Different responses of selenoproteins to the altered expression of selenoprotein W in chicken myoblasts
Haidong Yao†
a,
Wei Liu†ab,
Wenchao Zhao†a,
Ruifeng Fan†a,
Xia Zhao†a,
Pervez Ahmed Khoso†a,
Ziwei Zhang†*a and
Shiwen Xu†*a
aDepartment of Veterinary Medicine, Northeast Agricultural University, Harbin 150030, P. R. China. E-mail: shiwenxu@neau.edu.cn; zhangziwe@sina.com; Tel: +86 451 55190407
bThe Key Laboratory of Myocardial Ischemia, Harbin Medical University, Ministry of Education, Heilongjiang Province, P. R. China
First published on 19th November 2014
Abstract
The aim of the present study was to examine the role of Selenoprotein W (Sepw1) in modulating the expression of other selenoproteins. In the present study, we silenced and overexpressed the expression of Sepw1 in chicken myoblasts and subsequently treated the myoblasts with a reactive oxygen species (ROS) scavenger, N-acetyl-L-cysteine (NAC), and H2O2. Thereafter, the levels of expression of 25 selenoproteins and the activities of certain antioxidative enzymes, glutathione peroxidase (Gpx), superoxide dismutase (SOD), and catalase (CAT) were analyzed. In addition, principal component analysis (PCA) was used to define the most important parameters that could be used as key factors. The results indicated that as a highly expressed selenoprotein (only lower than Gpx1, Selk, Sels and Sep15), Sepw1 could interact with H2O2 (P < 0.05) and influence the expression of some selenoproteins (Gpx3, Gpx4, Txnrd1, Selt, Selh, Sepp1, Sels and Sep15, P < 0.05) and the sensitivity of the cells to H2O2. Both the overexpression and silencing of Sepw1 influenced the mRNA levels of selenoproteins. However, the responses of selenoproteins to altered Sepw1 expression were different. The results indicated that Sepw1 played a special role in H2O2 metabolism and may modulate the expression of certain selenoproteins through the redox pathway. Therefore, these results indicate that Sepw1 is an essential antioxidative selenoprotein in chicken myoblasts.
Introduction
As an essential micronutrient, selenium (Se) is primarily incorporated into selenoproteins as the amino acid selenocysteine (Sec) to perform its biological functions. In recent years, at least 25 selenoprotein genes in animals and humans have been identified.1,2 Among these selenoproteins, several families of selenoproteins have been cloned and partially characterized according to their function. The glutathione peroxidase (Gpx) family (including Gpx1–Gpx6), the thioredoxin reductase (Txnrd) family (including Txnrd1, Txnrd2 and Txnrd3) and the iodothyronine deiodinase (Dio) family (including Dio1, Dio2 and Dio3) have been the most extensively studied selenoprotein families in recent years.3 These typical selenoproteins play important roles in antioxidant defense, redox regulation, conversion of T4 to T3, and regulation in human disease.3,4 In addition, other selenoproteins, including selenophosphate synthetase 2 (SPS2), Seli, Selk, Selm, Sep15, Sepw1, Selh, Selv, Sepn1, Selo, Sepp1, Selr, Sels, and Selt, are also observed in mammals or chicks and have preserved functions.5,6 This finding suggests that these proteins' roles may include the regulation of intracellular calcium (Sepw1, Selk, Selm, Sepn1, Selt),7,8 the control of protein folding in the endoplasmic reticulum (ER) (Sep15, Sels, Selk),9,10 the synthesis of selenophosphate (SPS2), and Se transport (Sepp1).3 Although the functions of different types of selenoproteins are still not well-studied, the possible redox roles of selenoproteins have been studied the most.Se deficiency causes different forms of skeletal and cardiac muscle disease in mammals and humans.7 The abundant studies on the mechanism of Se deficiency disease have indicated that selenoproteins play important roles in skeletal muscle, especially in newborns.11 The first selenoprotein linked to muscular disorders is Sepw1. Therefore, Sepw1 is a good candidate for the study of Se deficiency disorders with respect to selenoproteins. The gene sequences of Sepw1 from different types of animals have been identified,12 and there are some differences between species. Sepw1 contains the 10CXXU13 (where C is Cys, U is the Sec) motif, which is conserved among various mammalian species and implies the ability to catalyze redox reactions. Of the species examined, chicken and fish Sepw1 does not have Cys37, which is necessary for the antioxidative function in rats.13 Sepw1 is highly expressed in the muscle and heart in chicks, sheep, monkeys, cows, calves and humans,12,14,15 but it is not detected in the rat heart.16 Due to the conserved 10CXXU13 motif, Sepw1 has been shown to serve as an antioxidative selenoprotein in different types of cells, including mouse embryonic neurons,13 C2C12 murine myoblasts,17 and chicken myoblasts.18 In contrast, in rat muscle cells, Wang19 indicated that the main function of Sepw1 is not in the antioxidative system because the depletion of Sepw1 could be compensated for by other intracellular antioxidative enzymes. Because there are numerous selenoproteins preserving the function of antioxidants,4 it is reasonable to hypothesize that the deficiency of Sepw1 may be compensated for by other antioxidative selenoproteins.19,20 However, while Sepw1 may not be important in some cell lines, Sepw1 deficiency induces oxidative damage in other cell lines.13,18,19 Whether this contradiction results from the different animal models or from other factors is unknown. Thus, the following questions arose: is there an interaction between Sepw1 and other antioxidative selenoproteins that may compensate for the missing function of Sepw1, and does Sepw1 have an irreplaceable role in different types of cells. In the present study, we examined the mRNA expression levels of selenoproteins following the silencing and overexpression of Sepw1 and discussed the roles of Sepw1 in chicken myoblasts.
Materials and methods
Cell culture
Primary cultures of chicken embryo-derived myoblasts were prepared as we previously described.18 Briefly, myoblasts were isolated from the pectoralis of 12 day old chicken embryos and digested with 0.1% collagenase I (Invitrogen, Carlsbad, CA). To release single cells, the suspension was triturated by gentle pipetting and filtered to remove large debris. The cell suspension was washed twice and subjected to density gradient centrifugation in three discontinuous layers of 20%, 30% and 55% Percoll (Pharmacia, Uppsala, Sweden). The cells at the 30% and 55% Percoll interface were harvested, rinsed twice, and re-suspended in proliferation medium (PM) that consisted of DMEM (Gibco, BRL), 10% horse serum (HS) (Hyclone, Logan, UT), and a 5% chicken embryo extract prepared from 11 day old embryos by our laboratory according to previously published procedures.21 Myoblasts were seeded in gelatin-coated (Sigma, St. Louis, Mo) 6-well culture plates (Jet, China) at a density of 4 × 105 cells per cm2 with PM. Myoblasts proliferated for 24 h in 5% CO2 at 37 °C and were induced to differentiate by replacing the PM with differentiation medium (DM) that consisted of Opti-MEM (Gibco, BRL) and 7.5% knockout serum replacement (KSR) (Gibco, BRL).Sepw1 RNA interference
The method and reagents are the same as in our prior study,18 where we demonstrated that the target siRNAs have no off-target effects. Chicken myoblasts were plated in 6-well plates at 70–80% confluence and transfected with 3 μL of 20 μM siRNAs and 3 μL of Lipofectamine RNAiMAX Reagent (Invitrogen) in 2 mL of Opti-MEM. After transfection for approximately 48 h, the cells were harvested for analysis.Cloning of the chicken Sepw1 cDNA and expression vector construction
The method was performed as described by our group.22 Briefly, total RNA was extracted from chicken skeletal muscle tissue samples (50 mg) using Trizol reagent (Invitrogen, China). First-strand cDNAs were synthesized using oligo dT primers and Superscript II reverse transcriptase (Invitrogen, China), according to the manufacturer's instructions. Unless stated otherwise, the resulting cDNA pool was used directly for PCR amplification. Using the chicken sequence from GenBank (accession no. GQ919055), primers were designed to amplify chicken Sepw1, as in the prior study.22 The PCR amplifications were performed using Taq DNA polymerase (Fermentas), and the PCR product was separated on a 1% agarose gel. The amplified DNA fragments were then purified from the agarose gels and subcloned into the pMD-18 T (TaKaRa, China) plasmid vector. The DNA fragment that contained the cDNA was released from the modified pMD-18 T plasmid by digestion with EcoRI and XhoI and was then sub-cloned into the mammalian expression vector pCDNA3.1 (+) (Invitrogen, China), yielding pCDNA3.1/Sepw1. The fragment insert was then sequenced (Huada, China). Transfections were performed using the X-tremeGENE HP DNA transfection reagent (Roche), following the manufacturer's instructions. A 3 μg sample of pCDNA3.1/Sepw1 that encoded chicken Sepw1 was added to 200 μL Opti-MEM (Invitrogen) before mixing with 3 μL of X-tremeGENE HP DNA transfection reagent. The plasmid DNA/X-tremeGENE HP mixture was incubated at room temperature for 25 min and then added into each well of the 6-well plates.After transfection for approximately 48 h, the cells were treated with 50 μM H2O2 in differentiation medium for 6 h and then were harvested for analysis. In the NAC group, cells were co-incubated with 2.5 mM N-acetyl-L-cysteine (NAC) for 6 h after transfection and were harvested for analysis.
Quantitative real-time PCR (qPCR) analysis of selenoprotein mRNA levels
The total RNA was isolated23 from individual wells of myoblasts using TRIzol reagent according to the manufacturer's instructions (Invitrogen, Shanghai, China). The dried RNA pellets were re-suspended in 50 μL of diethyl-pyrocarbonate-treated water. The concentration and purity of the total RNA were determined spectrophotometrically at 260/280 nm. First-strand cDNA was synthesized from 10 pg of total RNA using dN12 primers and the AccuPower® RocketScript™ RT PreMix (BIONEER) according to the manufacturer's instructions. Synthesized cDNA was diluted five times with sterile water and stored at −80 °C before use.Primer Premier Software (PREMIER Biosoft International, USA) was used to design specific primers for selenoproteins, GAPDH and β-actin based on known chicken sequences (Table 1). Standard PCR was first performed to confirm the specificity of the primers. The PCR products were electrophoresed on 2% agarose gels, extracted, cloned into the pMD18-T vector (TaKaRa, China), and sequenced. Quantitative real-time PCR was performed on a BIO-RAD C1000 Thermal Cycler (USA). Reactions were performed in a 20 μL reaction mixture containing 10 μL of AccuPower® 2X Greenstar qPCR Master Mix (BIONEER), 2 μL of diluted cDNA, 1 μL of each primer (10 μM) and 6 μL of PCR-grade water. The PCR procedure consisted of 95 °C for 30 s, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 60 °C for 30 s. The melting curve analysis showed only one peak for each PCR product. Electrophoresis was performed with the PCR products to verify primer specificity and product purity. A dissociation curve was run for each plate to confirm the production of a single product. The amplification efficiency for each gene was determined using the DART-PCR program.24 The mRNA relative abundance was calculated as previously described25 to account for gene-specific efficiencies and was normalized to the mean expression of GAPDH and β-actin.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Gpx1 | 5′-ACGGCGCATCTTCCAAAG-3′ | 5′-TGTTCCCCCAACCATTTCTC-3′ |
| Gpx2 | 5′-ACGGCACCAACGAGGAGAT-3′ | 5′-TTCAGGTAGGCGAAGACGG-3′ |
| Gpx3 | 5′-CCTGCAGTACCTCGAACTGA-3′ | 5′-CTTCAGTGCAGGGAG GATCT-3′ |
| Gpx4 | 5′-CTTCGTCTGCATCATCACCAA-3′ | 5′-TCGACGAGCTGAGTGTAATTCAC-3′ |
| Txnrd1 | 5′-TACGCCTCTGGGAAATTCGT-3′ | 5′-CTTGCAAGGCTTGTCCCAGTA-3′ |
| Txnrd2 | 5′-GCTCTTAAAGATGCCCAGCACTAC-3′ | 5′-GAACAGCTTGAGCCATCACAGA-3′ |
| Txnrd3 | 5′-CCTGGCAAAACGCTAGTTGT G-3′ | 5′-CGCACCATTACTGTGACATCTAGAC-3′ |
| Dio1 | 5′-GCGCTATACCACAGGCAGTA-3′ | 5′-GGTCTTGCAAATGTCACCAC-3′ |
| Dio2 | 5′-ATTTGCTGATCACGCTTCAG-3′ | 5′-GCTCAGAAACAGCACCATGT-3′ |
| Dio3 | 5′-CTGTGCATTCGCAAGAAGAT-3′ | 5′-GCCGACTTGAAGAAGTCCAG-3′ |
| Sepn1 | 5′-CAGGATCCATGCTGAGTTCCA-3′ | 5′-GAGAGGACGATGTAACCCGTAAAC-3′ |
| Selk | 5′-GAAGAGGGCCTCCAGGAAAT-3′ | 5′-CAGCCATTGGTGGTGGACTAG-3′ |
| Sels | 5′-GCGTCGCCATCTATCTCATCGT-3′ | 5′-TCTTCTGCCTTCGCTTCTGTTCTT-3′ |
| Sepw1 | 5′-TGGTGTGGGTCTGCTTTACG-3′ | 5′-CCAAAGCTGGAAGGTGCAA-3′ |
| Selt | 5′-AGGAG TACAT GCGGG TCATC A-3′ | 5′-GACAGACAGGAAGGATGCTATGTG-3′ |
| Selh | 5′-CATCGAGCACTGCCGTAG-3′ | 5′-GACACCTCGAAGCTGTTCCT-3′ |
| Selm | 5′-AAGAAGGACCACCCAGACCT-3′ | 5′-GCTGTCCTGTCTCCCTC ATC-3′ |
| Sep15 | 5′-ACTTGGCTTCTCCAGTAACTTGCT-3′ | 5′-GCCTACAGAATGGATCCAACTGA-3′ |
| Seli | 5′-TGCCAGCCTCTGAACTGGAT-3′ | 5′-TGCAAACCCAGACATCACCAT-3′ |
| Selu | 5′-GATGCTTTCAGGCTTCTTCC-3′ | 5′-CTGTCTTCCTGCTCCAATCA-3′ |
| Selpb | 5′-AGGCCAACAGTACCATGGAG-3′ | 5′-GTGGTGAGGATGGAGATGGT-3′ |
| Sepp1 | 5′-CCAAGTGGTCAGCATTCACATC-3′ | 5′-ATGACGACCACCCTCACGAT-3′ |
| Selo | 5′-CCAGCGTTAACCGGAATGAT-3′ | 5′-ATGCGCCTCCTGGATTTCT-3′ |
| Sepx1 | 5′-TGGCAAGTGTGGCAATGG-3′ | 5′-GAATTTGAGCGAGCTGCTGAAT-3′ |
| SPS2 | 5′-CGTTGGGTATCGGAACTGAC-3′ | 5′-CGTCCACCAGAGGGTAGAAA-3′ |
| β-Actin | 5′-CCGCTCTATGAA GGCTACGC-3′ | 5′-CTCTCG GCTGTGGTGGTGAA-3′ |
| GAPDH | 5′-AGAACATCATCCCAGCGT-3′ | 5′-AGCCTTCACTACCCTCTTG-3′ |
Detection of intracellular antioxidative enzymes
Detection of intracellular glutathione peroxidase (Gpx). The assay was performed using the Total Cellular Glutathione Peroxidase Assay Kit (Beyotime Biotechnology, China). Cells were treated with 0.02% EDTA and were washed and collected in PBS. Cells (1 × 106) were then lysed with 200 μL of cell lysis buffer (Beyotime Biotechnology, P0013). The lysate was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g at 4 °C for 10 min, and the supernatant was then assayed according to the kit's instructions. The assay was performed at 25 °C with Multimode Plate Readers (TECAN Infinite M200 PRO, Switzerland) at 340 nm.
000 g at 4 °C for 10 min, and the supernatant was then assayed according to the kit's instructions. The assay was performed at 25 °C with Multimode Plate Readers (TECAN Infinite M200 PRO, Switzerland) at 340 nm.
Detection of intracellular superoxide dismutase (SOD). The assay was performed using the Total Superoxide Dismutase Assay Kit (Beyotime Biotechnology, China). Briefly, 1 × 106 cells were collected and homogenized with PBS on ice. Lysates were used to determine the enzyme activity. The SOD assay was performed according to the manufacturer's instructions with Multimode Plate Readers (TECAN Infinite M200 PRO, Switzerland) at 450 nm. The catalase (CAT) assay was performed using the Catalase Assay Kit (Beyotime Biotechnology, China). Briefly, cell lysates were used to determine enzyme activity. The CAT assay was performed according to the manufacturer's instructions with Multimode Plate Readers (TECAN Infinite M200 PRO, Switzerland) at 520 nm.
The formation of malondialdehyde (MDA) was determined as an indicator of lipid peroxidation using the thiobarbituric acid assay26 (Beyotime Biotechnology, China). The MDA assay was performed according to the manufacturer's instructions with Multimode Plate Readers (TECAN Infinite M200 PRO, Switzerland) at 532 nm. The protein concentrations of the samples were measured using the Bradford method.27
Western blot analysis
Protein extracts were subjected to SDS–polyacrylamide gel electrophoresis under reducing conditions on 15% gels. Separated proteins were then transferred to nitrocellulose membranes using tank transfer for 1 h at 100 mA in Tris–glycine buffer containing 20% methanol. The membranes were blocked with 5% skim milk for 16–24 h and incubated overnight with diluted primary antibody against Sepw1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, production of polyclonal antibody by our lab), Gpx1 and Gpx4 (1
1000, production of polyclonal antibody by our lab), Gpx1 and Gpx4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Abcam, China), followed by a horseradish peroxidase (HRP)-conjugated secondary antibody against rabbit IgG (1
1000, Abcam, China), followed by a horseradish peroxidase (HRP)-conjugated secondary antibody against rabbit IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Santa Cruz Biotechnology, USA). To verify equal sample loading, the membrane was incubated with a monoclonal β-actin antibody (1
1000, Santa Cruz Biotechnology, USA). To verify equal sample loading, the membrane was incubated with a monoclonal β-actin antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Santa Cruz Biotechnology, USA), followed by an HRP-conjugated goat anti-mouse IgG (1
1000, Santa Cruz Biotechnology, USA), followed by an HRP-conjugated goat anti-mouse IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000). The signal was detected by X-ray films (TransGen Biotech Co., Beijing, China). The optical density (OD) of each band was determined using the Image VCD gel imaging system.
1000). The signal was detected by X-ray films (TransGen Biotech Co., Beijing, China). The optical density (OD) of each band was determined using the Image VCD gel imaging system.
Statistical analysis
Data analysis was performed using SPSS statistical software for Windows (Version 13; SPSS Inc, Chicago, IL). Differences between the means were assessed by Tukey's honestly significant difference test for post hoc multiple comparisons. Data are expressed as the means ± standard deviation. The differences were considered to be significant at P < 0.05.In addition, principal component analysis (PCA) was used to define the most important parameters that could be used as key factors for individual variations using the Statistics 6.0 program.
Results
The expression pattern of selenoproteins in chicken myoblasts
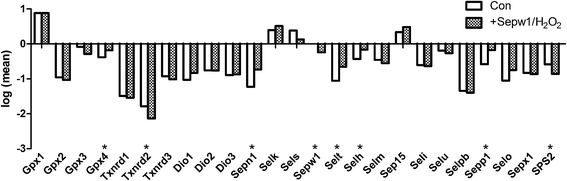
In the present study, we first examined the expression of selenoproteins in chicken myoblasts (Table 2). As shown in Fig. 1, under common conditions, the expression levels of the selenoproteins were different from one another. There were only four selenoproteins (Gpx1, Selk, Sels, and Sep15) that were expressed at higher levels than Sepw1. Compared with other selenoprotein families, the Rdx family (Sepw1, Selt, Selh, Selm, and Sep15) was relatively highly expressed. However, the expression of the Txnrd family (Txnrd1, Txnrd2, Txnrd3) and Dio family (Dio1, Dio2, Dio3) was relatively lower than other selenoproteins.| Con | Over-expression | siRNA | siRNA/NAC | Con/H2O2 | Over-expression/H2O2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | log(mean) | Mean | SD | log(mean) | Mean | SD | log(mean) | Mean | SD | log(mean) | Mean | SD | log(mean) | Mean | SD | log(mean) | |
| Gpx1 | 7.61 | 0.50 | 0.88 | 6.50 | 1.50 | 0.81 | 7.09 | 1.66 | 0.85 | 6.13 | 0.15 | 0.79 | 0.67 | 0.05 | −0.17 | 7.62 | 1.08 | 0.88 |
| Gpx2 | 0.11 | 0.03 | −0.96 | 0.09 | 0.02 | −1.05 | 0.14 | 0.01 | −0.84 | 0.03 | 0.01 | −1.55 | 2.87 | 0.12 | 0.46 | 0.09 | 0.02 | −1.03 |
| Gpx3 | 0.83 | 0.15 | −0.08 | 0.48 | 0.05 | −0.32 | 1.90 | 0.29 | 0.28 | 0.54 | 0.09 | −0.27 | 3.43 | 0.40 | 0.54 | 0.51 | 0.05 | −0.29 |
| Gpx4 | 0.42 | 0.06 | −0.38 | 0.67 | 0.06 | −0.18 | 1.25 | 0.17 | 0.10 | 0.59 | 0.17 | −0.23 | 2.98 | 0.25 | 0.47 | 0.65 | 0.04 | −0.19 |
| Txnrd1 | 0.03 | 0.01 | −1.49 | 0.03 | 0.00 | −1.54 | 0.05 | 0.01 | −1.29 | 0.01 | 0.00 | −1.84 | 2.16 | 0.22 | 0.33 | 0.03 | 0.00 | −1.54 |
| Txnrd2 | 0.02 | 0.00 | −1.79 | 0.01 | 0.00 | −1.85 | 0.01 | 0.00 | −2.11 | 0.00 | 0.00 | −2.43 | 0.51 | 0.10 | −0.29 | 0.01 | 0.00 | −2.14 |
| Txnrd3 | 0.12 | 0.03 | −0.93 | 0.14 | 0.03 | −0.85 | 0.07 | 0.01 | −1.16 | 0.05 | 0.01 | −1.30 | 5.80 | 0.06 | 0.76 | 0.10 | 0.01 | −1.02 |
| Dio1 | 0.09 | 0.01 | −1.03 | 0.13 | 0.01 | −0.87 | 0.07 | 0.03 | −1.14 | 0.04 | 0.01 | −1.43 | 3.54 | 0.46 | 0.55 | 0.15 | 0.02 | −0.84 |
| Dio2 | 0.17 | 0.02 | −0.76 | 0.18 | 0.05 | −0.75 | 0.17 | 0.02 | −0.77 | 0.09 | 0.02 | −1.04 | 6.61 | 0.16 | 0.82 | 0.17 | 0.04 | −0.76 |
| Dio3 | 0.13 | 0.01 | −0.89 | 0.15 | 0.04 | −0.82 | 0.20 | 0.07 | −0.70 | 0.04 | 0.01 | −1.39 | 1.76 | 0.02 | 0.25 | 0.13 | 0.05 | −0.87 |
| Sepn1 | 0.06 | 0.01 | −1.23 | 0.11 | 0.00 | −0.97 | 0.06 | 0.01 | −1.21 | 0.04 | 0.01 | −1.38 | 2.53 | 0.42 | 0.40 | 0.19 | 0.03 | −0.73 |
| Selk | 2.48 | 0.75 | 0.39 | 2.85 | 0.33 | 0.45 | 2.52 | 0.53 | 0.40 | 1.49 | 0.25 | 0.17 | 4.58 | 0.35 | 0.66 | 3.26 | 0.45 | 0.51 |
| Sels | 2.41 | 0.40 | 0.38 | 2.55 | 0.28 | 0.41 | 1.75 | 0.31 | 0.24 | 0.90 | 0.04 | −0.05 | 9.33 | 1.60 | 0.97 | 1.34 | 0.28 | 0.13 |
| Sepw1 | 1.00 | 0.00 | 0.00 | 6.08 | 0.69 | 0.78 | 0.36 | 0.06 | −0.45 | 0.26 | 0.10 | −0.58 | 0.33 | 0.06 | −0.48 | 0.57 | 0.06 | −0.24 |
| Selt | 0.09 | 0.02 | −1.06 | 0.18 | 0.00 | −0.75 | 0.22 | 0.02 | −0.67 | 0.09 | 0.01 | −1.04 | 0.42 | 0.10 | −0.37 | 0.22 | 0.02 | −0.66 |
| Selh | 0.37 | 0.01 | −0.43 | 0.68 | 0.05 | −0.17 | 0.77 | 0.10 | −0.11 | 0.53 | 0.15 | −0.28 | 0.96 | 0.22 | −0.02 | 0.68 | 0.09 | −0.16 |
| Selm | 0.35 | 0.03 | −0.46 | 0.54 | 0.04 | −0.27 | 0.28 | 0.07 | −0.56 | 0.36 | 0.05 | −0.44 | 2.75 | 0.52 | 0.44 | 0.28 | 0.04 | −0.55 |
| Sep15 | 2.18 | 0.28 | 0.34 | 1.67 | 0.30 | 0.22 | 0.63 | 0.06 | −0.20 | 0.31 | 0.09 | −0.50 | 9.61 | 0.80 | 0.98 | 3.03 | 0.67 | 0.48 |
| Seli | 0.25 | 0.04 | −0.61 | 0.28 | 0.07 | −0.55 | 0.25 | 0.04 | −0.60 | 0.12 | 0.02 | −0.92 | 11.37 | 2.18 | 1.06 | 0.23 | 0.04 | −0.64 |
| Selu | 0.64 | 0.06 | −0.20 | 0.80 | 0.24 | −0.10 | 0.78 | 0.04 | −0.11 | 0.42 | 0.10 | −0.38 | 15.49 | 3.53 | 1.19 | 0.54 | 0.11 | −0.27 |
| Selpb | 0.05 | 0.01 | −1.34 | 0.09 | 0.00 | −1.06 | 0.05 | 0.01 | −1.33 | 0.05 | 0.01 | −1.34 | 1.99 | 0.05 | 0.30 | 0.04 | 0.00 | −1.40 |
| Sepp1 | 0.26 | 0.04 | −0.58 | 0.41 | 0.12 | −0.39 | 0.44 | 0.02 | −0.35 | 0.79 | 0.18 | −0.10 | 8.40 | 0.35 | 0.92 | 0.66 | 0.08 | −0.18 |
| Selo | 0.09 | 0.02 | −1.05 | 0.16 | 0.04 | −0.80 | 0.08 | 0.01 | −1.12 | 0.02 | 0.00 | −1.61 | 4.53 | 0.61 | 0.66 | 0.18 | 0.06 | −0.75 |
| Sepx1 | 0.15 | 0.02 | −0.83 | 0.27 | 0.01 | −0.56 | 0.16 | 0.00 | −0.79 | 0.09 | 0.02 | −1.05 | 4.83 | 0.13 | 0.68 | 0.14 | 0.01 | −0.86 |
| SPS2 | 0.26 | 0.02 | −0.58 | 0.32 | 0.07 | −0.49 | 0.17 | 0.03 | −0.78 | 0.06 | 0.01 | −1.23 | 6.26 | 1.20 | 0.80 | 0.14 | 0.03 | −0.86 |
The expression of selenoproteins were influenced by H2O2 under oxidative conditions (Fig. 1). H2O2 primarily decreased the levels of Sepw1 and Gpx1, and increased the expression of other selenoproteins (P < 0.05). It has been shown that selenoproteins have the unique ability to trap H2O2. Because of the rapid oxidation kinetics of selenols, H2O2 in the cytosol reacts completely with Sec containing proteins to produce Sec-selenenic acid before it can react with other potential targets.28 The present study showed that different selenoproteins have a different response to H2O2 in myoblasts. Sepw1 and Gpx1 react mainly with H2O2, so the levels of Sepw1 and Gpx1 were predominantly consumed by H2O2. Other selenoproteins were primarily increased, which may suggest that they play protective or buffering roles. This is because most of selenoproteins can regulate oxidation–reduction homeostasis in cells,20 enhance the ability of cells to protect themselves against oxidative stress29 and decrease oxidative damage.30 Although many selenoproteins preserve their antioxidative function, their response to oxidative stress is different. In the present study, Sepw1 played a special role in H2O2 metabolism.
The effect of Sepw1 overexpression on selenoproteins levels
Over-expression of Sepw1 (Fig. 2) induced higher levels of Sepn1, Selt, Selh, Selm, Selpb and Sepx1 expression, but lower Gpx3 expression (P < 0.05). The increased expression of these antioxidative selenoproteins (Sepn1, Selh, and Sepx1) can regulate oxidation–reduction homeostasis in cells.3,20,31 Therefore, over-expression of Sepw1 may enhance the antioxidative ability of cells.After treatment with H2O2, selenoprotein expression in the overexpressing group was influenced (Fig. 3). Similar to the control/H2O2 group, Sepw1 levels were also reduced by H2O2 in cells overexpressing Sepw1. The expression of some selenoproteins (Gpx4, Txnrd2, Sepn1, Selt, Selh, and Sepp1) was increased (P < 0.05). Compared with the control group, there were fewer selenoproteins that showed increased expression with H2O2 in the Sepw1-overexpressing group. The overexpression of Sepw1 decreased the cells' oxidative stress response. Thus, we observed that overexpression of Sepw1 could enhance the levels of some selenoproteins and decrease its own expression to regulate oxidation–reduction homeostasis and decrease the cells' response to oxidative stress. Therefore, Sepw1 may play a crucial antioxidative function in chicken myoblasts.
The effect of Sepw1 deficiency on the selenoprotein expression
As an important antioxidative selenoprotein, we hypothesized that Sepw1 may influence the levels of other selenoproteins through the redox pathway. Several interaction may exist between Sepw1 and other selenoproteins, and the deficiency of Sepw1 may be compensated for by other antioxidative selenoproteins. Therefore, we silenced the expression of Sepw1 to test this hypothesis. Following the silencing of Sepw1 (Fig. 4), the expression of some selenoproteins varied. The expression of Gpx3, Gpx4, Txnrd1, Selt, Selh and Sepp1 was increased (P < 0.05); however, Sep15 and Sels expression was reduced (P < 0.05). Among the affected selenoproteins, Gpx3, Gpx4, Txnrd1, Selt, Selh, Sepp1 and Sels preserve their antioxidative function,7 which may replace the missing redox regulation role of Sepw1. However, our prior study showed that Sepw1 silencing in myoblasts induced higher levels of apoptosis and ROS production.18 Although there were increased levels of antioxidants (Gpx3, Gpx4, Txnrd1, Selt, and Sepp1), Sepw1 deficiency was not compensated for by these increased endogenous antioxidants.It was unclear whether exogenous antioxidants could influence the effect of Sepw1 on selenoprotein expression. In the present study, we treated the myoblasts with the antioxidant NAC to eliminate the increased ROS levels and reverse the oxidative damage induced by Sepw1 deficiency. The results showed that NAC treatment influenced the effect of Sepw1 on the regulation of selenoproteins (Fig. 5). In the NAC group, the effect of Sepw1 deficiency on the expression of Gpx3, Gpx4, Txnrd1, and Selt was eliminated by NAC (P > 0.05), but the effect on the expression of Sels, Sep15 and Sepp1 remained (P < 0.05). NAC treatment reduced the oxidative damage, but did not reverse all of the effects of Sepw1 deficiency. Therefore, chicken Sepw1 may play a crucial role in chicken myoblasts.
The protein levels of Sepw1, Gpx1 and Gpx4
In the present study, we also examined the protein levels of some characterized selenoproteins, namely Sepw1, Gpx1 and Gpx4 (Fig. 6). The results showed that, similar to the mRNA levels, the protein levels of Sepw1 increased in the over-expressing group but were decreased in the other groups. Compared with the control, the protein level of Gpx1 was decreased by H2O2 treatment. However, in the other groups, the protein level of Gpx1 was not influenced. In addition, the Gpx4 protein levels were increased in the siRNA and Con/H2O2 groups. The results indicated that the effect of the siRNA- and H2O2-induced oxidative stress on the expression of some characterized selenoproteins was similar at both the mRNA and the protein levels. | ||
| Fig. 6 The protein levels of Sepw1, Gpx1 and Gpx4. +Sepw1 and +W indicate the overexpressing groups; −Sepw1 and −W indicate the siRNA groups. | ||
Measurement of antioxidative enzymes and MDA levels
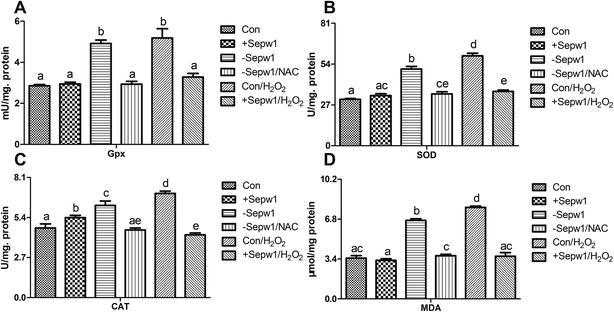
In the present study, we also examined the activities of certain crucial antioxidants, specifically Gpx, SOD, and CAT. As a typical oxidant, H2O2 also induced oxidative damage in chicken myoblasts. The results (Fig. 7) showed that compared with the control group, H2O2 treatment induced higher activities of Gpx, SOD, and CAT and increased MDA levels (P < 0.05). However, this effect was eliminated by overexpressing Sepw1 in cells (P > 0.05 or not). Therefore, Sepw1 reversed the H2O2-induced oxidative injury, which indicated that Sepw1 served as an antioxidant in chicken myoblasts.Similar to H2O2 treatment, Sepw1 deficiency also increased the activities of Gpx, SOD, and CAT and induced oxidative damage (higher MDA levels) (P < 0.05). In contrast, when treating these cells with NAC, the Sepw1-induced oxidative damage was reduced (P > 0.05), indicating that exogenous antioxidant could regulate the oxidative unbalance induced by Sepw1 deficiency.
Principal component analysis (PCA)
Using PCA, all of the parameters were distinguished on ordination plots corresponding to the first and second principle components (90.64% and 4.38%, respectively) (Fig. 8). Furthermore, the observed relationships among the parameters were confirmed and quantified according to Spearman's test (Table 3), which showed that selenoproteins had a negative correlation with Sepw1, but Gpx1 had a positive correlation with Sepw1. Selenoproteins had a strong positive correlation with component one, and Gpx1 and Sepw1 were negatively correlated with component one. Sepp1, Gpx4, and Gpx3 had a relatively strong positive correlation with component two, but only Selk, Selh, Gpx1 and Sepw1 had a negative correlation with component two (Table 4). Thus, Gpx1, Gpx3, Gpx4, Selk, Selh, Sepp1 and Sepw1 may play special roles in the response to oxidative stress or altered Sepw1 expression in myoblasts. | ||
| Fig. 8 Principal component analysis. The rotating components in space. Ordination plots corresponding to the first and second principle components were 90.64% and 4.38%, respectively. | ||
| Gpx1 | Gpx2 | Gpx3 | Gpx4 | Txnrd1 | Txnrd2 | Txnrd3 | Dio1 | Dio2 | Dio3 | Sepn1 | Selk | Sels | Sepw1 | Selt | Selh | Selm | Sep15 | Seli | Selu | Selpb | Sepp1 | Selo | Sepx1 | SPS2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gpx1 | 1.00 | −0.97 | −0.85 | −0.94 | −0.97 | −0.97 | −0.97 | −0.97 | −0.97 | −0.96 | −0.97 | −0.71 | −0.94 | 0.16 | −0.83 | −0.71 | −0.98 | −0.88 | −0.97 | −0.97 | −0.98 | −0.98 | −0.97 | −0.97 | −0.97 |
| Gpx2 | −0.97 | 1.00 | 0.90 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.98 | −0.24 | 0.89 | 0.72 | 0.99 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Gpx3 | −0.85 | 0.90 | 1.00 | 0.96 | 0.90 | 0.89 | 0.89 | 0.89 | 0.89 | 0.91 | 0.88 | 0.72 | 0.88 | −0.37 | 0.86 | 0.76 | 0.86 | 0.80 | 0.89 | 0.90 | 0.89 | 0.88 | 0.89 | 0.89 | 0.89 |
| Gpx4 | −0.94 | 0.96 | 0.96 | 1.00 | 0.96 | 0.95 | 0.96 | 0.96 | 0.96 | 0.97 | 0.95 | 0.80 | 0.93 | −0.27 | 0.93 | 0.85 | 0.94 | 0.88 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.95 |
| Txnrd1 | −0.97 | 1.00 | 0.90 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.82 | 0.98 | −0.24 | 0.88 | 0.72 | 0.99 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Txnrd2 | −0.97 | 1.00 | 0.89 | 0.95 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.98 | −0.22 | 0.88 | 0.71 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Txnrd3 | −0.97 | 1.00 | 0.89 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.98 | −0.22 | 0.88 | 0.71 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Dio1 | −0.97 | 1.00 | 0.89 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.84 | 0.98 | −0.22 | 0.89 | 0.72 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Dio2 | −0.97 | 1.00 | 0.89 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.98 | −0.23 | 0.88 | 0.71 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Dio3 | −0.96 | 1.00 | 0.91 | 0.97 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.85 | 0.99 | −0.22 | 0.90 | 0.74 | 0.99 | 0.96 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 |
| Sepn1 | −0.97 | 1.00 | 0.88 | 0.95 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.84 | 0.98 | −0.23 | 0.89 | 0.73 | 0.99 | 0.97 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Selk | −0.71 | 0.83 | 0.72 | 0.80 | 0.82 | 0.83 | 0.83 | 0.84 | 0.83 | 0.85 | 0.84 | 1.00 | 0.85 | −0.01 | 0.92 | 0.75 | 0.82 | 0.92 | 0.83 | 0.83 | 0.82 | 0.81 | 0.84 | 0.83 | 0.83 |
| Sels | −0.94 | 0.98 | 0.88 | 0.93 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.99 | 0.98 | 0.85 | 1.00 | −0.10 | 0.86 | 0.68 | 0.99 | 0.96 | 0.98 | 0.98 | 0.98 | 0.97 | 0.98 | 0.98 | 0.99 |
| Sepw1 | 0.16 | −0.24 | −0.37 | −0.27 | −0.24 | −0.22 | −0.22 | −0.22 | −0.23 | −0.22 | −0.23 | −0.01 | −0.10 | 1.00 | −0.15 | −0.05 | −0.15 | −0.19 | −0.23 | −0.22 | −0.21 | −0.26 | −0.22 | −0.21 | −0.21 |
| Selt | −0.83 | 0.89 | 0.86 | 0.93 | 0.88 | 0.88 | 0.88 | 0.89 | 0.88 | 0.90 | 0.89 | 0.92 | 0.86 | −0.15 | 1.00 | 0.93 | 0.87 | 0.88 | 0.88 | 0.88 | 0.88 | 0.88 | 0.89 | 0.88 | 0.88 |
| Selh | −0.71 | 0.72 | 0.76 | 0.85 | 0.72 | 0.71 | 0.71 | 0.72 | 0.71 | 0.74 | 0.73 | 0.75 | 0.68 | −0.05 | 0.93 | 1.00 | 0.70 | 0.66 | 0.71 | 0.72 | 0.71 | 0.72 | 0.72 | 0.72 | 0.71 |
| Selm | −0.98 | 0.99 | 0.86 | 0.94 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 0.82 | 0.99 | −0.15 | 0.87 | 0.70 | 1.00 | 0.95 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 |
| Sep15 | −0.88 | 0.96 | 0.80 | 0.88 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.97 | 0.92 | 0.96 | −0.19 | 0.88 | 0.66 | 0.95 | 1.00 | 0.96 | 0.96 | 0.96 | 0.95 | 0.96 | 0.96 | 0.96 |
| Seli | −0.97 | 1.00 | 0.89 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.98 | −0.23 | 0.88 | 0.71 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Selu | −0.97 | 1.00 | 0.90 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.98 | −0.22 | 0.88 | 0.72 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Selpb | −0.98 | 1.00 | 0.89 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.82 | 0.98 | −0.21 | 0.88 | 0.71 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Sepp1 | −0.98 | 1.00 | 0.88 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.81 | 0.97 | −0.26 | 0.88 | 0.72 | 0.99 | 0.95 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Selo | −0.97 | 1.00 | 0.89 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.84 | 0.98 | −0.22 | 0.89 | 0.72 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Sepx1 | −0.97 | 1.00 | 0.89 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.98 | −0.21 | 0.88 | 0.72 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| SPS2 | −0.97 | 1.00 | 0.89 | 0.95 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 0.99 | −0.21 | 0.88 | 0.71 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Component | Dio3 | Sepx1 | Selm | Selo | SPS2 | Sepn1 | Dio1 | Selu | Selpb | Gpx2 | Seli | Txnrd3 | Dio2 | Txnrd2 | Txnrd1 | Sels | Sepp1 | 15-Sep | Gpx1 | Gpx4 | Selt | Selk | Gpx3 | Selh | Sepw1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a Rotating convergence after three iteration. | |||||||||||||||||||||||||
| 1 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.96 | −0.96 | 0.96 | 0.93 | 0.88 | 0.87 | 0.78 | |
| 2 | 0.14 | 0.14 | 0.15 | 0.14 | 0.15 | 0.15 | 0.15 | 0.15 | 0.16 | 0.16 | 0.16 | 0.16 | 0.16 | 0.17 | 0.18 | −0.12 | 0.17 | −0.16 | 0.29 | −0.13 | −0.96 | ||||
Discussion
Sepw1 preserves the antioxidative function in different types of cells.12,18 Through redox activity, Sepw1 may regulate the cell cycle32–34 and the development of muscle,17 interact with 14-3-3 protein,32 regulate the level of GSH in human neuronal cells,35 and compensate for the missing function of other selenoproteins, such as Selt in murine fibroblast cells.20 However, because there are so many selenoproteins and enzymes preserving the function of antioxidants,4 it is reasonable to hypothesize that Sepw1 deficiency may be compensated for by the other antioxidative selenoproteins.19,20 In the present study, we first examined the antioxidative function of Sepw1 and then discussed the possible interactions between Sepw1 and selenoproteins. The results showed that, as a highly expressed selenoprotein in myoblasts, Sepw1 has a crucial antioxidative function. When the level of Sepw1 was altered, the mRNA levels of some selenoproteins were influenced, which may rely on the levels of ROS.Oxidative stress always occurs during the unbalance between antioxidative ability and ROS levels. Cells can defend against excessive ROS by non-enzymatic small molecule antioxidants, such as glutathione, and enzymes that include SOD, CAT and Gpx. In addition, different types of selenoproteins also preserve the antioxidative role. Approximately 25 selenoprotein genes have been identified in mammals2 and chicks.6 Among these selenoproteins, there is a group of antioxidative selenoproteins, including the Gpx family (Gpx1, Gpx2 Gpx3 and Gpx4), the Txnrd family (Txnrd1, Txnrd2 and Txnrd3),7 and other identified or possible antioxidative selenoproteins, such as Sepw1, Selk, Sepn1, Sepp1, Sels,3 Sepx1, Selh, and Selo.31 Under common conditions, these antioxidants can regulate the susceptibility of the cell to oxidative injury6 and reduce the oxidative damage induced by some oxidants, such as H2O2.36 These selenoproteins, such as Gpx, Sepn1, Selh, Selm, Sels, and Sepw1, can metabolize H2O2 and regulate several cellular responses. In skeletal muscles, Sepw1 is highly expressed and plays a crucial role in this tissue. However, the exact role of Sepw1 required more study. In the present study, we first examined the possible antioxidative function of chicken Sepw1 in myoblasts. Under common conditions, the mRNA level of Sepw1 was higher than most selenoproteins, except Gpx1, Selk, Sels and Sep15. This higher level may also allow it to serve as an important antioxidant in myoblasts. After treating myoblasts with H2O2, the levels of Sepw1 and Gpx1 were reduced in the cells, which indicated that Sepw1 might play a crucial role in metabolizing H2O2 in chicken myoblasts. The rapid reaction of Sepw1 and Gpx1 with H2O2 may act to limit the duration and distance of H2O2 signals, which decreased the cells' response to H2O2.28,30 In the overexpressing cells, H2O2 decreased Sepw1 levels but only increased a few selenoproteins (5 selenoproteins) compared with the control cells (21 selenoproteins), which showed that overexpressed Sepw1 decreased the cells' response to H2O2. In addition, Sepw1 overexpression could reduce the H2O2-induced oxidative damage.37 In addition, the increased expression of certain antioxidative selenoproteins may regulate oxidation–reduction homeostasis in cells,20 enhance the ability of cells to protect against oxidative stress29 and decrease the oxidative damage and the concentration of H2O2.30 In contrast, Sepw1 deficiency induced increased Gpx, SOD, CAT and MDA levels, and elevated ROS levels and apoptosis.18 It showed that Sepw1 deficiency decreased the ability of myoblasts to buffer the exogenous H2O2 and increased the sensitivity of cells to H2O2. Therefore, similar to its mammalian homolog in different cells or organs,13,19,35 chicken Sepw1 plays important role in antioxidative functions in myoblasts. In addition, the principal components analysis also supported this point, indicating that Sepw1 and Gpx1 may play special roles in the response to oxidative stress in chicken myoblasts. Therefore, chicken Sepw1 also preserves antioxidative function.
Selenoproteins can respond to oxidative stress and change their expression levels to balance or reverse an oxidants-induced unbalance.30,31,38 However, in response to oxidative stress, selenoproteins possess different expression patterns. In the heart, ROS can induce higher levels of Gpx3 and Gpx4, but not Gpx1, to minimize intracellular oxidative damage.39 In irradiated normal human fibroblasts, elevated ROS increased the mRNA levels of Txnrd1 and Sepp1.40 In addition, the expression of Drosophila Selr was also elevated in response to several agents that cause oxidative stress, such as H2O2.41 However, in cultured myotubes, oxidative stress did not influence the mRNA levels of Sepn1, Sepp1 and Sels.42 Oxidative stress induced by Selt deficiency only elevated Sepw1 expression in murine fibroblast cells, but did not influence the levels of Txnrd1, Gpx1, Gpx4, and Sep15.20 The decrease in Sepp1 was associated with lower Gpx activity in malignant tissue.43 Although the expression patterns of selenoproteins responding to oxidative stress in chicken myoblasts has not been identified, there must be some interaction between selenoproteins and oxidative stress. In the present study, by silencing the expression of Sepw1 and treating with H2O2, we simulated the conditions of oxidative stress in myoblasts. Although the treatment with H2O2 or Sepw1 deficiency induced higher ROS levels, apoptosis18 and oxidative stress in myoblasts, the effect on the expression of selenoproteins was different. Exogenous H2O2 induced higher levels of most of the selenoproteins, but decreased Gpx1 and Sepw1. However, Sepw1 silencing induced higher expression of Gpx3, Gpx4, TrxR1, Selt, Selh and Sepp1 and lower expressions of Sels and Sep15 in myoblasts, which showed that the response of selenoproteins to oxidative stress induced by exogenous H2O2 or Sepw1 silencing was different. Thus, Sepw1 deficiency influenced fewer selenoproteins (6 increased selenoproteins and 2 decreased selenoproteins) compared to H2O2 treatment (22 increased selenoproteins and 2 decreased selenoproteins), but more selenoproteins than the deficiency of Selt (1 increased selenoprotein). This showed that 50 μM H2O2 induced higher ROS levels than Sepw1 deficiency in chicken myoblasts.18 In addition, the effects of Sepw1 on the expressions of other selenoproteins was influenced by treatment with the antioxidant, NCA. The effects of Sepw1 deficiency on the expression of Gpx3, Gpx4, Txnrd1, Selt, and Sepp1 were eliminated by NAC, but the effects on the expressions of Sels, Sep15 and Sepp1 remained. Therefore, the different responses of selenoproteins to Sepw1 deficiency or H2O2 may be dependent on the ROS levels.
The responses of selenoproteins to the overexpression of other selenoproteins have also been analyzed in previous studies.44 In Selm overexpression rats, the activities of SOD and GPx were higher than the wild type rats.29 In Gpx1 overexpression rats, the activities of Gpx1 and Gpx4 were increased in liver and muscles and the activities of thioredoxin reductase were increased in liver but not in muscles, but activities of SOD were decreased in these organs.45 In Txnrd1 overexpression HEK-293 cells, Gpx activity was decreased in limited Se medium, but not in normal medium.46 Therefore, in response to selenoprotein overexpression, the expression patterns of other selenoproteins were different. In the present study, Sepw1 overexpression also affected the expression of other selenoproteins. Sepw1 overexpression level increased the expression of Sepn1, Selt, Selh, Selm, Selpb and Sepx1, and reduced the responses of selenoproteins to H2O2. Compared with these prior studies, the present study examined more selenoproteins in response to altered selenoprotein expression. Thus we demonstrated that altering Sepw1 expression could influence the mRNA levels of selenoproteins in chicken myoblast.
In summary, under common conditions, Sepw1 was expressed at relatively higher levels than most selenoproteins in myoblasts. Sepw1 and Gpx1 react mainly with H2O2, so the levels of Sepw1 and Gpx1 were primarily consumed by H2O2. However, the expression of other selenoproteins was increased and may play a protective or buffering role in chicken myoblasts. In the present study, Sepw1 played special role in H2O2 metabolism and may modulate the expressions of some selenoproteins through the redox pathway. Therefore, Sepw1 is an essential antioxidative selenoprotein in chicken myoblast.
Conflict of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This study was supported by the National Natural Science Foundation of China (31272626) and the International (Regional) Cooperation and Exchange Projects of the National Natural Science Foundation of China (31320103920); and the Doctoral Fund of the Ministry of Education of China (20122325110018). We are thankful for the support of the Key Laboratory of Myocardial Ischemia, Harbin Medical University. The authors thank the Elsevier English Language Editing System to correct grammatical, spelling, and other common errors.References
- A. V. Lobanov, D. L. Hatfield and V. N. Gladyshev, Genome Biol., 2008, 9, R62 CrossRef.
- G. V. Kryukov, S. Castellano, S. V. Novoselov, A. V. Lobanov, O. Zehtab, R. Guigo and V. N. Gladyshev, Science, 2003, 300, 1439–1443 CrossRef CAS.
- A. C. Pappas, E. Zoidis, P. F. Surai and G. Zervas, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2008, 151, 361–372 CrossRef CAS.
- F. P. Bellinger, A. V. Raman, M. A. Reeves and M. J. Berry, Biochem. J., 2009, 422, 11–22 CrossRef CAS.
- A. Dikiy, S. V. Novoselov, D. E. Fomenko, A. Sengupta, B. A. Carlson, R. L. Cerny, K. Ginalski, N. V. Grishin, D. L. Hatfield and V. N. Gladyshev, Biochemistry, 2007, 46, 6871–6882 CrossRef CAS.
- M. Mariotti, P. G. Ridge, Y. Zhang, A. V. Lobanov, T. H. Pringle, R. Guigo, D. L. Hatfield and V. N. Gladyshev, PLoS One, 2012, 7, e33066 CAS.
- A. Lescure, M. Rederstorff, A. Krol, P. Guicheney and V. Allamand, Biochim. Biophys. Acta, 2009, 1790, 1569–1574 CrossRef CAS.
- M. A. Reeves, F. P. Bellinger and M. J. Berry, Antioxid. Redox Signaling, 2010, 12, 809–818 CrossRef CAS.
- S. Du, J. Zhou, Y. Jia and K. Huang, Arch. Biochem. Biophys., 2010, 502, 137–143 CrossRef CAS.
- S. Du, H. Liu and K. Huang, Biochim. Biophys. Acta, 2010, 1800, 511–517 CrossRef CAS.
- E. N. Terry, J. J. Michal, C. E. Hostetler and R. L. Kincaid, Livest. Sci., 2009, 124, 21–25 CrossRef.
- P. D. Whanger, Biochim. Biophys. Acta, 2009, 1790, 1448–1452 CrossRef CAS.
- Y. W. Chung, D. Jeong, O. J. Noh, Y. H. Park, S. I. Kang, M. G. Lee, T. H. Lee, M. B. Yim and I. Y. Kim, Mol. Cells, 2009, 27, 609–613 CrossRef CAS.
- M. Rederstorff, A. Krol and A. Lescure, Cell. Mol. Life Sci., 2006, 63, 52–59 CrossRef CAS.
- B. R. Ou, M. J. Jiang, C. H. Lin, Y. C. Liang, K. J. Lee and J. Y. Yeh, Biometals, 2011, 24, 323–333 CrossRef CAS.
- P. D. Whanger, CMLS, Cell. Mol. Life Sci., 2000, 57, 1846–1852 CrossRef CAS.
- J. Loflin, N. Lopez, P. D. Whanger and C. Kioussi, J. Inorg. Biochem., 2006, 100, 1679–1684 CrossRef CAS.
- H. D. Yao, Q. Wu, Z. W. Zhang, S. Li, X. L. Wang, X. G. Lei and S. W. Xu, Biochim. Biophys. Acta, 2013, 1830, 3112–3120 CrossRef CAS.
- X.-L. Wang, C.-P. Yang, K. Xu and O.-J. Qin, Biochemistry, 2010, 75, 201–207 CAS.
- A. Sengupta, B. A. Carlson, V. M. Labunskyy, V. N. Gladyshev and D. L. Hatfield, Biochem. Cell Biol., 2009, 87, 953–961 CrossRef CAS.
- G. Shefer and Z. Yablonka-Reuveni, Methods Mol. Biol., 2005, 290, 281–304 Search PubMed.
- Y. H. Han, Z. W. Zhang, C. Shao, S. Li, S. W. Xu and X. L. Wang, Biol. Trace Elem. Res., 2012, 148, 61–68 CrossRef CAS.
- H. D. Yao, Q. Wu, Z. W. Zhang, J. L. Zhang, S. Li, J. Q. Huang, F. Z. Ren, S. W. Xu, X. L. Wang and X. G. Lei, J. Nutr., 2013, 143, 613–619 CrossRef CAS.
- S. N. Peirson, J. N. Butler and R. G. Foster, Nucleic Acids Res., 2003, 31, e73 CrossRef.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS.
- H. Esterbauer and K. H. Cheeseman, Methods Enzymol., 1990, 186, 407–421 CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- C. C. Winterbourn and M. B. Hampton, Free Radical Biol. Med., 2008, 45, 549–561 CrossRef CAS.
- D. Y. Hwang, J. S. Sin, M. S. Kim, S. Y. Yim, Y. K. Kim, C. K. Kim, B. G. Kim, S. B. Shim, S. W. Jee, S. H. Lee, C. J. Bae, B. C. Lee, M. K. Jang, J. S. Cho and K. R. Chae, Int. J. Mol. Med., 2008, 21, 169–179 CAS.
- W. C. Hawkes and Z. Alkan, Biol. Trace Elem. Res., 2010, 134, 235–251 CrossRef CAS.
- M. A. Reeves and P. R. Hoffmann, Cell. Mol. Life Sci., 2009, 66, 2457–2478 CrossRef CAS.
- W. C. Hawkes, T. T. Wang, Z. Alkan, B. D. Richter and K. Dawson, Biol. Trace Elem. Res., 2009, 131, 229–244 CrossRef CAS.
- W. C. Hawkes and Z. Alkan, Biochem. Biophys. Res. Commun., 2011, 413, 36–40 CrossRef CAS.
- W. C. Hawkes and Z. Alkan, J. Biol. Chem., 2012, 287, 27371–27379 CrossRef CAS.
- Y. J. Kim, Y. G. Chai and J. C. Ryu, Biochem. Biophys. Res. Commun., 2005, 330, 1095–1102 CrossRef CAS.
- H. Nishida, H. Ichikawa and T. Konishi, J. Pharmacol. Sci., 2007, 105, 342–352 CrossRef CAS.
- Y. H. Han, Z. W. Zhang, J. Su, B. Zhang, S. Li and S. W. Xu, Biol. Trace Elem. Res., 2012, 147, 395–402 CrossRef CAS.
- J. Lu and A. Holmgren, J. Biol. Chem., 2009, 284, 723–727 CrossRef CAS.
- F. W. Hoffmann, A. S. Hashimoto, B. C. Lee, A. H. Rose, R. V. Shohet and P. R. Hoffmann, Arch. Biochem. Biophys., 2011, 512, 38–44 CrossRef CAS.
- J. C. Eckers, A. L. Kalen, W. Xiao, E. H. Sarsour and P. C. Goswami, Int. J. Radiat. Oncol., Biol., Phys., 2013, 87, 619–625 CrossRef CAS.
- G. V. Kryukov, R. A. Kumar, A. Koc, Z. Sun and V. N. Gladyshev, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4245–4250 CrossRef CAS.
- J. J. Dowling, S. Arbogast, J. Hur, D. D. Nelson, A. McEvoy, T. Waugh, I. Marty, J. Lunardi, S. V. Brooks, J. Y. Kuwada and A. Ferreiro, Brain, 2012, 135, 1115–1127 CrossRef.
- Y. Saito and K. Takahashi, Eur. J. Biochem., 2002, 269, 5746–5751 CrossRef CAS.
- Z. Touat-Hamici, Y. Legrain, A. L. Bulteau and L. Chavatte, J. Biol. Chem., 2014, 289, 14750–14761 CrossRef CAS.
- J. P. McClung, C. A. Roneker, W. Mu, D. J. Lisk, P. Langlais, F. Liu and X. G. Lei, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8852–8857 CrossRef CAS.
- I. Nalvarte, A. E. Damdimopoulos, C. Nystom, T. Nordman, A. Miranda-Vizuete, J. M. Olsson, L. Eriksson, M. Bjornstedt, E. S. Arner and G. Spyrou, J. Biol. Chem., 2004, 279, 54510–54517 CrossRef CAS.
Footnote |
| † All authors have read the manuscript and agreed to submit it in its current form for consideration for publication in the journal. |
| This journal is © The Royal Society of Chemistry 2014 |