Peptide arrays for detecting naphthenic acids in oil sands process affected water†
Kamaljit Kaur*ab,
Subir Bhattacharjeecd,
Rajesh G. Pillaic,
Sahar Ahmedae and
Sarfuddin Azmia
aFaculty of Pharmacy and Pharmaceutical Sciences, University of Alberta, Edmonton, Alberta T6G 2E1, Canada. E-mail: kkaur@ualberta.ca; Tel: +1 780-492-8917
bChapman University School of Pharmacy (CUSP), Harry and Diane Rinker Health Science Campus, Chapman University, Irvine, CA 92618-1908, USA
cDepartment of Mechanical Engineering, University of Alberta, Edmonton, Alberta T6G 2G8, Canada
dWater Planet Engineering, 721 Glasgow Ave, Inglewood, California 90301, USA
eMedicinal Chemistry Department, Faculty of Pharmacy, Assiut University, Assiut 71526, Egypt
First published on 4th November 2014
Abstract
Naphthenic acids (NAs) are water-soluble components of petroleum. The characterization and quantification of NAs by analytical methods have proved quite challenging, whilst the toxic effects of these water-soluble compounds on a variety of organisms adversely affecting reproduction and steroid production is becoming apparent. In this study, we report a fluorescence-based competitive binding method for rapid sensing of the presence of NAs using cellulosic peptide array strips as sensors. The peptide array was designed from sequences derived from the estrogen receptor (ER). Several of these peptides were able to detect the presence of NAs at low micromolar (∼5 mg L−1) levels in different oil sands process affected water samples. The specific binding of one of the peptides, peptide 17 (EGXVEIFDXLLATS) with NAs was evaluated using isothermal titration calorimetry. The results show that peptide 17 interacts strongly with NAs with an apparent binding constant (Ka) of 96 × 106 M−1, and may bind NAs in a similar fashion as ER interacts with estrogen. Finally, the data support that the peptides displaying high affinity for NAs can be used for developing disposable peptide-based sensor arrays for NA detection in oil sands process affected water samples.
Introduction
The worldwide demand for oil is increasing while conventional supplies are diminishing, necessitating the exploration of other unconventional sources of oil. Alberta's oil sands developments represent a significant source of unconventional oil.1,2 The extraction process utilizes large volumes of water to separate the bitumen from the oil sands.3 There are two variations of the extraction process used in Alberta, namely, mining based extraction, which uses low temperature extraction by “washing” the oil bearing sand with large volumes of water, and thermal or in situ extraction, where steam at around 240 °C is injected underground to reduce the viscosity of the oil, following which the oil and water are recovered and separated at the surface. As a result of these processes, large volumes of oil sands process affected water (OSPW) are created.In surface mining operations, the process-affected water is stored in large tailing ponds and is recycled back into the extraction process.4–6 In thermal extraction, the water is often treated and recycled to make steam, but a small fraction of the wastewater is discarded at the end of the process as “blowdown water”, which is often injected into deep wells. The OSPW from surface mining operations (henceforth referred to as sm-OSPW) has resulted in over a billion cubic meters of tailings water held in tailings ponds. There have been considerable concerns about the amount of organic matter present in these water bodies, and the ensuing problems that could be engendered by the release of these compounds into the surface water systems.5,6 Although thermal or in situ extraction produces significantly lower volumes of wastewater compared to mining, the high temperature contact of the petroleum and water solubilizes large amounts of organic matter in the water that comes in contact with the bitumen. We will refer to this wastewater as in situ oil sands process-affected water (is-OSPW). This wastewater usually contains large amounts of organic matter such as naphthenic acids.
Naphthenic acids (NAs) are a group of alkyl-substituted saturated acyclic and cyclic carboxylic acids that can account for up to 4% (by weight) of raw petroleum (Scheme 1).7,8 These organic acids occur naturally in bitumen and become solubilized in wastewater streams generated during petroleum extraction. NA concentrations in some petroleum recovery wastewaters are as high as 110–120 mg L−1.9 NAs are known to cause toxicity to a variety of organisms, adversely affecting reproduction and steroid production.10,11 For example, these compounds are known to bind to the estrogen receptor or ER (weak agonists) and antagonize the androgen receptor.11–13 Due to the adverse aquatic environmental effects of NAs, interest in developing sensitive and robust analytical methods for detection of NAs has grown in recent years.
 | ||
| Scheme 1 Proposed structures of a few representative naphthenic acids (R = alkyl group).11 | ||
Different analytical techniques have been used to characterize NAs, including mass spectrometric (MS) analysis using electrospray (ES-MS),14 gas chromatography (GC-MS),15,16 Fourier transform ion cyclotron resonance (FTICR-MS),17 and thermal conversion/elemental analysis-isotope ratio (TC/EA-IRMS).7 These analytical techniques require heavy instrumentation, skilled operators, are time consuming, and expensive. Here we propose a simple assay using peptide arrays as molecular recognition elements for the detection of NAs in contaminated water samples. We hypothesize that peptide sequences derived from ER18 may exhibit strong interaction with NAs.
Accordingly in this study, we describe the development of a peptide-array based sensing method with peptides (14-mer) derived from the ER sequence (Fig. 1) for the selective detection of NAs. Peptides showing high binding to NAs are used to develop a competitive binding assay to detect and estimate the amount of NAs in commercial NAs solution as well as two types of oil sands process affected water samples (sm-OSPW and is-OSPW). Isothermal titration calorimetry (ITC) is used to validate the specific interaction between a high binding peptide 17 and NAs. The results demonstrate that NAs binds to selected peptides immobilized on cellulose membrane which can be potentially used as sensing strips for detecting NAs in oil sands process affected water samples.
Materials and methods
Chemicals and materials
Fmoc L-amino acids were purchased from NovaBiochem (San Diego, CA) while O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluoro phosphate (HCTU) and 1-hydroxybenzotriazole (HOBt) were obtained from Chem-Impex International Inc., USA. Naphthenic acids (NAs, Technical grade), sucrose (99%), invertase (baker's yeast), N-(1-naphthyl)ethylenediamine dihydrochloride (EDAN, >98%), N,N′-diisopropylcarbodiimide (DIC, >98%), N,N-dimethylformamide (DMF, 99%), N-methyl morpholine (NMM, 99%), trifluoroacetic acid (TFA, 99%) and isopropyl alcohol (IPA, 99.9%) were purchased from Sigma-Aldrich and used as received.The field samples of oil sands process affected water (OSPW) were obtained from major producers of bitumen from the Athabasca oil sands region in Alberta, Canada. These samples include OSPW collected from surface mining operations (sm-OSPW) and the boiler blowdown water from an in situ surface treatment plant (is-OSPW) earmarked for deep well disposal. The is-OSPW sample was collected hot and nitrogen sealed, following which it was cooled and transferred to our laboratory.
After each binding experiment, the bound EDAN–NAs were removed from the membrane by washing with IPA for 3 h by shaking on an automatic shaker. For the 2nd library, the membrane was incubated with different concentrations of EDAN–NAs adduct (25–200 μM) in 35% IPA/water for 30 min, and the remaining experiment was performed as described above. Different incubation times (30 min–4 h) were attempted, and the results suggest that 30 min incubation time was optimal. This was selected because incubation beyond this point did not affect the relative fluorescence intensity. On the other hand, it was found that the increase in the percentage of IPA decreased the fluorescence signal possibly due to a slower kinetics of peptide and EDAN–NAs binding at higher IPA concentrations. However, 35% IPA was the minimal amount required to ensure solubility of the EDAN–NAs adduct. The data for five high binding peptides were fitted using one binding site equation to obtain apparent Kd values as shown in Fig. S5.†
Prior to each ITC run, the solutions were stirred and degased using ThermoVac accessory for 10 minutes to remove any air bubbles. After degasing, commercial naphthenic acid solutions (10–50 μM, where 5 mg mL−1 is approximately 20 μM with m/z ranging between 150–350) were kept inside the calorimetric cell and peptide solutions (100–200 μM) were injected to the cell via titration syringe in aliquots of 10 μL. The injection duration was 20 s with an initial delay of 60 seconds and a wait time of 160 s between the injections. The stirring speed of the ITC syringe paddle was set at 307 revolutions per minute (rpm). The heat of reaction associated with each injection was integrated using the software developed by MicroCal LLC. Control experiments were performed with blank IPA/water (35% v/v) solution to find the heat of dilutions of peptide and NAs, and were subtracted from the heats determined in the corresponding peptide–NAs binding experiments. At least two trials for each ITC experiments were performed and showed good reproducibility.
Results and discussion
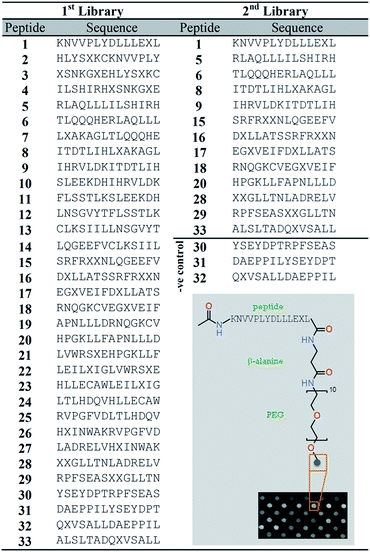
Synthesis and screening of peptide arrays
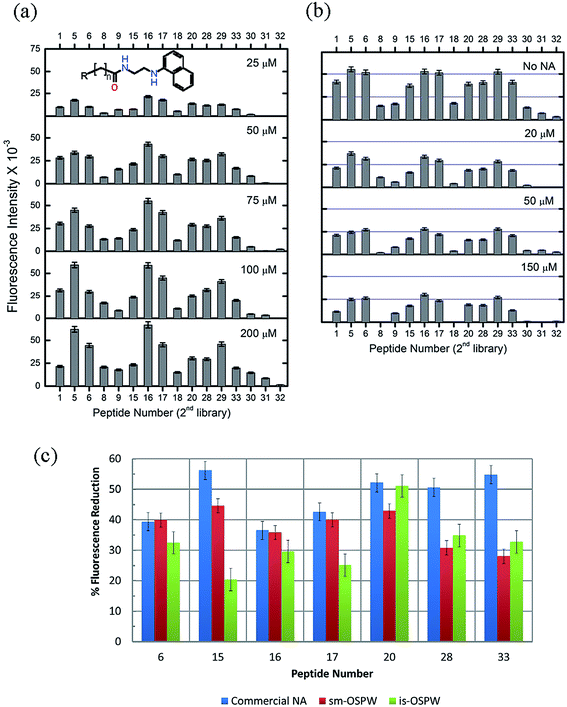
Peptide arrays offer a promising alternative approach that could be used to estimate the amount of NA mixtures in OSPW. A peptide array consisting of 33 overlapping sequences from ER, referred to as 1st library, was synthesized on a cellulose membrane with PEG and β-alanine as spacers (Fig. 1).21,22 The peptides were synthesized by conjugation of amino acids to the free amino group of β-alanine residues using SPOT synthesis.19 Commercial NAs labelled with fluorescent N-(1-naphthyl)ethylenediamine dihydrochloride (EDAN) were synthesized (Fig. S1†). The peptide array (1st library) was incubated with these labelled NAs (henceforth called EDAN–NAs) at room temperature for 30 minutes. After washing, the membrane was imaged using a fluorescence imager at λex/em 353/440 nm (Fig. S4†). Thirteen peptides from 1st library showed strong binding with the EDAN–NAs. These 13 peptides, along with peptides 30–32 (which showed negligible binding, and used here as negative controls) from the 1st library were chosen for further experiments (2nd library, Fig. 1).The binding of the peptides in the 2nd library to EDAN–NAs was evaluated at five different concentrations (25–200 μM). Fig. 2a shows that all peptides except for the negative controls bind the EDAN–NAs with different affinities even at low concentration of EDAN–NAs (25 μM). The binding isotherms of the EDAN–NAs to the peptides were fitted to the experimental data, indicating that peptides bind to EDAN–NAs with an apparent binding constant (Kd) in the low micromolar range (Fig. S5†). For each peptide, the fluorescence signal reaches a plateau as the amount of EDAN–NAs is increased, suggesting saturation in peptide binding above ca. 100 μM.
Competitive binding of peptides and EDAN–NAs
A competitive binding assay was performed on the 2nd library by incubating the peptide arrays for 30 minutes with 50 μM EDAN–NAs in the presence of commercial NAs at various concentrations (0–150 μM) in 35% IPA/water solution, followed by fluorescence measurement of the library. The presence of NAs as a competitor at concentrations of 20 μM (5 mg L−1) or higher decreased the binding of EDAN–NAs to the library, lowering the resulting fluorescence intensity (Fig. 2b). This suggests that the NAs participate in the binding to the peptides, and the hydrophobic portion of NAs is involved in the interaction between NAs and the immobilized peptides.Next, we used this competitive binding assay to determine the presence of NAs in two different petroleum wastewater samples obtained from oil sands (bitumen) extraction processes practiced in Alberta, Canada. These wastewaters are generally referred to as oil sands process affected water (OSPW).8,16,23 The first sample was obtained from the tailings pond of a surface mining based operation8 (sm-OSPW), whereas the second sample was obtained from an in situ thermal recovery operation16,23 (is-OSPW).
The dissolved organic carbon concentration in these water samples were adjusted by dilution to measure between 20–120 mg L−1 (total organic carbon). The pH of both water samples were also adjusted to ca. 8. The NAs are weak acids with pKa around 5. Considering ionizable organic acids are 100% deprotonated when the pH is 2 units above the pKa, we expect that there won't be any significant difference in the nature of NAs and their binding to the peptide before and after these pH adjustments. The 2nd library was then incubated with 50 μM EDAN–NAs and the water samples, followed by washing and measurement of fluorescence. The percent fluorescence reduction for seven representative peptides from the 2nd library using a commercial NAs standard, the sm-OSPW, and the is-OSPW samples are compared in Fig. 2c.
The competitive binding experiment using these water samples showed decrease in fluorescence response due to EDAN–NAs binding (Fig. 2c and S4†). The data for both sm-OSPW and is-OSPW samples show good correlation with the presence of NAs, indicating that the dissolved organic molecules present in these water samples have similar interactions with the peptide array as observed with commercial NAs. Since the fluorescence reduction for the sm-OSPW and is-OSPW samples is less than that for the commercial NAs, we can estimate that the amount of NAs in these field samples is less than 20 μM (Fig. 2c). Concentrated solutions of sm-OSPW samples (10×) were also evaluated using the competitive binding experiment and these showed further reduction in fluorescence intensity (Fig. S6†). The results presented here suggest that the developed array acts as a presence/absence detector of NAs in water samples down to 5 mg L−1 (∼20 μM, Fig. 2b) sensitivity.
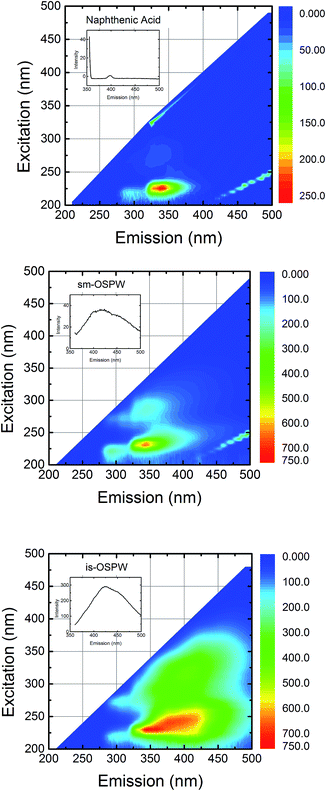
The fluorescence of the dissolved organics in the water samples may interfere with the competitive binding assay results. To verify this, the fluorescence excitation emission matrix (EEM) spectra of the water samples were obtained (Fig. 3). While the emission intensities of commercial NAs and sm-OSPW at 440 nm are low, the intensity of is-OSPW is significantly high. This interference could lead to a less significant fluorescence intensity reduction for is-OSPW (Fig. 2c), even though more NAs could be present in this sample. From these results, we conclude that seven peptides (6, 15, 16, 17, 20, 28, and 33) show measurable response with ≥30% reduction in fluorescence intensity due to the dissolved organic matter in the samples (Fig. 2c). In general, these results suggest that the water samples most likely contain a similar class of compounds that are recognized by these peptides.
Interaction of soluble peptides with naphthenic acids
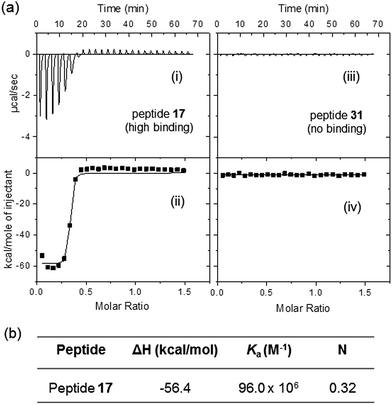
To further validate the specific binding of peptides derived from ER and NAs, the interaction between selected soluble peptides and NAs was characterized using ITC.24,25 A high binding peptide, peptide 17, was chosen to evaluate specific interaction with NAs whereas peptide 31 (low-binding) was selected as a negative control peptide (Fig. S7†). To perform ITC titrations, fixed volumes (10 μL) of peptide solution (17 or 31, 200 μM) were added to NA solution (25 μM) kept inside the calorimetric cell. The top panels represent the raw ITC thermograms showing the heat of reaction associated with each injection (Fig. 4a) and the bottom panels show the corresponding titration curves showing integrated heat values as a function of [peptide] to [NAs] molar ratio in the calorimetric cell. The integrated heat values were fitted to the ‘one set of sites’ binding model using Origin software to obtain the titration curve for peptide 17 binding to NAs.As expected, peptide 17 shows strong interaction and associated heat change whereas peptide 31 displayed negligible changes when titrated against NAs under similar conditions. The thermodynamic parameters for the peptide–NAs interactions (Fig. 4b) obtained from one-site binding model yielded an apparent binding constant (Ka) of 96 × 106 M−1, and the N for the interaction was found to be 0.3 suggesting that three naphthenic acid molecules interact with each peptide molecule. Notably, soluble (unbound) peptides were used for ITC experiments whereas those used for binding studies shown in Fig. S5† were peptides bound (covalently) to the cellulose membrane. It is possible that soluble, unbound peptides may have better affinity to NAs. Furthermore, the apparent binding constant values derived from ITC experiments correspond to the direct interaction between peptide and NAs in the absence of any fluorescent label. Since we have used a heterogeneous mixture of NAs, the thermodynamic parameters derived from ITC experiments are only approximate. These results demonstrate that ITC can be used as a very valuable and sensitive tool to study the thermodynamics of peptide–naphthenic acid interactions. For more accurate thermodynamic results, individual NAs can be separated in high purity and tested for their interaction with these ER peptides. Finally, the ITC response of peptide 17 is consistent with fluorescence results of the immobilized peptide and support strong interaction with NAs.
Mechanism of peptide–NAs binding
The three dimensional structure of estrogen bound ER (PDB 1A52)18 was used to excise the structure of estrogen and surrounding sequences as shown in Fig. 5. It shows that estrogen binds with three helical regions (H1–H3) and one β-sheet segment of the receptor. Comparing the high binding sequences obtained from the binding assay with the sequences in ER that flank estrogen bound to the receptor, we observe that peptides 17 (helical), 20 (β-sheet), 22 (helical), 28 (helical) belong to the estrogen binding sequences. It is, however, possible that these sequences behave differently when excised from a complete protein.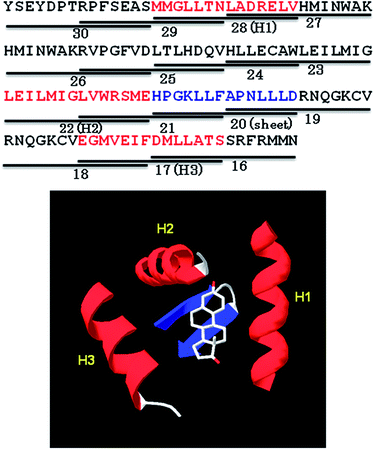 | ||
| Fig. 5 Top: Sequence of a portion of ER highlighting peptides 16–30 (14-mers) used in the peptide array herein. Peptides 17 (H3), 20 (sheet), 22 (H2), and 28 (H1) from the peptide array contain residues that interact with estrogen based on the 3D structure of estrogen bound receptor. Of these four peptides, the peptides 17, 20 and 28 showed high binding to EDAN–NAs. Bottom: The 3D structure of ER (PDB 1A52)18 showing estrogen interacting with three helical regions and one sheet. | ||
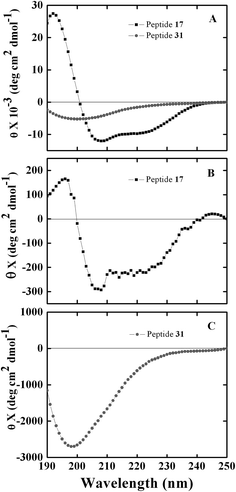
We evaluated the solution conformation of soluble peptides 17 (high-binding) and 31 (low-binding) using circular dichroism (CD) spectroscopy. The CD spectra for the peptides were obtained in 2,2,2-trifluororethanol (40% TFE), a secondary structure promoting solvent26 as well as in isopropanol (35% IPA) that was used for all peptide–NAs interaction experiments. In TFE, peptide 17 showed significant helical structure as indicated by the appearance of distinct negative band at 207 nm (θ = −10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 936 deg cm2 dmol−1), a negative shoulder near 222 nm, and a positive band at 195 nm, whereas, peptide 31 displayed negative minimum at ∼195 nm characteristic of random coil structure (Fig. 6). Similarly in 35% IPA, the CD spectra of peptide 17 showed negative bands at ∼207 nm (θ = −289 deg cm2 dmol−1) and 222 nm, and a positive band at ∼195 nm suggesting the peptide remains helical in IPA. However, the helicity was much less compared to that in secondary structure stabilizing medium TFE. CD spectra of peptide 31 showed no helical or sheet conformation in either environment. The helical conformation of peptide 17 and its high affinity toward NAs supports our conjecture that peptide 17 binds to NAs in a similar fashion as ER interacts with estrogen. In addition, our results show that three of the ER derived peptides 17, 20 and 28 display high binding to NAs. Consecutive peptides 15, 16, and 17 were also able to bind as well as compete for EDAN–NAs suggesting that the molecular recognition segment must be common in these overlapping sequences.
936 deg cm2 dmol−1), a negative shoulder near 222 nm, and a positive band at 195 nm, whereas, peptide 31 displayed negative minimum at ∼195 nm characteristic of random coil structure (Fig. 6). Similarly in 35% IPA, the CD spectra of peptide 17 showed negative bands at ∼207 nm (θ = −289 deg cm2 dmol−1) and 222 nm, and a positive band at ∼195 nm suggesting the peptide remains helical in IPA. However, the helicity was much less compared to that in secondary structure stabilizing medium TFE. CD spectra of peptide 31 showed no helical or sheet conformation in either environment. The helical conformation of peptide 17 and its high affinity toward NAs supports our conjecture that peptide 17 binds to NAs in a similar fashion as ER interacts with estrogen. In addition, our results show that three of the ER derived peptides 17, 20 and 28 display high binding to NAs. Consecutive peptides 15, 16, and 17 were also able to bind as well as compete for EDAN–NAs suggesting that the molecular recognition segment must be common in these overlapping sequences.
To summarize, we demonstrate that peptide arrays synthesized using fragments of ER on cellulose membranes using SPOT® synthesis can be used to detect the presence of NAs in water samples. Several peptides, including a few from the estrogen binding region of the estrogen receptor, displayed enhanced binding to NAs. These peptides act as a sensing platform for binding fluorescently labelled adducts of NAs (EDAN–NAs) as well as competitively binding NAs from water samples in the presence of EDAN–NAs. They can detect NAs at micromolar concentrations (∼5 mg L−1) in petroleum process affected water samples. Such peptide arrays on cellulose membrane can potentially be used directly for water quality analysis in fields or natural water bodies. More detailed experiments are required to confirm the selectivity of this method in the presence of other contaminants in water that may exhibit similar affinities as NAs to these ER peptides.
Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and NSERC-Industrial Research Chair program of SB. The infrastructure support from the Canada Foundation for Innovation (CFI) is also acknowledged. RGP thank financial support from NSERC-Industrial Research Chair program.Notes and references
- E. A. R. E. Ministry of Energy, Alberta, C. A. http://www.energy.alberta.ca/Org/Publications/AR2010.pdf and a. 23.04.2011.
- S. S. Leung, M. D. MacKinnon and R. E. Smith, Aquat. Toxicol., 2003, 62, 11–26 CrossRef CAS.
- Z. J. Z. Jacob Masliyah, Z. Xu, J. Czarnecki and H. Hamza, Can. J. Chem. Eng., 2004, 82, 628–654 CrossRef.
- L. F. Del Rio, A. K. Hadwin, L. J. Pinto, M. D. MacKinnon and M. M. Moore, J. Appl. Microbiol., 2006, 101, 1049–1061 CrossRef CAS PubMed.
- J. P. Giesy, J. C. Anderson and S. B. Wiseman, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 951–952 CrossRef CAS PubMed.
- E. W. Allen, J. Environ. Eng. Sci., 2008, 7, 123–138 CrossRef CAS.
- J. M. Ahad, H. Pakdel, M. M. Savard, M. C. Simard and A. Smirnoff, Anal. Chem., 2012, 84, 10419–10425 CrossRef CAS PubMed.
- D. M. Grewer, R. F. Young, R. M. Whittal and P. M. Fedorak, Sci. Total Environ., 2010, 408, 5997–6010 CrossRef CAS PubMed.
- J. V. Headley and D. W. McMartin, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2004, 39, 1989–2010 CrossRef PubMed.
- R. J. Kavanagh, R. A. Frank, K. Burnison, R. F. Young, P. M. Fedorak, K. R. Solomon and G. Van Der Kraak, Aquat. Toxicol., 2012, 116–117, 34–42 CrossRef CAS PubMed.
- K. V. Thomas, K. Langford, K. Petersen, A. J. Smith and K. E. Tollefsen, Environ. Sci. Technol., 2009, 43, 8066–8071 CrossRef CAS PubMed.
- Y. He, S. B. Wiseman, M. Hecker, X. Zhang, N. Wang, L. A. Perez, P. D. Jones, M. G. El-Din, J. W. Martin and J. P. Giesy, Environ. Sci. Technol., 2011, 45, 6268–6274 CrossRef CAS PubMed.
- S. J. Rowland, C. E. West, D. Jones, A. G. Scarlett, R. A. Frank and L. M. Hewitt, Environ. Sci. Technol., 2011, 45, 9806–9815 CrossRef CAS PubMed.
- J. V. Headley, K. M. Peru, M. P. Barrow and P. J. Derrick, Anal. Chem., 2007, 79, 6222–6229 CrossRef CAS PubMed.
- D. Jones, C. E. West, A. G. Scarlett, R. A. Frank and S. J. Rowland, J. Chromatogr. A, 2012, 1247, 171–175 CrossRef CAS PubMed.
- M. A. Petersen and H. Grade, Ind. Eng. Chem. Res., 2011, 50, 12217–12224 CrossRef CAS.
- M. P. Barrow, J. V. Headley, K. M. Peru and P. J. Derrick, J. Chromatogr. A., 2004, 1058, 51–59 CrossRef CAS PubMed.
- D. M. Tanenbaum, Y. Wang, S. P. Williams and P. B. Sigler, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 5998–6003 CrossRef CAS.
- R. Frank, J. Immunol. Methods, 2002, 267, 13–26 CrossRef CAS.
- I. Wadso and R. N. Goldberg, Pure Appl. Chem., 2001, 73, 1625–1639 CrossRef CAS.
- S. Ahmed, A. S. Mathews, N. Byeon, A. Lavasanifar and K. Kaur, Anal. Chem., 2010, 82, 7533–7541 CrossRef CAS PubMed.
- R. Soudy, S. Ahmed and K. Kaur, ACS Comb. Sci., 2012, 14, 590–599 CrossRef CAS PubMed.
- A. Maiti, M. Sadrezadeh, S. G. Thakurta, D. J. Pernitsky and S. Bhattacharjee, Energy Fuels, 2012, 26, 5604–5612 CrossRef CAS.
- W. F. Tan, L. K. Koopal and W. Norde, Environ. Sci. Technol., 2008, 43, 591–596 CrossRef.
- T. Abraham, R. N. A. H. Lewis, R. S. Hodges and R. N. McElhaney, Biochemistry, 2005, 44, 11279–11285 CrossRef CAS PubMed.
- D. Roccatano, G. Colombo, M. Fioroni and A. E. Mark, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12179–12184 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the synthesis of immobilized peptides (peptide array), EDAN–NAs, soluble peptides and their characterization using fluorescence imaging, MALDI-TOF MS, HPLC and UV spectroscopy. See DOI: 10.1039/c4ra10981c |
| This journal is © The Royal Society of Chemistry 2014 |