Open Access Article
Open Access ArticleDinuclear ruthenium(II) antimicrobial agents that selectively target polysomes in vivo
Fangfei
Li
a,
Elizabeth J.
Harry
b,
Amy L.
Bottomley
b,
Michael D.
Edstein
c,
Geoffrey W.
Birrell
c,
Clifford E.
Woodward
a,
F. Richard
Keene
*def and
J. Grant
Collins
*a
aSchool of Physical, Environmental and Mathematical Sciences, University of New South Wales, Australian Defence Force Academy, Canberra, ACT 2600, Australia. E-mail: g.collins@adfa.edu.au
bThe ithree institute, University of Technology Sydney, PO Box 123, Broadway, NSW 2007, Australia
cDepartment of Drug Evaluation, Australian Army Malaria Institute, Gallipoli Barracks, Enoggera, QLD 4051, Australia
dCentre for Biodiscovery and Molecular Development of Therapeutics, James Cook University, Townsville, QLD 4811, Australia
eSchool of Pharmacy and Molecular Sciences, James Cook University, Townsville, QLD 4811, Australia. E-mail: richard.keene@jcu.edu.au
fSchool of Chemistry and Physics, University of Adelaide, Adelaide, SA 5066, Australia
First published on 25th November 2013
Abstract
Wide-field fluorescence microscopy at high magnification was used to study the intracellular binding site of Rubb16 in Escherichia coli. Upon incubation of E. coli cells at the minimum inhibitory concentration, Rubb16 localised at ribosomes with no significant DNA binding observed. Furthermore, Rubb16 condensed the ribosomes when they existed as polysomes. It is postulated that the condensation of polysomes would halt protein production, and thereby inhibit bacterial growth. The results of this study indicate that the family of inert dinuclear ruthenium complexes Rubbn selectively target RNA over DNA in vivo. Selective RNA targeting could be advantageous for the development of therapeutic agents, and because of differences in ribosome structure between bacteria and eukaryotic cells, the Rubbn complexes could be selectively toxic to bacteria. In support of this hypothesis, the toxicity of Rubb16 was found to be significantly less to liver and kidney cell lines than against a range of bacteria.
Introduction
There has been significant interest in the biological properties of inert ruthenium(II) complexes that contain polypyridyl ligands, primarily due to their nucleic acid binding ability.1–6 More recently, research has focused on the anticancer activity of these complexes and the site(s) of accumulation of the ruthenium complexes within eukaryotic cells.7–14 In addition to the studies with eukaryotic cells, there has also been considerable interest in the antimicrobial properties of inert polypyridylruthenium(II) complexes.15–19 As with eukaryotic cells, DNA binding is generally suggested or implied as the possible intra-cellular target. Indeed, in the only intra-bacterial localisation study reported to date (to our knowledge), Gill et al. showed that the rigidly-linked dinuclear ruthenium(II) complex [(phen)2Ru-(μ-tpphz)-Ru(phen)2]4+ (phen = 1,10-phenanthroline; tpphz = tetrapyrido [3,2-a:2′,3′-c:3′′,2′′-h:2′′′,3′′′-j] phenazine) bound chromosomal DNA in Staphylococcus aureus.9 However, toxicity has always been the major concern for the DNA-targeting metal complexes. Due to the lack of variation in the structure of DNA between bacteria and eukaryotic cells, DNA is unlikely to provide the selectivity required for development of a clinically-useful antimicrobial drug. By contrast, ribosomal RNA (rRNA), the most abundant form of RNA in both eukaryotes and bacteria,20,21 is an essential component of ribosomes which exhibit significant differences in bacteria and eukaryotic cells.22 While many antimicrobial drugs in current clinical use target bacterial ribosomes, few if any directly target bacterial chromosomal DNA without causing severe toxicity and side effects.23–25 Therefore, RNA binding could be advantageous for the development of metal complexes as new antimicrobial agents.We have recently demonstrated that a series of dinuclear ruthenium(II) complexes which contain a flexible methylene chain in the bridge [{Ru(phen)2}2(μ-bbn)]4+ {“Rubbn”, where bbn = bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane – see Fig. 1} exhibit excellent antimicrobial activity against both Gram positive and Gram negative bacteria, and retain their activity against drug-resistant strains such as methicillin-resistant S. aureus (MRSA).17–19 Similarly to other dinuclear inert ruthenium(II) complexes that contain polypyridyl ligands, the Rubbn complexes bind DNA and RNA relatively strongly in vitro.26–28 However, the Rubbn complexes show a distinct preference for non-duplex structures, e.g. bulges and hairpin loops.27,28 RNA contains a larger proportion of non-duplex type structures than DNA, forming complicated three-dimensional structures comprising of loops, bulges, pseudo-knots and turns.5 Consequently, we postulated that the Rubbn complexes would preferentially bind RNA, rather than DNA, in bacterial cells and could have significant clinical potential as novel antimicrobial agents with good selectivity and low toxicity.
 | ||
| Fig. 1 The structure of the dinuclear polypyridylruthenium(II) complexes Rubbn, where n = 2, 5, 7, 10, 12 and 16. | ||
Among the Rubbn compounds, Rubb16 has shown the best antimicrobial activity against bacteria with the most rapid and efficient cellular uptake.18 In one of our earlier studies, confocal microscopy was used to examine Escherichia coli that had been incubated with Rubb16.18 Although the magnification and resolution was relatively low, the results suggested that the ruthenium complex localised within the bacterium, but in a manner that was not consistent with chromosomal DNA binding. Consequently, in the present study we aimed to determine the intracellular binding site of Rubb16 by wide-field fluorescence microscopy at high magnification. The results indicate that Rubb16 localises at ribosomes in E. coli, selectively binding RNA of ribosomes, most likely as 70S ribosome and polysomes. It is the first time that a synthetic metal complex (not including metal-based derivatives of existing antibiotics) has shown in vivo RNA binding activity in bacteria, with the ribosome/polysome localisation being visualised via fluorescence microscopy. The cytotoxicity of Rubb16 against liver and kidney cells (where drugs usually accumulate) was also investigated, and the results provide evidence of the selectivity of Rubb16 for bacterial cells over eukaryotic cells.
Experimental
Materials
Rubb16 was synthesised as described previously.29 Luria Broth base was purchased from BD Difco. SYTO 9 Green fluorescent nucleic acid stain and the DNA stain DAPI (4′,6-diamidino-2-phenylindole) were obtained from Molecular Probes, Invitrogen. Rifampicin and chloramphenicol were purchased from Sigma-Aldrich.Bacterial strains and growth conditions
E. coli MG1665 was used in this study. The bacterial strain was grown on Luria Broth (LB) agar plates at 37 °C. A bacterial culture was obtained by inoculating bacteria in LB media and incubating overnight in a shaking incubator in a water bath at 37 °C. The overnight culture was then diluted to a suspension with an optical density of approximately 0.05 at 600 nm (OD600). A bacterial log-phase culture was obtained by continuing the incubation of this suspension for approximately 2 h until the OD600 reached 0.5.MIC assay
The minimum inhibitory concentration (MIC) was determined by the broth microdilution method as outlined in the CLSI guidelines.30 The overnight bacterial culture was diluted to a concentration of 4–8 × 105 cfu mL−1. Rubb16 was serially diluted in LB media on a sterile 96-well plate with a final volume of 100 μL. The bacterial suspension (100 μL) was added to each well and the final concentration range of Rubb16 ranged from 0.125 to 128 μg mL−1. The plate was then placed in a static incubator at 37 °C for 14–16 h before the MIC results were read.Drug treatment and staining protocols
A Rubb16 stock solution was prepared in Milli-Q water at a concentration of 128 μg mL−1. In the time-course movie and SYTO 9 competition experiments, the log-phase bacterial culture was added to Rubb16 at MIC and incubated for 1 h. For the other assays, the incubation time was 15 min and the concentration of Rubb16 ranged from MIC to 4× MIC as described. After incubation with Rubb16, the cells were washed twice with phosphate buffer solution before further treatment or preparation for slides.The concentrations of SYTO 9 and DAPI used in the co-localisation assays were 1 μM and 20 μg mL−1, respectively. The E. coli cells were incubated at room temperature for 15–30 min before being loaded onto agarose pads on slides for microscopy.
Rifampicin and chloramphenicol were initially dissolved in 20 μL of ethanol and then diluted in Milli-Q water to a concentration of 2 mg mL−1 and 20 mg mL−1, respectively. Bacterial cells in the log phase of growth were treated with either of the inhibitors at a concentration of 128 μg mL−1 for 30 min before Rubb16 was added.
Live cell microscopy
All live cell microscopy was performed by placing cells on 2% (w/v) agarose pads (prepared with identical media to that in which the cells were grown) within a 65 μL Gene Frame (Thermo Fisher Scientific). Time-course movies and luminescent images were obtained using a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss) as described previously.31,32 SYTO 9 and DAPI fluorescence and Rubb16 phosphorescence were visualised with filter sets 488009, 02 and 488015 (Carl Zeiss), respectively. Time-lapse studies were performed using a Zeiss Axioplan 2 fluorescence microscope with a Zeiss heated stage equipped with an objective heater, heatable universal mounting frame and an achromatic condenser, set at 37 °C. The movie was set for 20 min at 2 min intervals with an exposure time of 200 ms. Images were analysed and processed using AxioVision version 4.5 (Carl Zeiss).Rubb16 phosphorescence quenching by SYTO 9 and rifampicin
The effect of SYTO 9 or rifampicin on the Rubb16 phosphorescence was determined by monitoring the Rubb16 emission upon addition of SYTO 9 (0.03–4 μM) or rifampicin (0.125–512 μg mL−1) to a Rubb16 solution at constant concentration (4 μg mL−1; final volume of 200 μL).Cytotoxicity of Rubb12 and Rubb16 in mammalian cell lines
The in vitro cytotoxicity of Rubb16 was assessed against the BHK (baby hamster kidney) cell line and two human cell lines – HEK-293 (embryonic kidney) and HEP-G2 (liver carcinoma) – using the alamar Blue cytotoxicity assay.33Results
Localisation of Rubb16
The MIC of Rubb16 against E. coli MG1665 was determined to be 4 μg mL−1. An initial microscopy experiment was carried out to confirm the effect of Rubb16 at MIC on the growth of the bacteria. E. coli cells were incubated with Rubb16 at 37 °C for 1 hour. The treated cells were then prepared and imaged using time-lapse fluorescence microscopy over a 20 minute time-course. During the movie, no elongation of the bacteria was observed in Rubb16-treated samples while untreated control bacteria exhibited normal elongation, confirming that Rubb16 inhibited the growth of the bacteria at MIC (data not shown).E. coli cells were then incubated with Rubb16 at MIC (4 μg mL−1), 2× MIC (8 μg mL−1) and 4× MIC (16 μg mL−1) at 37 °C for 1 hour, washed twice and loaded on agarose pads on slides for fluorescence microscopy. The phase-contrast and luminescence images are shown in Fig. 2. At MIC, orange phosphorescent “spots” indicate that Rubb16 predominantly accumulated at the cell poles and in the middle of the cell at the cylindrical wall. Each of the Rubb16 spots is approximately 0.3 μm (3000 Å) in diameter, and interestingly, the spots located at the cell poles appear to be at the point of maximum curvature. This localisation pattern was also observed at 2× and 4× MIC, but with additional spots in most cells (data not shown). Previous NMR studies have shown that the Rubbn family of complexes bind strongly to both DNA and RNA.26–28 For example, Rubbn complexes bind chromosomal DNA in S. aureus cells (albeit rather weakly).28 However, the localisation of Rubb16 shown in Fig. 2 suggests that the major accumulation site is not chromosomal DNA, which generally occupies a central region of a non-dividing E. coli cell with the poles generally devoid of DNA.34 The DNA-“selective” dye DAPI (which binds DNA 100-fold more strongly than RNA and has a 3-fold higher fluorescence quantum yield when bound to DNA than to RNA) was used to track the DNA distribution in E. coli cells.35
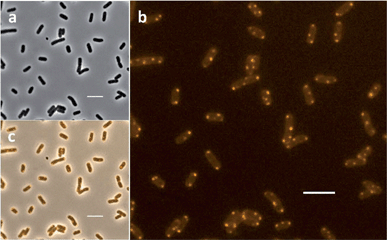 | ||
| Fig. 2 Rubb16 localisation in E. coli MG1665 cells at MIC – 4 μg mL−1. The fluorescence microscopy images are: (a), phase-contrast; (b), phosphorescence; and (c), merged. Scale bar = 5 μm. | ||
Co-staining with DAPI
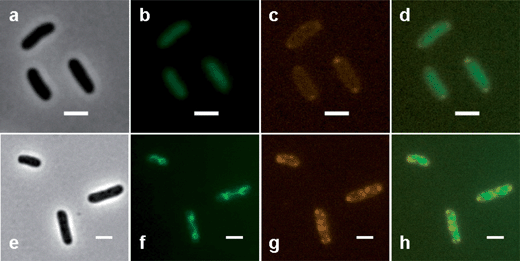
DAPI-stained E. coli cells that had been previously treated with Rubb16 are shown in Fig. 3. The localisation pattern of Rubb16 is consistent with that in bacteria without DAPI staining. At MIC, two to three relatively faint Rubb16 phosphorescence spots are observed and the nucleoid appears to occupy the rest of the cell. Alternatively, five to six spots are observed at 4× MIC and the nucleoid appears to have condensed and become strand-like in appearance (see Fig. 3). At both MIC and 4× MIC there was no significant overlap of DAPI and Rubb16 luminescence, suggesting that chromosomal DNA is not the major localisation site for Rubb16.Competition assay with SYTO 9
Given that Rubb16 has a high affinity for nucleic acids, the lack of Rubb16 phosphorescence localised at chromosomal DNA suggests that RNA is the major in vivo binding target for Rubb16. SYTO 9 is a fluorescent dye that binds RNA more strongly than DNA (although it has a higher fluorescence quantum yield when bound to DNA). Gill et al. have utilised SYTO 9 to visualise RNA in the nucleolus of eukaryotic cells.9 SYTO 9 was used in the present study to further investigate the localisation of Rubb16 with respect to the nucleic acid-enriched regions of the bacteria.E. coli cells at the mid-log phase of growth were incubated with Rubb16 at MIC for 1 hour and then stained with SYTO 9. After 15 minutes of exposure to SYTO 9, the Rubb16 phosphorescence decreased significantly (shown in Fig. 4). After 30 minutes, no Rubb16 phosphorescence was observed. In control experiments, it was shown that SYTO 9 does not quench the Rubb16 phosphorescence, even at concentrations four-times higher than that used in experiments where no Rubb16 phosphorescence was observed. Instead, SYTO 9 appears to be displacing Rubb16 from its binding site.
The effect of chloramphenicol and rifampicin on Rubb16 localisation
The DAPI co-localisation assays indicated that DNA is not the major target, while the competitive binding with SYTO 9 suggests binding of Rubb16 to RNA in the cytoplasm. The polar and central localisation of the Rubb16 foci at MIC are within the regions of E. coli cells normally occupied by ribosomes.36,37 This suggests that rRNA within the ribosomes may be a primary binding target for Rubb16, although the volume of the foci may appear to be too small to account for all the ribosomes in the cell. Therefore we conducted further localisation studies of Rubb16 in E. coli, wherein the population and cellular distribution of RNA and ribosomes were intentionally disrupted by the use of antibiotics.It has been reported that antibiotics which inhibit either transcription or translation can affect the morphology and distribution of the nucleoid as well as the population of RNAs and ribosomes in bacterial cells.36–38 Chloramphenicol and rifampicin are two antibiotics that have been commonly used to bring about such effects. Chloramphenicol inhibits translation by binding to the bacterial ribosome and preventing protein chain elongation via the inhibition of peptidyl transferase.36,37 On the other hand, rifampicin halts transcription initiation by inhibiting RNA polymerase (RNAP).37 It has been previously shown that when exponentially-growing E. coli cells are treated with chloramphenicol, the nucleoids become more condensed both radially and axially. Subsequently, the ribosomes then expand to into the space made available by the nucleoid contraction.36,37 On the other hand, it has been established that in bacterial cells treated with rifampicin (for incubation times ≥ 30 minutes), the nucleoid material expands so that it is distributed throughout the entire cell.37,38 It was also shown that the ribosomes become distributed throughout the cellular space, rather than being localised at the polar regions.
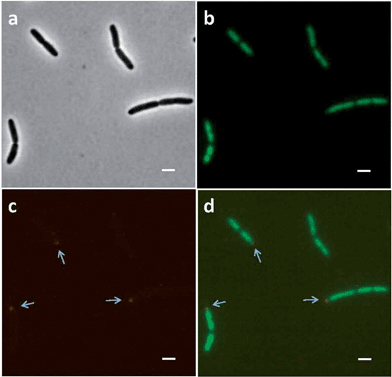
Using the same experimental conditions as Bakshi et al.,37E. coli cells treated with 128 μg mL−1 chloramphenicol for 30 minutes were then incubated with Rubb16 at MIC for 15 minutes. After chloramphenicol treatment, the DAPI-stained nucleoid contracted, while the Rubb16 luminescence was mainly located at the two cell end-caps (Fig. 5a–d). In comparison with chloramphenicol-untreated bacteria (shown again for convenience in Fig. 5e–g), the Rubb16 phosphorescence in the chloramphenicol-treated cells displayed no bright foci but was instead more uniform in intensity, and occupied most of the end-cap volumes. Indeed, the Rubb16 distribution was entirely consistent with that of ribosomes in chloramphenicol-treated cells observed in previous studies.37 This strongly suggests that Rubb16 co-localises with the rRNA in the ribosomes of these cells.
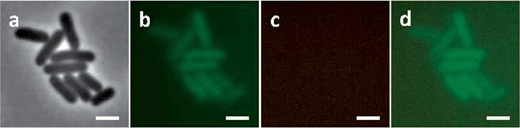
E. coli cells were also treated with 128 μg mL−1 rifampicin for 30 minutes37 and then incubated with Rubb16 at MIC for 15 minutes. As with previous studies, we noted that the nucleoid in rifampicin-treated cells expanded to fill the entire cell (Fig. 6b). Surprisingly, no phosphorescence of Rubb16 was subsequently observed. In control experiments, it was shown that rifampicin only quenches the Rubb16 phosphorescence by 30% at 128 μg mL−1. Consequently, it is probable that the ribosome-bound Rubb16 was now dispersed evenly throughout the whole cell region and the weak phosphorescence of Rubb16 was beyond the detection limit of the fluorescence microscope. Hence, the incubation concentration of Rubb16 was boosted to 4× MIC; however, once again no phosphorescence was detected (Fig. 6). A second control experiment was conducted whereby E. coli cells were incubated with Rubb16 at 4× MIC for 15 minutes before the addition of rifampicin. The results are shown in Fig. 7. Remarkably, the observed Rubb16 phosphorescence was the same as if the cells had not been treated with rifampicin. Furthermore, the nucleoid also appeared to be condensed and essentially identical to that in untreated cells.
Cytotoxicity assay
We have demonstrated that the Rubbn complexes are significantly less toxic to red blood cells and a human leukaemia cell line than against bacteria in our earlier work.17 However, to further explore the potential selectivity of the Rubbn complexes between bacteria and eukaryotic cells, we evaluated the cytotoxicity of Rubb16 towards liver and kidney cell lines. Cell lines from these organs were selected because the liver and kidney are the major sites of drug metabolism.39 The results are summarised in Table 1. The cytotoxicity of Rubb16 was compared with its MICs against bacterial cell strains of (Gram positive) S. aureus and (Gram negative) E. coli shown as a selectivity index (SI = IC50/MIC). The IC50 values of Rubb16 against liver and kidney cells are significantly higher than the MICs against a range of pathogenic bacteria, including MRSA and Pseudomonas aeruginosa.17 It is noteworthy that Rubb16 showed a better selectivity to Gram positive bacteria than their Gram negative counterparts.| BHK | HEK-293 | HEP-G2 | |
|---|---|---|---|
| IC50 (μM) | 210 ± 60 | 172 ± 65 | 106 ± 29 |
| SI (S. aureus) | 350 | 287 | 177 |
| SI (E. coli) | 84 | 69 | 42 |
Discussion
A bacterial ribosome consists of a small 30S subunit and a large 50S subunit, with both components comprised of rRNA and protein. These two subunits associate together with mRNA to form the intact 70S ribosome. A number of ribosomes attached to mRNA form an anionic polysome chain that can efficiently synthesise protein. It has been shown that around 80% of bacterial ribosomes are active in protein synthesis independent of growth rate and conditions.37,40 In growing bacterial cells, most of the active ribosomes occur in polysomes.41,42 A theoretical model of cellular organisation in E. coli, proposed by Mondal et al., purports that ribosomes segregate to the cylindrical wall and polar regions of a bacterial cell to sample the space left unoccupied by the DNA in order to maximise their translational entropy.43Treating E. coli cells with Rubb16 (at MIC) gave rise to small bright foci in regions where polysomes are expected to be located (Fig 2 and Fig. 5e–g). In experiments where the E. coli cells were pre-treated with chloramphenicol no bright foci were observed, instead a more uniform and diffuse Rubb16 phosphorescence filled the end cap regions not occupied by the nucleoid. This suggests that Rubb16 is bound to the rRNA in the polysomes of these cells. It could be expected that the binding of Rubb16 would be similar in cells untreated by chloramphenicol. However, the Rubb16 phosphorescent foci in untreated cells at MIC do not appear to have sufficient volume to account for the number of ribosomes in a typical cell. This paradox can be explained by the occurrence of polysome aggregation. Precipitation of polysomes with multivalent cations (e.g. Cu2+, Zn2+ and Ca2+) is a well-known phenomenon44 and provides a plausible explanation for the findings in this study. An estimate of the total volume of the small foci at MIC indicates that they represent 30–50% of the ribosomes.45 Hence we suggest that when untreated E. coli cells are exposed to Rubb16 at MIC, a proportion (30–50%) of the ribosomes condense and the subsequent high concentration of Rubb16 gives rise to intense luminescent foci, which dominate the image. The remaining non-aggregated polysomes give rise to a lower intensity luminescence. To confirm this, further image analysis of this system was performed to enhance the luminescence of the Rubb16 outside of the foci. Fig. 8 shows that the foci are part of a greater volume of Rubb16 phosphorescence, which has a lower intensity than the foci, but is still much greater than that of the nucleoid and the background solution. These regions of secondary intensity have a volume and location that is consistent with the expected distribution of non-aggregated polysomes. Treatment of E. coli with chloramphenicol is known to give rise to a reduction in the percentage of ribosomes that exist as polysomes.42 Furthermore, as noted earlier, chloramphenicol condenses the nucleoid and allows polysomes to expand into a greater volume, thus reducing their concentration. Taken together, this would lead to diminished polysome aggregation upon addition of Rubb16. Indeed it appears that, when cells are treated with chloramphenicol, the polysomes remain largely dispersed in the cytoplasm, as indicated by the relatively uniform intensity of the Rubb16 phosphorescence.
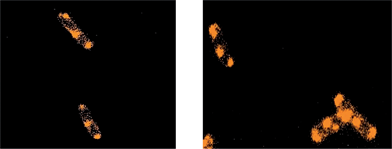 | ||
| Fig. 8 Rubb16 localisation in E. coli MG1665 cells at MIC (left) and 2× MIC (right), with the image re-processed to enhance the luminescence of the Rubb16 outside of the foci. | ||
At higher concentrations (4× MIC) the ruthenium complex could also condense the chromosomal DNA, as shown by staining with DAPI. DNA condensation in the presence of multivalent cationic species is well established.46 At 4× MIC of Rubb16, the rapid and high uptake of Rubb16 would allow some of the ruthenium complex to also bind at secondary lower affinity sites such as the chromosomal DNA. It was puzzling that no significant phosphorescence of the Rubb16 was observed from the nucleoid region at the higher concentration of Rubb16. It is likely that the Rubb16 concentration in the nucleoid remained too low to be observed as a phosphorescence image, despite it causing contraction of the chromosomal DNA. Based upon the number of nucleic acid base pairs and the volume available, the charge density on chromosomal DNA in an E. coli bacterium is about 100-fold less than that of a 70S ribosome (without considering the negative charge of the proteins).47 This is consistent with the relatively lower binding to chromosomal DNA that was previously observed with S. aureus.28
It is noteworthy that at 4× MIC the Rubb16 phosphorescence remained localised and did not expand into the available cellular volume despite nucleoid contraction. Instead, an increased number of localised spots appeared in each cell, compared with cells incubated with Rubb16 at MIC (Fig. 3g and h). An estimate of the total volume of the foci in this case reveals that they could account for approximately 70–100% of the ribosomes. We also observed faint orange phosphorescence in the areas surrounding the condensed nucleoid (see Fig. 3g and h). This could indicate Rubb16 binding to nascent RNA that was localised near the nucleoid before being incorporated into ribosomes.51
The accumulation of ribosomes into polysomal chains gives rise to a significant negative electrostatic potential, which strongly attracts the cationic Rubb16. It is possible that electrostatic correlations cause polysome chains to collapse, via a mechanism similar to that which drives DNA condensation by multivalent cations.46 Polysome aggregates would be expected to nucleate in regions where they have a high concentration. Furthermore, the negatively-charged inner membrane would be likely to provide a scaffold for nucleation due to cooperative adsorption. This explains why the bright foci tended to appear bound to the membrane at the polar end caps, and in the equatorial regions for larger cells (wherein the nucleoid displays two lobes). Interestingly, fainter surface-bound spots were also observed within the cells, indicating areas where secondary nucleation had occurred. Over time, it is probable that these smaller aggregates will diminish through some coarsening mechanism, such as Ostwald ripening, to give rise to just a few major sites of aggregation.52
Polysomes in the cytosol are degraded in a matter of minutes, but their population is replenished by mRNA formation through transcription in the nucleoid. Rifampicin halts transcription through inhibition of RNAP, which co-localises with chromosomal DNA. Therefore, rifampicin does not directly compete with Rubb16 binding. Pre-treatment of E. coli cells with rifampicin led to no measurable detection of Rubb16, even at 4× MIC. During the 30 minutes of rifampicin incubation before the addition of Rubb16, the polysome population would have completely degraded and the 70S ribosomes dissociated into the free 30S and 50S subunits.37,42 These smaller particles are better able to mix with the nucleoid material and the latter is able to expand into the cell cavity. In this environment Rubb16 would compete less effectively with higher concentrations of other cations to neutralise the ribosomal material. The higher mixing entropy and lower charge density favour a dispersed ribosomal phase relative to the aggregated one (even in the presence of Rubb16). This leads to a negligible Rubb16 phosphorescence intensity throughout the cell, due to the lack of ribosomal aggregation.
When rifampicin was added to cells previously incubated with 4× MIC Rubb16, the polysome aggregates which initially formed remained intact, as indicated by the persistent bright foci. That is, the subsequent treatment with rifampicin did not lead to degradation of the polysomal aggregates. The implication is that the formation of condensed aggregates lends some protection to the polysomes from the natural degradation mechanisms in the cell. This was possibly due to large enzymatic molecules finding it difficult to penetrate into the aggregates. On the other hand, the nucleoid was also unaffected by the addition of rifampicin – that is, it did not expand into the cellular cavity, as was the case where rifampicin was added first. The retained compactness of the nucleoid may have been due to the stability of a nucleoid condensed by the addition of 4× MIC Rubb16. Nucleoid compaction may have also negated the effect of the rifampicin on transcription, thus providing an alternative (or additional) cause for the continued presence of polysome aggregates.
Treatment of the cells with the dye SYTO 9, after incubation with Rubb16, gave rise to a surprising diminution of the Rubb16 phosphorescence (Fig. 4). Because of their chemical inertness, Rubbn complexes bind non-duplex nucleic acid structures reversibly as minor groove binders via electrostatic interaction with the negative charge of the sugar phosphate backbone.27,28 As a small cationic dye that binds strongly to RNA via intercalation, SYTO 9 is likely to diffuse within the aggregates and slowly displace Rubb16 from the ribosomes. Presumably the polysomal aggregates subsequently re-dissolve due to the decreasing concentration of bound Rubb16. The dissociated Rubb16 then diffuses out through the cytoplasmic membrane, reducing its total concentration in the cytosol. While SYTO 9 appears to compete with Rubb16 for binding sites on the ribosome, chloramphenicol does not displace the ruthenium complex. As SYTO 9 is a non-specific RNA binding agent, but chloramphenicol selectively binds at the 23S rRNA of the 50S subunit,53 the results tentatively suggest Rubb16 localises at the 30S ribosomal subunit.
We have previously demonstrated that the Rubbn family of complexes are highly active antimicrobial agents. In this study we have also demonstrated that Rubb16 was 40- to 350-fold less toxic to liver and kidney cells compared to several strains of bacteria, indicating a significant selective toxicity for bacteria over eukaryotic cells. In our earlier studies, we have shown that Rubb16 accumulates primarily in the mitochondria in eukaryotes.10 It has been reported that antibiotics targeting the ribosome may show a certain degree of toxicity towards eukaryotes, such as aminoglycosides which cause ototoxicity and nephrotoxicity.54 Although those antibiotics generally have no effect on the cytoplasmic ribosomes in eukaryotes, their toxicity is triggered by the inhibition of mitochondrial ribosomes (mitoribosomes).54 Compared to cytoplasmic ribosomes, mitoribosomes are considered to be more similar to bacterial ribosomes and they contain some sequences that can bind ribosome-targeting antibiotics.54 However, there are still considerable differences in the physical properties of mitochondrial 55S and bacterial 70S ribosomes: the latter contain around 65% RNA, whereas RNA only makes up 33% of the former.55,56 Moreover, 55S ribosomes have a larger mass and physical dimensions than their 70S bacterial counter-parts.57 In addition, a high population of polysomes are found in bacteria while mitoribosomes only appear in mitopolysomes occasionally.41,58 All of these factors suggest that the absolute charge density in mitoribosomes is expected to be lower than that in bacterial ribosomes, which would result in a weaker binding affinity of Rubb16. Finally, it has been estimated that in bacteria 15% of 70S ribosomes are membrane-associated.59 By contrast, in mitochondria most 55S ribosomes are associated with the inner mitochondrial membrane.58 Thus, although Rubb16 accumulates in mitochondria in eukaryotes, mitoribosomes would be unlikely to be condensed by Rubb16. This is especially so, given that the uptake of ruthenium complex in eukaryotes is much less and slower than that in bacteria.10,18 This hypothesis is consistent with the relatively low cytotoxicity against liver and kidney cells of Rubb16 compared to its MIC values.
Conclusions
The results of this study show that Rubb16 – a dinuclear polypyridylruthenium(II) complex which contains a flexible methylene chain in the bridge – known to bind non-duplex nucleic acid structures in vitro, preferentially binds bacterial RNA in vivo and accumulates in bacterial ribosomes. The specific targeting and condensation of the polysomes would enable Rubb16 to halt translation, thus interrupting protein synthesis in actively growing bacterial cells. Rapidly growing bacteria are expected to contain a larger population of polysomes due to the increased requirement of cellular proteins. More importantly, Rubb16 has significant potential as a broad-spectrum antibiotic and is expected to be effective against drug-resistant strains – as has been shown.17 Since the antimicrobial action appears to be controlled by electrostatic interactions with negatively-charged intracellular structures that are essential to all bacteria, mutations are unlikely to combat its activity unless they reduce the cellular uptake. Notably, Rubb16 showed selective toxicity against bacteria over eukaryotic cells, and it appears to result from its RNA binding and localisation in ribosomes with high negative charge density. Furthermore, the modular design and general ease with which the structure of the ruthenium complexes can be modified will facilitate the optimisation of the selective antimicrobial activity of these compounds.Acknowledgements
We are grateful to the Australian Research Council for funding support and Fangfei Li thanks the China Scholarship Council for a PhD scholarship. FRK acknowledges the award of a Visiting Professorship from UNSW Canberra (ADFA). The opinions expressed are those of the authors and do not necessarily reflect those of the Australian Defence Force or any extant Australian Defence Force policy.References
- B. Nordén, P. Lincoln, B. Akerman and E. Tuite, Met. Ions Biol. Syst., 1996, 33, 177 Search PubMed.
- K. E. Erkkila, D. T. Odom and J. K. Barton, Chem. Rev., 1999, 99, 2777 CrossRef CAS PubMed.
- C. Metcalfe and J. A. Thomas, Chem. Soc. Rev., 2003, 32, 215 RSC.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565 RSC.
- F. R. Keene, J. A. Smith and J. G. Collins, Coord. Chem. Rev., 2009, 253, 2021 CrossRef CAS.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179 RSC.
- C. A. Puckett, R. J. Ernst and J. K. Barton, Dalton Trans., 2010, 39, 1159 RSC.
- C. A. Puckett and J. K. Barton, Biochemistry, 2008, 47, 11711 CrossRef CAS PubMed.
- M. R. Gill, J. Garcia-Lara, S. J. Foster, C. Smythe, G. Battaglia and J. A. Thomas, Nat. Chem., 2009, 1, 662 CrossRef CAS PubMed.
- M. J. Pisani, P. D. Fromm, Y. Mulyana, R. J. Clarke, H. Korner, K. Heimann, J. G. Collins and F. R. Keene, ChemMedChem, 2011, 6, 848 CrossRef CAS PubMed.
- M. Matson, F. R. Svensson, B. Nordén and P. Lincoln, J. Phys. Chem. B, 2011, 115, 1706 CrossRef CAS PubMed.
- F. R. Svensson, J. Andersson, H. L. Åmand and P. Lincoln, J. Biol. Inorg. Chem., 2012, 17, 565 CrossRef CAS PubMed.
- T. Chen, Y. Liu, W.-J. Zheng, J. Liu and Y.-S. Wong, Inorg. Chem., 2010, 49, 6366 CrossRef CAS PubMed.
- M. R. Gill, H. Derrat, C. G. W. Smythe, G. Battaglia and J. A. Thomas, ChemBioChem, 2011, 12, 877 CrossRef CAS PubMed.
- F. P. Dwyer, E. C. Gyarfas, W. P. Rogers and J. H. Koch, Nature, 1952, 170, 190 CrossRef CAS PubMed.
- F. P. Dwyer, I. K. Reid, A. Shulman, G. M. Laycock and S. Dixson, Aust. J. Exp. Biol. Med. Sci., 1969, 47, 203 CrossRef CAS PubMed.
- F. Li, Y. Mulyana, M. Feterl, J. M. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032 RSC.
- F. Li, M. Feterl, Y. Mulyana, J. M. Warner, J. G. Collins and F. R. Keene, J. Antimicrob. Chemother., 2012, 67, 2686 CrossRef CAS PubMed.
- M. Pandrala, F. Li, M. Feterl, Y. Mulyana, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2013, 42, 4686 RSC.
- C. Peano, A. Pietrelli, C. Consolandi, E. Rossi, L. Petiti, L. Tagliabue, G. D. Bellis and P. Landini, Microb. Inf. Exp., 2013, 3, 1 CrossRef CAS PubMed.
- H. Lodish, A. Berk, S. L. Zipursky, P. Matsudaira, D. Baltimore and J. Darnell, Molecular Cell Biology, W. H. Freeman, New York, 4th edn, 2000 Search PubMed.
- S. Klinge, F. Voigts-Hoffmann, M. Leibundgut and N. Ban, Trends Biochem. Sci., 2012, 37, 189 CrossRef CAS PubMed.
- J. Poehlsgaard and S. Douthwaite, Nat. Rev. Microbiol., 2005, 3, 870 CrossRef CAS PubMed.
- T. Lambert, Rev. sci. tech. Off. int. epiz., 2012, 31, 57 CAS.
- L. Ming, Med. Res. Rev., 2003, 23, 697 CrossRef CAS PubMed.
- C. B. Spillane, J. A. Smith, D. P. Buck, J. G. Collins and F. R. Keene, Dalton Trans., 2007, 5290 RSC.
- D. P. Buck, J. A. Paul, M. Pisani, J. G. Collins and F. R. Keene, Aust. J. Chem., 2010, 63, 1365 CrossRef CAS.
- F. Li, D. K. Weber, J. L. Morgan, J. G. Collins and F. R. Keene, Dalton Trans., 2012, 41, 6528 RSC.
- Y. Mulyana, D. K. Weber, D. P. Buck, C. A. Motti, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 1510 RSC.
- P. A. Wayne and Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement M100–S19, C.L.S.I., USA, 2009 Search PubMed.
- P. C. Peters, M. D. Migocki, C. Thoni and E. J. Harry, Mol. Microbiol., 2007, 64, 487 CrossRef CAS PubMed.
- S. Moriya, R. A. Rashid, C. D. Andrade Rodrigues and E. J. Harry, Mol. Microbiol., 2010, 76, 634 CrossRef CAS PubMed.
- J. O'Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 267, 5421 CrossRef.
- E. Harry, L. Monahan and L. Thompson, Int. Rev. Cytol., 2006, 253, 27 CrossRef CAS PubMed.
- F. A. Tanious, J. M. Veal, H. Buczak, L. S. Ratmeyer and W. D. Wilson, Biochemistry, 1992, 31, 3103 CrossRef CAS PubMed.
- J. Mascarenhas, M. H. W. Weber and P. L. Graumann, EMBO Rep., 2001, 2, 685 CrossRef CAS PubMed.
- S. Bakshi, A. Siryaporn, M. Goulian and J. C. Weisshaar, Mol. Microbiol., 2012, 85, 21 CrossRef CAS PubMed.
- J. E. Cabrera, C. Cagliero, S. Quan, C. L. Squires and D. J. Jin, J. Bacteriol., 2009, 191, 4180 CrossRef CAS PubMed.
- H. L. Liston, J. S. Markowitz and C. L. DeVane, J. Clin. Psychopharmacol., 2001, 21, 500 CrossRef CAS PubMed.
- H. Bremer and P. P. Dennis, in Escherichia coli and Salmonella: Cellular and Molecular Biology, ASM Press, Washington DC, 2nd edn, 1996, pp. 1553–1569 Search PubMed.
- L. A. Phillips and R. M. Franklin, Cold Spring Harbor Symp. Quant. Biol., 1969, 34, 243 CrossRef CAS PubMed.
- H. L. Ennis, Antimicrob. Agents Chemother., 1972, 1, 197 CrossRef CAS PubMed.
- J. Mondal, B. P. Bratton, Y. Li, A. Yethiraj and J. C. Weisshaar, Biophys. J., 2011, 100, 2605 CrossRef CAS PubMed.
- A. O. Jackson and B. A. Larkins, Plant Physiol., 1976, 57, 5 CrossRef CAS PubMed.
- These calculations are based upon the radius of a 70S ribosome being 10 nm, and hence have an approximate volume of 3.14 × 103 nm3. As there are approximately 20
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ribosomes in a single E. coli cell (http://www.bscb.org/?url=softcell/ribo), the total volume of all the ribosomes would be around 0.06 μm3. Given the diameter of a single Rubb16 phosphorescent spot is 0.3 μm, the volume of a single condensation spot is approximately 0.01 μm3.
000 ribosomes in a single E. coli cell (http://www.bscb.org/?url=softcell/ribo), the total volume of all the ribosomes would be around 0.06 μm3. Given the diameter of a single Rubb16 phosphorescent spot is 0.3 μm, the volume of a single condensation spot is approximately 0.01 μm3. - V. B. Teif and K. Bohinc, Prog. Biophys. Mol. Biol., 2011, 105, 208 CrossRef CAS PubMed.
- The chromosomal DNA in an E. coli bacterium has a compact volume of ∼1 μm3 and contains 4.6 × 106 base pairs.48 Therefore the nucleic acid density (DNA) at the nucleoid region of E.coli is nearly 107 nucleotides per μm3. The volume of an E. coli 70S ribosome is 3.4 × 106 Å3, which contains 4560 nucleotides.49,50 Consequently the nucleic acid density (RNA) at a single 70S ribosome in E.coli is about 109 nucleotides per μm3.
- T. Romantsov, I. Fishov and O. Krichevsky, Biophys. J., 2007, 92, 2875 CrossRef CAS PubMed.
- J. Zhu, J. Struct. Biol., 1997, 118, 197 CrossRef CAS PubMed.
- B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, Molecular Biology of the Cell, Garland Science, New York, 4th edn, 2002 Search PubMed.
- D. J. Jin, C. Cagliero and Y. N. Zhou, Chem. Rev., 2013, 113, 8662 CrossRef CAS PubMed.
- W. Ostwald, Zeitschrift für Physikalische Chemie, 1897, 22, 289 CAS.
- L. McCoy, Y. Xie and Y. Tor, Wiley Interdiscip. Rev.: RNA, 2011, 2, 209 CrossRef CAS PubMed.
- E. C. Bottger, Cell. Mol. Life Sci., 2007, 64, 791 CrossRef CAS PubMed.
- E. C. Koc, W. Burkhart, K. Blackburn, M. B. Moyer, D. M. Schlatzer, A. Moseley and L. L. Spremulli, J. Biol. Chem., 2001, 276, 43958 CrossRef CAS PubMed.
- D. Ridgway, G. Broderick, A. Lopez-Campistrous, M. Ru'aini, P. Winter, M. Hamilton, P. Boulanger, A. Kovalenko and M. J. Ellison, Biophys. J., 2008, 94, 3748 CrossRef CAS PubMed.
- M. G. Hamilton and T. W. O'Brien, Biochemistry, 1974, 13, 5400 CrossRef CAS PubMed.
- J. J. Curgy, Biol. Cell, 1985, 54, 1 CrossRef CAS PubMed.
- J. O. Ortiz, F. Forster, J. Kurner, A. A. Linaroudis and W. Baumeister, J. Struct. Biol., 2006, 156, 334 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |