the extraordinary meta-stability of the individual quadruplex conformations is of any biological significance, especially since activities such as quadruplex-resolving helicases and quadruplex binders might strongly influence the kinetics and thermodynamics of quadruplex formation in vivo.
Investigation of individual conformations
We next addressed the nature of the two different anti-parallel and parallel conformations adopted at low and high K+ concentrations. Thermal stabilities of the anti-parallel quadruplex investigated at 1 mM K+ and 25 mM K+ were independent of the concentration of the oligonucleotide (Fig. S7†), indicating that the anti-parallel topology is adopted by an intramolecular structure such as a basket- or chair-like conformation with the oligonucleotide folding back on itself as depicted in Fig. 1b. Due to the high stability of the quadruplex structure under high salt conditions, we could not use melting temperatures in order to deduce the molecularity of the parallel conformer (Fig. S3d†).However, in the past we have successfully employed EPR spectroscopy to probe the human telomeric quadruplex sequence and studied the topology of the individual quadruplexes as well as in the context of the extended telomeric sequence in buffer solution, cell extracts and in frog oocytes.33–35,45,46 Since two-frequency EPR spectroscopy approaches, namely Double Electron Electron Resonance (DEER) is based on the dipolar coupling of two or more electron spins, the technique is ideally suited to elucidate the stoichiometry of the quadruplex structures in the parallel and anti-parallel form. For an intramolecular quadruplex one expects one spin-label per object, whereas in a multimeric arrangement two or four spin-labels are in nanometer proximity to each other, which can easily be detected by DEER spectroscopy (Fig. 6a). We utilized site-directed spin labeling in order to attach a single nitroxide spin-label to the last thymine from the 5′ end of d[(G4CT)3G4] during solid phase DNA synthesis (Fig. 6b and c). Prior to EPR measurements we confirmed by CD spectroscopy that the spin-label does not affect the structural transition (Fig. S8†). The labeling efficiency, i.e. the number of intact spin labels per oligomer was determined to be 75% (see ESI†).
Based on the different modulation depths of the DEER curves (Fig. 6d) the number of spin-labels per quadruplex was calculated.47,48 Taking the labeling efficiency into account the number n of oligomers per quadruplex was determined. The DEER measurements indicated a single spin-label per quadruplex for d[(G4CT)3G4] in the presence of 0 mM (n = 1.1) and 1 mM (n = 1.1) KCl confirming an intramolecular folding at low K+ concentrations. In contrast, the measurement of d[(G4CT)3G4] with 500 mM KCl shows a significant increase in modulation depth corresponding to n = 3.9, which clearly indicates the presence of a tetrameric quadruplex.
In addition to the results obtained by CD and EPR we confirmed the presence of different quadruplex species in an EMSA. Prior to electrophoresis we again verified that the oligonucleotide shows conformational switching in TBE buffer (Fig. S9a†). 5′-radiolabeled d[(G4CT)3G4] was folded in the presence of increasing concentrations of KCl and run on a 16% native polyacrylamide gel in 1xTBE supplemented with 100 mM KCl (Fig. S9b†). When the quadruplex was folded in the absence of K+ and in 1 mM K+ we observed a high mobility band on the gel corresponding to the unfolded oligonucleotide and the anti-parallel conformer. A slower migrating band corresponding to the parallel conformer appears at 100 mM KCl and increases with increasing KCl to 500 mM. Retention of the band at 500 mM K+ in comparison to the anti-parallel conformer at 1 mM K+ indicates formation of a multimeric structure as observed by EPR.
Furthermore, we also employed AUC to distinguish between the different quadruplex species and assess conformational changes (Fig. S10a and b†). We measured different fractions of faster and slower sedimenting species upon increasing KCl concentration. d[(G4CT)3G4] folded in the presence of 1 mM K+ contained a single sedimenting species with the lowest sedimentation coefficient (s-value) of all species. Samples prepared with 100 mM K+ showed a slow sedimenting species and a second, smaller peak for a larger, faster sedimenting species. At 500 mM K+ the situation is reversed, the peak for the slower sedimenting species has decreased while the peak for the faster sedimenting species increased and broadened, indicating multimeric structures. When different temperature gradients were applied during folding of d[(G4CT)3G4] the ratio between the faster and slower sedimenting species in the presence of 500 mM K+ changed (Fig. S10c†). Slow cooling favored the formation of the slower sedimenting species (monomeric), whereas immediate transfer to ice for folding yielded more of the faster sedimenting species (oligomeric). This is in agreement with the results obtained by CD spectroscopy described above (Fig. S6†).
Finally we measured the 1H-NMR spectra of d[(G4CT)3G4] in the presence of 1 mM, 200 mM and 500 mM KCl. At a concentration of 1 mM KCl NMR spectra displayed 16 imino proton signals between 11 and 12 ppm, demonstrating the presence of a well-defined quadruplex conformation. This finding suggests the participation of all four guanosines of each G-tract in tetrad formation. The NMR spectra showed that additional proton signals appear between 10 and 11 ppm upon increasing the KCl concentration to 200 mM KCl or 500 mM (Fig. S11†). However the signals at high KCl are less well defined, likely due to signal overlap of an increased number of non-equivalent imino protons and the high salt concentration.
Influence of loop sequence composition
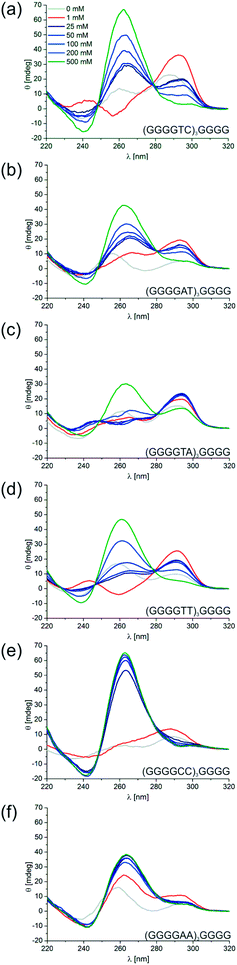
To the best of our knowledge the observed cation selectivity and structural polymorphism are unique among the many quadruplex sequences studied so far. This motivated us to investigate the influence of sequence variations on these remarkable properties. First we varied the loop sequence and analyzed the structural transition and its dependence on K+ by CD spectroscopy. The oligonucleotides d[(G4TC)3G4], d[(G4T2)3G4], d[(G4TA)3G4] and d[(G4AT)3G4] all showed the presence of different quadruplex topologies and a more or less pronounced transition from the anti-parallel to the parallel conformation with increasing concentrations of KCl similar to the structural transition observed for d[(G4CT)3G4] (Fig. 7a–d), likewise none were significantly stabilized by NaCl (Fig. S14b–f†). Inversion of the loop sequence in d[(G4TC)3G4] shifts the equilibrium more to the parallel conformation at lower K+ concentrations, at 500 mM K+ the spectrum shows a strong signal for a parallel quadruplex. Replacement of C in the loop with A again shows a more pronounced parallel fold at lower K+ concentrations for d[(G4AT)3G4], while d[(G4TA)3G4] is present as a mixture of both species over the entire K+ range tested. A structural change from the anti-parallel to the parallel topology could also be observed for the Oxytricha telomeric G-quadruplex d[(G4T2)3G4]. In contrast d[(G4A2)3G4] and d[(G4C2)3G4] exclusively adopted the parallel conformation (Fig. 7e and f). This indicates that a T in the loop sequence is necessary for the stabilization of the anti-parallel conformer at lower K+ concentrations, however in all combinations tested the anti-parallel spectrum was most distinct for the initially studied sequence d[(G4CT)3G4]. Next we examined the influence of loop lengths on the degree of the structural transitions in dependence to K+ by varying the length of the loop sequence from one to four thymidines. We found that a short loop sequence in d[(G4T)3G4] favored the parallel fold in KCl (Fig. S12a†) as well as in NaCl solution (Fig. S14g†). This is in agreement with previous studies by Neidle and co-workers on loop length-dependent folding of G-quadruplexes, who noted a preference for quadruplexes with short loops (1–2 nt) to fold in the parallel conformation with lateral loops as the linker length is unfavorable for diagonal crossing of a tetrad and hence the formation of an anti-parallel topology.49 As mentioned earlier with d[(G4T2)3G4], structural switching was observed (Fig. 7d and S12b†). Increasing the loop size in d[(G4T3)3G4] lead to a mostly anti-parallel topology with only minor structural transition to the parallel conformer at high K+ (Fig. S12c†). Notably, d[(G4T3)3G4] shows a strong signal of an anti-parallel quadruplex even in the complete absence of metal cations. When further increasing the loop size in d[(G4T4)3G4] the anti-parallel fold was observed exclusively (Fig. S12d†). Both d[(G4T3)3G4] and d[(G4T4)3G4] are also stabilized in the anti-parallel conformation in the presence of NaCl (Fig. S14i and j†). In conclusion, while a short 1 nt loop only promotes the parallel fold and long loops of 3–4 nt promote the anti-parallel fold, a loop of two nucleotides enables a structural transition between both topologies depending on the concentration of stabilizing cations. The investigated loop variations follow a general trend, hence with the presented insights it should be able to fine-tune quadruplex equilibria and K+-dependent structure switching for the desired purposes.Finally we also investigated the influence of the G-tract length on K+ dependent topological switching. CD spectroscopy showed that a shorter G-tract in d[(G3CT)3G3] favored the all parallel topology and no switching to the anti-parallel conformation could be observed (Fig. S13a†). Na+ did not stabilize a quadruplex fold (Fig. S14k†). In contrast, a longer G-tract in d[(G5CT)3G5] resulted in a strong anti-parallel spectrum even in the absence of K+ ions (Fig. S13b†). Increasing K+ concentration promoted a structural transition, however retaining predominantly the anti-parallel fold with only an emerging shoulder at 270 nm. Likewise, the spectrum in the presence of Na+ shows predominantly an anti-parallel fold (Fig. S14l†).
Occurrence of (G4CT)3G4 in bacterial genomes
The sequence (G4CT)3G4 was initially identified when antigenic variation was studied in the human pathogen Treponema pallidum.21 Its connection with hypervariable protein products pointed at a role of quadruplex formation in genetic recombination mechanisms as has been described before in Neisseria species.19,20 When we recognized that the sequence behaved very unusually with respect to its propensity to fold into quadruplex conformations, we investigated the occurrence of this G-rich motif in genomic sequence data. We screened bacterial genomes for occurrence of (G4CT)3G4 using BLAST50 (http://blast.ncbi.nlm.nih.gov), searching only completely sequenced bacterial genomes, see ESI.† The sequence is not exclusive to Treponema, but in total present 58 times in a variety of bacterial strains, among others in Burkholderia, Frankia, Salmonella, Shigella and Geobacter strains, see Tables S1–3.† In a BLAST search the Expect value (E value) describes the number of hits expected to be found simply by chance in a particular sequence space; which is dependent on the effective sequence space searched and the length of the query sequence. For illustration, the probability of finding a certain sequence just once by chance in a particular sequence space is described by an E value of 1. The sequence space searched in this study was 52.2 billion nt; the 58 hits found scored an E value of 0.003, meaning that only 0.3% of a sequence space was searched that would randomly yield one perfect match. The sequence (G4CT)3G4 was therefore found significantly more often in bacterial genomes than expected by chance. In addition, the sequence occurs as well in some eukaryotic genomes. For example, the motif occurs several times in the human genome, both within as well as outside transcribed sequences (E value 0.002). For a detailed list of all sites in the human genome see Table S4.†In bacteria we found the sequence to be equally distributed between being part of ORFs, appearing in reverse complementary orientation to ORFs, and being located within the untranslated region (UTR or intergenically) between two genes, as is the case in the original context in Treponema pallidum.21 Interestingly, in 15 cases we found longer repeat sequences of G4CTn, with the longest containing n = 15 repetitions located in the UTR between genes annotated as diguanylate cyclase and aspartate-semialdehyde dehydrogenase in Verrucosispora maris. When the sequence is located within ORFs it encodes glycine rich stretches of amino acids of the type, GWn, GLn or GAn, found for example within the potassium-efflux system protein KefC in Salmonella enterica species (Table S1†). When the sequence is located in the UTR it is most often localized within in the distance of 200 bp from either neighboring gene (Table S3†). In a computational study Chowdhury and co-workers identified putative quadruplex forming sequences within 18 prokaryotes and found an enrichment of such sequences in regulatory regions, which they defined as the region of 200 bp upstream of coding regions.3 This arrangement is the case for 8 out of 9 genera with putative (G4CT)-quadruplexes located intergenically, with an average distance of ∼84 bp from the start of the following coding region. These ORFs encode among a number of unknown proteins two-component system response regulators, the previously mentioned diguanylate cyclase and a pyrophosphokinase.
Conclusions
In this study we characterized the sequence d[(G4CT)3G4] that occurs widely in bacterial genomes. It displays remarkable properties such as a pronounced cation selectivity, folding kinetics with very high activation energies, and a continuous K+-dependent structural transition from a monomolecular anti-parallel topology at low K+ to a four-stranded parallel topology at 500 mM K+. d[(G4CT)3G4] was the only sequence of the studied G-rich oligonucleotides to adopt exclusively the anti-parallel fold at low K+ and at the same time completely converting into the all-parallel topology with increasing concentrations of stabilizing cations. We found that while Na+ and Li+ alone did not stabilize either of the two conformations, addition of either cation to increase ionic strength was sufficient to switch the conformations once initial K+ was available. This definitive requirement for the presence of K+ in order to fold quadruplexes even in the presence of high concentrations of other cations is remarkable and so-far not reported for any other DNA sequence. Further analysis of the sequence requirements of the structural transition showed that thymidines in the loops seemed to be necessary. All 2 nt loop sequences with at least a single thymine could adopt multiple topologies whereas sequences lacking thymidines always showed a parallel quadruplex conformation. The variation of loop length determined that loop sizes of 1 to 2 nt promoted conformational switching. Furthermore, G-tract variations showed that a shortened G-tract of 3 guanines showed the parallel conformation but increasing G-tract length to 5 guanines promoted the anti-parallel topology. With sequence variations possessing K+-dependent conformational polymorphism, Na+ alone was in most cases unable to induce pronounced quadruplex formation. Hence we were able to identify several additional sequences that display a so-far unknown, remarkable selectivity for K+. For further characterization of the nature of the adopted conformations we employed EPR and AUC as analytical methods well-suited to study changes in stoichiometry during the observed structural transition. We demonstrated that d[(G4CT)3G4] is an intramolecular quadruplex in the anti-parallel topology and tetramolecular in the parallel conformation.Several open questions remain regarding a potential role of the studied sequence in cellular processes. Although it seems likely that in Treponema pallidum the motif is involved in genetic recombination events that result in antigenic variation in other species,19,20 the sequence is often associated with intracellular proteins where antigenic variation processes are very unlikely to play a role. Alternatively, an involvement in regulation of gene expression could be discussed. Whether the peculiar structural transitions of the sequence motif upon K+ variation contributes to some mechanism that responds to conditional changes of the bacterial environment is at this point very speculative. However it is intriguing to note that in Salmonella the motif is found associated with two genes coding for K+-transport proteins. Gene expression and dependence on intracellular K+-concentration studies are necessary and under way.
Apart from potential roles in cellular processes, the investigated quadruplex sequence might be utilized as a building block in DNA nanotechnology applications.51 In particular, the possibility of switching between drastically different conformations and being able to control the molecularity of well-defined complexes makes quadruplexes such as the investigated sequence well-suited for building functional DNA nano-objects. We are currently exploring the possibility of exploiting the described K+-dependent tetramerization of DNA sequences for nanotechnological purposes.
Acknowledgements
We thank Žarko Kulić for help with NMR analysis, Marius Schmid and Helmut Coelfen for help with AUC experiments.Notes and references
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed.
- J. L. Huppert, Chem. Soc. Rev., 2008, 37, 1375–1384 RSC.
- P. Rawal, V. B. Kummarasetti, J. Ravindran, N. Kumar, K. Halder, R. Sharma, M. Mukerji, S. K. Das and S. Chowdhury, Genome Res., 2006, 16, 644–655 CrossRef CAS PubMed.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2005, 33, 2908–2916 CrossRef CAS PubMed.
- J. A. Capra, K. Paeschke, M. Singh and V. A. Zakian, PLoS Comput. Biol., 2010, 6, e1000861 Search PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed.
- A. Siddiqui-Jain, C. L. Grand, D. J. Bearss and L. H. Hurley, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11593–11598 CrossRef CAS PubMed.
- K. I. McLuckie, Z. A. Waller, D. A. Sanders, D. Alves, R. Rodriguez, J. Dash, G. J. McKenzie, A. R. Venkitaraman and S. Balasubramanian, J. Am. Chem. Soc., 2011, 133, 2658–2663 CrossRef CAS PubMed.
- S. Cogoi and L. E. Xodo, Nucleic Acids Res., 2006, 34, 2536–2549 CrossRef CAS PubMed.
- A. Joachimi, A. Benz and J. S. Hartig, Bioorg. Med. Chem., 2009, 17, 6811–6815 CrossRef CAS PubMed.
- K. Halder, M. Wieland and J. S. Hartig, Nucleic Acids Res., 2009, 37, 6811–6817 CrossRef CAS PubMed.
- K. Halder, E. Largy, M. Benzler, M. P. Teulade-Fichou and J. S. Hartig, ChemBioChem, 2011, 12, 1663–1668 CrossRef CAS PubMed.
- R. Halder, J. F. Riou, M. P. Teulade-Fichou, T. Frickey and J. S. Hartig, BMC Res. Notes, 2012, 5, 138 CrossRef CAS PubMed.
- M. Wieland and J. S. Hartig, Chem. Biol., 2007, 14, 757–763 CrossRef CAS PubMed.
- T. Endoh, Y. Kawasaki and N. Sugimoto, Angew. Chem., 2013, 52, 5522–5526 CrossRef CAS PubMed.
- T. Endoh and N. Sugimoto, Anal. Chem., 2013, 85, 11435–11439 CrossRef CAS PubMed.
- N. Beaume, R. Pathak, V. K. Yadav, S. Kota, H. S. Misra, H. K. Gautam and S. Chowdhury, Nucleic Acids Res., 2013, 41, 76–89 CrossRef CAS PubMed.
- L. A. Cahoon and H. S. Seifert, Science, 2009, 325, 764–767 CrossRef CAS PubMed.
- L. A. Cahoon and H. S. Seifert, PLoS Pathog., 2013, 9, e1003074 CAS.
- L. Giacani, S. L. Brandt, M. Puray-Chavez, T. B. Reid, C. Godornes, B. J. Molini, M. Benzler, J. S. Hartig, S. A. Lukehart and A. Centurion-Lara, J. Bacteriol., 2012, 194, 4208–4225 CrossRef CAS PubMed.
- O. Kikin, L. D'Antonio and P. S. Bagga, Nucleic Acids Res., 2006, 34, W676–W682 CrossRef CAS PubMed.
- P. A. Rachwal, T. Brown and K. R. Fox, FEBS Lett., 2007, 581, 1657–1660 CrossRef CAS PubMed.
- P. A. Rachwal, I. S. Findlow, J. M. Werner, T. Brown and K. R. Fox, Nucleic Acids Res., 2007, 35, 4214–4222 CrossRef CAS PubMed.
- P. A. Rachwal, T. Brown and K. R. Fox, Biochemistry, 2007, 46, 3036–3044 CrossRef CAS PubMed.
- G. N. Parkinson, M. P. Lee and S. Neidle, Nature, 2002, 417, 876–880 CrossRef CAS PubMed.
- Y. Wang and D. J. Patel, Structure, 1993, 1, 263–282 CrossRef CAS.
- J. Li, J. J. Correia, L. Wang, J. O. Trent and J. B. Chaires, Nucleic Acids Res., 2005, 33, 4649–4659 CrossRef CAS PubMed.
- A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Nucleic Acids Res., 2006, 34, 2723–2735 CrossRef CAS PubMed.
- J. Dai, M. Carver and D. Yang, Biochimie, 2008, 90, 1172–1183 CrossRef CAS PubMed.
- L. Ying, J. J. Green, H. Li, D. Klenerman and S. Balasubramanian, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14629–14634 CrossRef CAS PubMed.
- I. N. Rujan, J. C. Meleney and P. H. Bolton, Nucleic Acids Res., 2005, 33, 2022–2031 CrossRef CAS PubMed.
- V. Singh, M. Azarkh, T. E. Exner, J. S. Hartig and M. Drescher, Angew. Chem., 2009, 48, 9728–9730 CrossRef CAS PubMed.
- M. Azarkh, V. Singh, O. Okle, D. R. Dietrich, J. S. Hartig and M. Drescher, ChemPhysChem, 2012, 13, 1444–1447 CrossRef CAS PubMed.
- V. Singh, M. Azarkh, M. Drescher and J. S. Hartig, Chem. Commun., 2012, 48, 8258–8260 RSC.
- F. M. Chen, Biochemistry, 1992, 31, 3769–3776 CrossRef CAS.
- T. Miura and G. J. Thomas Jr, Biochemistry, 1994, 33, 7848–7856 CrossRef CAS.
- T. Miura, J. M. Benevides and G. J. Thomas Jr, J. Mol. Biol., 1995, 248, 233–238 CAS.
- D. Miyoshi, A. Nakao, T. Toda and N. Sugimoto, FEBS Lett., 2001, 496, 128–133 CrossRef CAS.
- D. Miyoshi, A. Nakao and N. Sugimoto, Nucleic Acids Res., 2003, 31, 1156–1163 CrossRef CAS PubMed.
- J. Kypr, I. Kejnovska, D. Renciuk and M. Vorlickova, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- L. Shabala, J. Bowman, J. Brown, T. Ross, T. McMeekin and S. Shabala, Environ. Microbiol., 2009, 11, 137–148 CrossRef CAS PubMed.
- W. Epstein and S. G. Schultz, J. Gen. Physiol., 1965, 49, 221–234 CrossRef CAS.
- J. L. Mergny, A. De Cian, A. Ghelab, B. Sacca and L. Lacroix, Nucleic Acids Res., 2005, 33, 81–94 CrossRef CAS PubMed.
- M. Azarkh, V. Singh, O. Okle, I. T. Seemann, D. R. Dietrich, J. S. Hartig and M. Drescher, Nat. Protoc., 2013, 8, 131–147 CrossRef CAS PubMed.
- M. Azarkh, O. Okle, V. Singh, I. T. Seemann, J. S. Hartig, D. R. Dietrich and M. Drescher, ChemBioChem, 2011, 12, 1992–1995 CrossRef CAS PubMed.
- D. Hilger, H. Jung, E. Padan, C. Wegener, K. P. Vogel, H. J. Steinhoff and G. Jeschke, Biophys. J., 2005, 89, 1328–1338 CrossRef CAS PubMed.
- A. K. Upadhyay, P. P. Borbat, J. Wang, J. H. Freed and D. E. Edmondson, Biochemistry, 2008, 47, 1554–1566 CrossRef CAS PubMed.
- P. Hazel, G. N. Parkinson and S. Neidle, Nucleic Acids Res., 2006, 34, 2117–2127 CrossRef CAS PubMed.
- S. F. Altschul, T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS PubMed.
- I. T. Seemann, V. Singh, M. Azarkh, M. Drescher and J. S. Hartig, J. Am. Chem. Soc., 2011, 133, 4706–4709 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–14 and Tables S1–4 as described in text; experimental details. See DOI: 10.1039/c4sc00440j |
| This journal is © The Royal Society of Chemistry 2014 |