Suppressing Pseudomonas aeruginosa adhesion via non-fouling polymer brushes†
Abstract
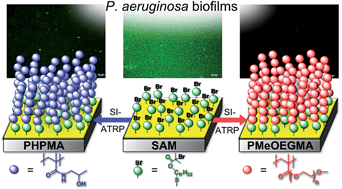
In the current study, well-defined polymer brushes are shown as an effective surface modification to resist biofilm formation from opportunistic pathogens. Poly[oligo(ethylene glycol)methyl ether methacrylate] (poly(MeOEGMA)) and poly[N-(2-hydroxypropyl)methacrylamide] (poly(HPMA)) brushes were grown by surface initiated atom transfer radical polymerization (SI-ATRP) and subsequently characterized by Fourier-transform infrared (FTIR) spectroscopy, X-ray photoelectron spectroscopy (XPS) and dynamic water contact angle measurements. Their remarkable resistance to protein fouling after long term contact with biological media was evidenced by surface plasmon resonance spectroscopy. Challenging these brushes with an environmental strain of Pseudomonas aeruginosa in mineral media as well as a casein–soja–pepton–agar (CASO) medium resulted in no biofilm formation, while a decrease of the biofilm formation by 70% (poly(HPMA)) and 90% (poly(MeOEGMA)) was observed when the medium was rich in nutrients and proteins (fetal bovine serum). In contrast to the antibiotic sensitive strains, biofilm formation was observed using an antibiotic multi-resistant P. aeruginosa strain on both brushes. Protein fouling was fully prevented on both types of brushes, which might challenge the proposed mechanism of biofilm formation mediated by a pre-formed conditioning film of proteins. The resistance to biofilm formation and the possibility to precisely control their growth and functionalities makes these brushes ((poly(HPMA) and (poly(MeOEGMA)) promising candidates for surface modification of various biomaterials as well as platforms for basic studies into the mechanisms of bacteria fouling.

 Please wait while we load your content...
Please wait while we load your content...