Synthesis and quantitative structure–activity relationship (QSAR) studies of novel rosin-based diamide insecticides†
Jian Li*a,
Yanqing Gao*b,
Shibin Shangc,
Xiaoping Raoc,
Jie Songd and
Zongde Wange
aCollege of Forestry, Northwest Agriculture and Forestry University, Yangling, Shaanxi 712100, People’s Republic of China. E-mail: ericlee99@nwsuaf.edu.cn; gaoyanqinggc@163.com; Fax: +86-029-87082392; Tel: +86-029-87082392
bResearch & Development Center of Biorational Pesticide, College of Plant Protection, Northwest Agriculture and Forestry University, Yangling, Shaanxi 712100, People’s Republic of China
cInstitute of Chemical Industry of Forest Products, Chinese Academy of Forestry, Nanjing, Jiangsu 210042, People’s Republic of China
dDepartment of Chemistry and Biochemistry, University of Michigan-Flint, Flint, Michigan 48502, USA
eCollege of Forestry, Jiangxi Agriculture University, Nanchang, Jiangxi 330045, People’s Republic of China
First published on 21st October 2014
Abstract
In a continuing effort to develop novel natural product-based insecticidal agents endowed with low mammalian toxicity, two series of rosin-based diamides have been synthesized, and their insecticidal activities against Plutella xylostella and Mythimna separata were evaluated. Most of the synthesized compounds exhibited moderate to significant insecticidal activity. Among them, thiadiazole-containing diamides 6a–n displayed better activity than others, especially compounds 6f and 6n which exhibited excellent insecticidal activity against P. xylostella, with LC50 values of 0.223 and 0.214 mg L−1, respectively, which approximate to, or are lower than that of, the control flubendiamide (0.222 mg L−1). The preliminary structure–activity relationship analysis indicated that rosin-based diamides with electron-withdrawing groups on the benzene ring showed better insecticidal activity than those with electron-donating groups. Via a best multilinear regression analysis, the generated quantitative structure–activity relationship (QSAR) model (R2 = 0.9566) revealed a strong correlation of insecticidal activity against P. xylostella with molecular structures of these compounds. These consequences can be expected to instruct the design and development of new rosin-based insecticides.
1. Introduction
Synthetic insecticides that protect crops from plant insects and diseases are still playing an important role in current agricultural practice.1 However, the continual application and excessive use of all these conventional insecticides over the years has led to environmental problems and the undesired development of resistance to pests.2,3 Therefore, the search of potential insecticides with new targets and low mammalian toxicity remains desirable in the field of crop protection.4,5Diamides constitute a new class of insecticides for the control of the lepidopteron pest.6,7 Studies have shown that diamides are activators of the ryanodine receptors (RyRs) which regulate Ca2+ release from intracellular stores located in the sarcoplasmic reticulum.8 Owing to their exceptional activity, unique mode of action, and low mammalian toxicity, diamides have received considerable attention.9 So far, three diamide insecticides, i.e., flubendiamide, chlorantraniliprole (Rynaxypyr) and cyantraniliprole (Cyazypyr) have been discovered and commercialized (Fig. 1).10–12 However, the chemical synthesis of these diamide insecticides is time-consuming and expensive because of their complicated molecular structure and chiral centers. As a feasible solution, a class of secondary metabolites from natural sources can be chemically modified with similar structures and chiral centers.
Plant secondary metabolites are obtained in the process of coevolution between plants and the environment.13 Currently, a range of secondary metabolites from natural sources such as flavonoids, alkaloids and terpenoids have been developed as the lead compounds for the preparation of potent insecticides.14 With the advantage of a good environmental profile and the rare development of resistance, these botanical insecticides have been considered as attractive alternatives to synthetic agrochemicals for pest management.
Rosin, a major component of secretion from pine trees, is considered as one of the most abundant and popular natural terpenoid resources in China.15 The broad-spectrum biological activities of rosin and its derivatives, such as antitumor, antibacterial, antivirus, and hormone regulation have been well documented.16 Even so, investigations reporting on the systematic study of the quantitative structure–activity relationship (QSAR) of rosin-based insecticidal agents are few. Due to the rising cost, synthesizing all possible compounds and screening a few candidates from thousands of compounds has become economically and practically impossible. One approach to improve this time-consuming and expensive process is to apply QSAR. QSAR methodology is an essential tool, which enables the calculation of numerous quantitative descriptors solely on the basis of molecular structural information. Based on the constructed QSAR models, some vital features responsible for insecticidal activity can be identified, and the potential target sites can even be speculated. Meanwhile, the use of the computational approaches and methodologies of the QSAR study may provide further guidance for the design of novel potent insecticides.17–22
In order to obtain novel natural product-based insecticides, a series of rosin-based diamides were synthesized on the basis of molecular similarity. In addition, the thiadiazole heterocyclic group, an important pharmacophore, was introduced into the backbone structure of the diamides for designing insecticides with higher activity. The insecticidal activities of the title compounds against diamondback moth (P. xylostella) and oriental armyworm (M. separata) were evaluated. Moreover, the QSAR was also performed on all of the title compounds using the Gaussian and CODESSA software packages, which can account for their structural features responsible for the insecticidal activity. This exploration is expected to improve the application of rosin as an insecticide material.
2. Experimental sections
2.1 Synthetic procedures and characterisations
Gum rosin (grade one) was obtained from a commercial source (Wu Zhou Pine Chemicals Ltd., Guangxi, China) and used without further purification. All other chemicals used were of reagent grade. The IR spectra were taken on a Nicolet IS10 FT-IR (Nicolet, Madison, USA) spectrophotometer. The 1H NMR spectra were recorded on a Bruker AV-300 (Bruker, Karlsruhe, Germany) nuclear magnetic resonance spectrometer with either CDCl3 or DMSO-d6 as the solvent and TMS as an internal standard. The MS spectra were taken on an Agilent-5973 (Agilent, Santa Clara, USA) spectrophotometer. The melting points were determined using XT-5 (Saiao, Beijing, China) melting point apparatus. The elemental analysis (C, H, and N) was carried out on a Vario EL-III (Elementar, Hanau, Germany) elemental analyzer and the results were in good agreement with the calculated values. All reactions were traced by thin layer chromatography (TLC).![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (CDCl3 δ/ppm 300 MHz), 5.50 (s, H, C
O). 1H NMR (CDCl3 δ/ppm 300 MHz), 5.50 (s, H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–); 2.52–1.41 (m, 5H, –CH–); 1.86–1.24 (m, 14H, –CH2–); 1.27–1.10 (m, 12H, CH3). ESI-MS m/z = 412 [M + H] +. Anal. calcd for C23H32Cl2O2: C, 67.15; H, 7.84. Found: C, 67.21; H, 7.89.
CH–); 2.52–1.41 (m, 5H, –CH–); 1.86–1.24 (m, 14H, –CH2–); 1.27–1.10 (m, 12H, CH3). ESI-MS m/z = 412 [M + H] +. Anal. calcd for C23H32Cl2O2: C, 67.15; H, 7.84. Found: C, 67.21; H, 7.89.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (CDCl3 δ/ppm 300 MHz), 5.78 (m, 4H, CONH2); 5.49 (s, H, C
O). 1H NMR (CDCl3 δ/ppm 300 MHz), 5.78 (m, 4H, CONH2); 5.49 (s, H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–); 2.61 (m, H, –CH–(Me)2); 2.42 (s, H, –CH–C
CH–); 2.61 (m, H, –CH–(Me)2); 2.42 (s, H, –CH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–); 2.12–1.43 (m, 3H, –CH–); 1.82–1.26 (m, 14H, –CH2–); 1.34–1.06 (m, 12H, CH3). ESI-MS m/z = 373 [M + H]+. Anal. calcd for C23H36N2O2: C, 74.15; H, 9.74; N, 7.52. Found: C, 74.11; H, 9.69; N, 7.56.
O–); 2.12–1.43 (m, 3H, –CH–); 1.82–1.26 (m, 14H, –CH2–); 1.34–1.06 (m, 12H, CH3). ESI-MS m/z = 373 [M + H]+. Anal. calcd for C23H36N2O2: C, 74.15; H, 9.74; N, 7.52. Found: C, 74.11; H, 9.69; N, 7.56.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10] gave the fourteen resulting derivatives, 4a to 4n. The example data of compounds 4a and 4b are shown as follows, whereas data of compounds 4c–n can be found in the ESI.†
10] gave the fourteen resulting derivatives, 4a to 4n. The example data of compounds 4a and 4b are shown as follows, whereas data of compounds 4c–n can be found in the ESI.†
Data for compound 4a. Yield: 7.77 g (70.4%); white powder; m.p. 140.8–141.6 °C. IR (cm−1): 3363, 3259 (N–H); 2926, 2854 (–CH3, –CH2); 1659 (N–C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 738, 697 (Ar–H). 1H NMR (DMSO-d6 δ/ppm 300 MHz): 9.61, 9.06 (m, 2H, CONH–); 7.61–6.96 (m, 10H, Ar–H); 5.29 (s, H, C
O); 738, 697 (Ar–H). 1H NMR (DMSO-d6 δ/ppm 300 MHz): 9.61, 9.06 (m, 2H, CONH–); 7.61–6.96 (m, 10H, Ar–H); 5.29 (s, H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–); 4.71–4.30 (m, 4H, Ar–CH2–); 2.57 (s, H, –CH–C
CH–); 4.71–4.30 (m, 4H, Ar–CH2–); 2.57 (s, H, –CH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–); 2.01–1.83 (m, 3H, –CH–); 1.82–1.24 (m, 14H, –CH2–); 1.53 (m, H, –CH–(Me)2); 1.14–0.61 (m, 12H, CH3). ESI-MS m/z = 553 [M + H]+. Anal. calcd for C37H48N2O2: C, 80.39; H, 8.75; N, 5.07. Found: C, 80.27; H, 9.00; N, 4.86.
O–); 2.01–1.83 (m, 3H, –CH–); 1.82–1.24 (m, 14H, –CH2–); 1.53 (m, H, –CH–(Me)2); 1.14–0.61 (m, 12H, CH3). ESI-MS m/z = 553 [M + H]+. Anal. calcd for C37H48N2O2: C, 80.39; H, 8.75; N, 5.07. Found: C, 80.27; H, 9.00; N, 4.86.
Data for compound 4b. Yield: 7.55 g (72%); white powder; m.p. 135.6–137.4 °C. IR (cm−1): 3354 (N–H); 2961, 2861 (–CH3, –CH2); 1663 (N–C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 754, 691 (Ar–H). 1H NMR (DMSO-d6 δ/ppm 300 MHz): 8.08, 7.95 (m, 2H, CONH–); 7.29–7.20 (m, 10H, Ar–H); 5.25 (s, H, C
O); 754, 691 (Ar–H). 1H NMR (DMSO-d6 δ/ppm 300 MHz): 8.08, 7.95 (m, 2H, CONH–); 7.29–7.20 (m, 10H, Ar–H); 5.25 (s, H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–); 2.68 (s, H, –CH–C
CH–); 2.68 (s, H, –CH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–); 2.50–1.28 (m, 14H, –CH2–); 2.11–1.90 (m, 3H, –CH–); 1.57 (m, H, –CH–(Me)2); 1.17–0.55 (m, 12H, CH3). ESI-MS m/z = 525 [M + H]+. Anal. calcd for C35H44N2O2: C, 80.11; H, 8.45; N, 5.34. Found: C, 79.90; H, 8.54; N, 5.08.
O–); 2.50–1.28 (m, 14H, –CH2–); 2.11–1.90 (m, 3H, –CH–); 1.57 (m, H, –CH–(Me)2); 1.17–0.55 (m, 12H, CH3). ESI-MS m/z = 525 [M + H]+. Anal. calcd for C35H44N2O2: C, 80.11; H, 8.45; N, 5.34. Found: C, 79.90; H, 8.54; N, 5.08.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1099 (C–S–C). 1H NMR (DMSO-d6 δ/ppm 300 MHz), 5.44 (s, H, C
N); 1099 (C–S–C). 1H NMR (DMSO-d6 δ/ppm 300 MHz), 5.44 (s, H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–); 4.00 (m, 4H, –NH2); 2.74–1.78 (m, 5H, –CH–); 1.80–1.25 (m, 14H, –CH2–); 1.33–1.05 (m, 12H, CH3). ESI-MS m/z = 485 [M + H]+. Anal. calcd for C25H36N6S2: C, 61.95; H, 7.49; N, 17.34. Found: C, 61.93; H, 7.51; N, 17.05.
CH–); 4.00 (m, 4H, –NH2); 2.74–1.78 (m, 5H, –CH–); 1.80–1.25 (m, 14H, –CH2–); 1.33–1.05 (m, 12H, CH3). ESI-MS m/z = 485 [M + H]+. Anal. calcd for C25H36N6S2: C, 61.95; H, 7.49; N, 17.34. Found: C, 61.93; H, 7.51; N, 17.05.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5] gave the fourteen resulting derivatives, 6a to 6n. The example data of compounds 6a and 6b are shown as follows, whereas data of compounds 6c–n can be found in the ESI.†
5] gave the fourteen resulting derivatives, 6a to 6n. The example data of compounds 6a and 6b are shown as follows, whereas data of compounds 6c–n can be found in the ESI.†
Data for compound 6a. Yield: 3.98 g (70%); white powder; m.p. 235.9–236.7 °C. IR (cm−1): 3370, 3159 (N–H); 2926, 2854 (–CH3, –CH2); 1660 (N–C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1633 (C
O); 1633 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1080 (C–S–C). 1H NMR (DMSO-d6 δ/ppm 300 MHz), 8.01, 7.95 (s, 2H, –CONH–); 5.44 (s, H, C
N); 1080 (C–S–C). 1H NMR (DMSO-d6 δ/ppm 300 MHz), 8.01, 7.95 (s, 2H, –CONH–); 5.44 (s, H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–); 2.80–1.80 (m, 5H, –CH–); 2.02, 1.95 (m, 6H, –COCH3); 1.86–1.24 (m, 14H, –CH2–); 1.34–0.98 (m, 12H, –CH3). ESI-MS m/z = 569 [M + H]+. Anal. calcd for C29H40N6O2S2: C, 61.24; H, 7.09; N, 14.78. Found: C, 61.33; H, 7.01; N, 14.75.
CH–); 2.80–1.80 (m, 5H, –CH–); 2.02, 1.95 (m, 6H, –COCH3); 1.86–1.24 (m, 14H, –CH2–); 1.34–0.98 (m, 12H, –CH3). ESI-MS m/z = 569 [M + H]+. Anal. calcd for C29H40N6O2S2: C, 61.24; H, 7.09; N, 14.78. Found: C, 61.33; H, 7.01; N, 14.75.
Data for compound 6b. Yield: 4.50 g (65%); white powder; m.p. 222.3–223.2 °C. IR (cm−1): 3329, 3100 (N–H); 2961, 2880 (–CH3, –CH2); 1659 (N–C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1632 (C
O); 1632 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1079 (C–S–C); 751, 690 (Ar–H). 1H NMR (DMSO-d6 δ/ppm 300 MHz): 8.18, 8.02 (m, 2H, –CONH–); 7.59–7.44 (m, 10H, Ar–H); 5.25 (s, H, C
N); 1079 (C–S–C); 751, 690 (Ar–H). 1H NMR (DMSO-d6 δ/ppm 300 MHz): 8.18, 8.02 (m, 2H, –CONH–); 7.59–7.44 (m, 10H, Ar–H); 5.25 (s, H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–); 2.81–1.79 (m, 5H, –CH–); 1.95–1.28 (m, 14H, –CH2–); 1.37–0.55 (m, 12H, CH3). ESI-MS m/z = 693 [M + H]+. Anal. calcd for C39H44N6O2S2: C, 67.60; H, 6.40; N, 12.13. Found: C, 67.63; H, 6.41; N, 12.05.
CH–); 2.81–1.79 (m, 5H, –CH–); 1.95–1.28 (m, 14H, –CH2–); 1.37–0.55 (m, 12H, CH3). ESI-MS m/z = 693 [M + H]+. Anal. calcd for C39H44N6O2S2: C, 67.60; H, 6.40; N, 12.13. Found: C, 67.63; H, 6.41; N, 12.05.
2.2 Biological assay
The P. xylostella and M. separata used in the bioassay were provided by the research & development center of biorational pesticide, Northwest A&F University. The bioassays were performed in an artificial greenhouse with a constant temperature (25 ± 0.5 °C), relative humidity (75 ± 5%), and photoperiod (L/D = 14/10). All test compounds were dissolved in DMSO and diluted with sterile water to obtain required concentrations. Assessments were made on a dead/alive basis, and mortality rates were corrected using Abbott’s formula. Evaluations were based on a percentage scale of 0–100, where 0 = no activity and 100 = complete eradication. The standard deviations of the tested biological values were ±5%. LC50 values were calculated by probit analysis. For comparative purposes, the commercial product flubendiamide was tested under the same conditions.25,262.3 Building and validation of the QSAR model
Firstly, the optimal conformers with the lowest energy of the title compounds were performed at the DFT/6-31G (d) level using a Gaussian 03W package of programs.30 Secondly, the calculated results were changed into a form compatible with CODESSA 2.7.1533 using Ampac 9.1.3.34 Finally, all the molecular descriptors involved in these compounds were calculated by CODESSA 2.7.15. In order to find out which structural features play an important role in insecticidal activity against P. xylostella, the best multiple regression analysis was selected to generate the QSAR model equation. In this equation, the statistical criteria were indicated by the squared correction coefficient (R2), the squared standard error of the estimates (S2), and the Fisher significance ratio (F). Tested LC50 values were converted into the corresponding log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 and used as dependent variables in the QSAR studies. The quality of the final model was determined using both an internal validation and the “leave-one-out” cross-validation methods.
LC50 and used as dependent variables in the QSAR studies. The quality of the final model was determined using both an internal validation and the “leave-one-out” cross-validation methods.
3. Results and discussion
3.1 Synthesis
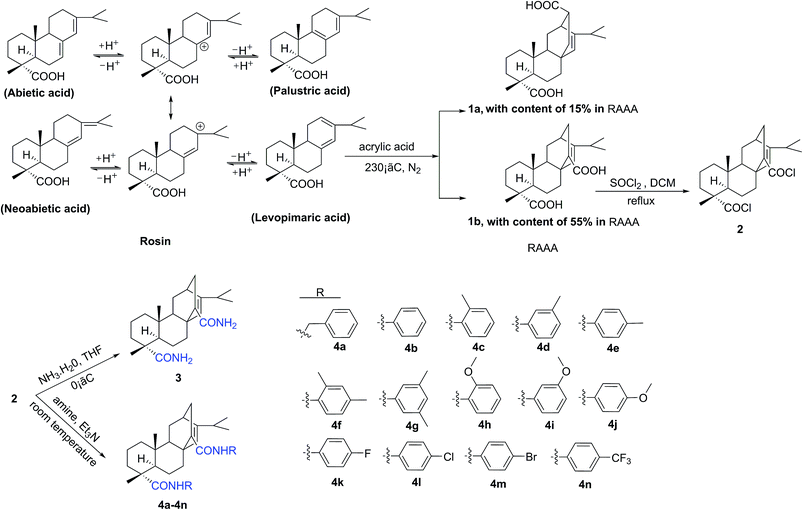
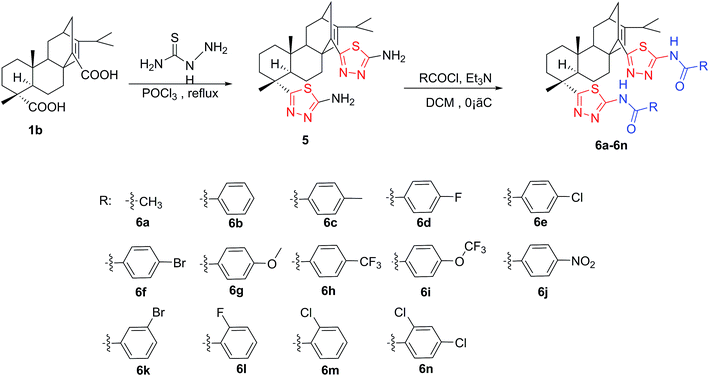
The syntheses of two series of rosin-based diamide derivatives developed in the present work are illustrated in Scheme 1 and 2 respectively. As a dicarboxylic acid, the RAAA was obtained by the Diels–Alder addition reaction between rosin and acrylic acid; the content of the RAAA in the products was 70%. Additional purifications were required to remove other reactants and separate the two isomers 1a and 1b from the RAAA. According to the literature, the target isomer 1b was separated by recrystallization with an overall yield of 95%.15 Acrylopimaryl chloride (2) was prepared from the reaction of 1b with thionyl chloride and used without purification. The target compounds 3 and 4a–n were prepared by a simple and convenient three-step procedure starting from rosin. At room temperature, compounds 4a–n were synthesized in high yields (60–90%) by the reaction of acrylopimaryl chloride (2) with various aromatic amines in DCM using Et3N as the acid acceptor. However, compound 3 was prepared by the reaction of NH3·H2O with acrylopimaryl chloride (2) at 0 °C in THF with a satisfactory yield (70%). As shown in Scheme 2, compound 1b and thiosemicarbazide refluxed in POCl3 to give the corresponding thiadiazole substituted amine 5 via a cyclization reaction. The reaction of 5 with various chlorides can give thiadiazole-containing amides, 6a to 6n. The structures of the title compounds were well-characterized by IR, 1H NMR, MS, and elemental analysis.3.2 Biological activity and structure–activity relationships
| No. | Compound | Larvicidal activity (%) at a concentration of (mg L−1) | LC50 | y = a + bx | R2 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 LC50 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 5 | 2.5 | 1 | 0.5 | 0.25 | 0.1 | 0.05 | ||||||
| 1 | 3 | 100 | 67 | 40 | 20 | 0 | — | — | — | 2.759 | y = −1.210 + 2.746x | 0.985 | 0.441 |
| 2 | 4a | 100 | 73 | 40 | 7 | 0 | — | — | — | 2.949 | y = −1.672 + 3.560x | 1.000 | 0.470 |
| 3 | 4b | 100 | 73 | 40 | 7 | 0 | — | — | — | 2.949 | y = −1.672 + 3.560x | 1.000 | 0.470 |
| 4 | 4c | 100 | 77 | 40 | 7 | 0 | — | — | — | 2.879 | y = −1.681 + 3.659x | 1.000 | 0.459 |
| 5 | 4d | 100 | 73 | 37 | 10 | 0 | — | — | — | 2.923 | y = −1.563 + 3.355 x | 0.992 | 0.466 |
| 6 | 4e | 100 | 77 | 37 | 10 | 0 | — | — | — | 2.853 | y = −1.567 + 3.443x | 0.986 | 0.455 |
| 7 | 4f | 100 | 80 | 57 | 7 | 0 | — | — | — | 2.506 | y = −1.463 + 3.667x | 0.974 | 0.399 |
| 8 | 4g | 100 | 73 | 40 | 7 | 0 | — | — | — | 2.949 | y = −1.672 + 3.560x | 1.000 | 0.470 |
| 9 | 4h | 70 | 40 | 17 | 3 | 0 | — | — | — | 6.193 | y = −1.948 + 2.460x | 0.998 | 0.792 |
| 10 | 4i | 70 | 37 | 17 | 3 | 0 | — | — | — | 6.379 | y = −1.968 + 2.446x | 0.995 | 0.805 |
| 11 | 4j | 67 | 40 | 20 | 3 | 0 | — | — | — | 6.339 | y = −1.837 + 2.290x | 0.998 | 0.802 |
| 12 | 4k | 100 | 83 | 50 | 23 | 3 | 0 | — | — | 2.131 | y = −0.924 + 2.813x | 0.983 | 0.329 |
| 13 | 4l | 100 | 80 | 50 | 20 | 3 | 0 | — | — | 2.246 | y = −0.988 + 2.812x | 0.993 | 0.351 |
| 14 | 4m | 100 | 77 | 50 | 23 | 3 | 0 | — | — | 2.244 | y = −0.931 + 2.652x | 0.980 | 0.351 |
| 15 | 4n | 100 | 77 | 50 | 27 | 3 | 0 | — | — | 2.185 | y = −0.875 + 2.578x | 0.964 | 0.339 |
| 16 | 6a | 100 | 87 | 50 | 30 | 3 | 0 | — | — | 2.076 | y = −0.922 + 2.906x | 0.982 | 0.317 |
| 17 | 6b | 100 | 87 | 57 | 30 | 3 | 0 | — | — | 1.982 | y = −0.874 + 2.942x | 0.992 | 0.297 |
| 18 | 6c | 100 | 90 | 57 | 27 | 3 | 0 | — | — | 1.879 | y = −0.816 + 2.977x | 0.983 | 0.274 |
| 19 | 6d | 100 | 90 | 70 | 57 | 23 | 7 | 0 | — | 1.096 | y = −0.084 + 2.115x | 0.972 | 0.040 |
| 20 | 6e | 100 | 90 | 70 | 53 | 23 | 7 | 0 | — | 1.123 | y = −0.107 + 2.125x | 0.981 | 0.050 |
| 21 | 6f | 100 | 100 | 90 | 80 | 77 | 60 | 33 | 7 | 0.223 | y = 1.004 + 1.543x | 0.953 | −0.652 |
| 22 | 6g | 100 | 70 | 40 | 10 | 0 | — | — | — | 2.927 | y = −1.516 + 3.250x | 1.000 | 0.466 |
| 23 | 6h | 100 | 90 | 70 | 50 | 20 | 7 | 0 | — | 1.182 | y = −0.155 + 2.136x | 0.974 | 0.073 |
| 24 | 6i | 100 | 67 | 37 | 10 | 0 | — | — | — | 3.068 | y = −1.559 + 3.201x | 0.999 | 0.487 |
| 25 | 6j | 100 | 73 | 40 | 7 | 0 | — | — | — | 2.949 | y = −1.672 + 3.560x | 1.000 | 0.470 |
| 26 | 6k | 100 | 90 | 70 | 53 | 23 | 7 | 0 | — | 1.123 | y = −0.107 + 2.125x | 0.981 | 0.050 |
| 27 | 6l | 100 | 90 | 67 | 50 | 20 | 7 | 0 | — | 1.211 | y = −0.178 + 2.147x | 0.983 | 0.083 |
| 28 | 6m | 100 | 90 | 70 | 57 | 23 | 7 | 0 | — | 1.096 | y = −0.084 + 2.115x | 0.972 | 0.040 |
| 29 | 6n | 100 | 100 | 90 | 80 | 77 | 60 | 33 | 13 | 0.214 | y = 1.001 + 1. 494x | 0.966 | −0.670 |
| Flubendiamide | 100 | 100 | 97 | 80 | 73 | 60 | 30 | 10 | 0.222 | y = 1.125 + 1.722x | 0.977 | −0.654 | |
| Compd | Larvicidal activity (%) at a concentration of (mg L−1) | ||
|---|---|---|---|
| 50 | 20 | 10 | |
| 3 | 40 | 0 | — |
| 4a | 30 | 0 | — |
| 4b | 40 | 0 | — |
| 4c | 40 | 0 | — |
| 4d | 37 | 0 | — |
| 4e | 37 | 0 | — |
| 4f | 27 | 0 | — |
| 4g | 40 | 0 | — |
| 4h | 17 | 0 | — |
| 4i | 17 | 0 | — |
| 4j | 20 | 0 | — |
| 4k | 40 | 0 | — |
| 4l | 30 | 0 | — |
| 4m | 40 | 0 | — |
| 4n | 40 | 0 | — |
| 6a | 37 | 0 | — |
| 6b | 37 | 0 | — |
| 6c | 20 | 0 | — |
| 6d | 40 | 0 | — |
| 6e | 17 | 0 | — |
| 6f | 17 | 0 | — |
| 6g | 20 | 0 | — |
| 6h | 40 | 0 | — |
| 6i | 30 | 0 | — |
| 6j | 40 | 0 | — |
| 6k | 40 | 0 | — |
| 6l | 37 | 0 | — |
| 6m | 37 | 0 | — |
| 6n | 30 | 0 | — |
| Flubendiamide | 100 | 100 | 100 |
3.3 QSAR
There are many regression approaches available for CODESSA 2.7.15 software, such as best multi-linear regression, multi-linear regression, principal component analysis, partial least-square regression, and heuristic regression.36 Taking into account the number of samples and descriptors used in this study, the best multi-linear regression was selected for developing the QSAR model. The “breaking point” rule for the determination of the number of the descriptors was employed as described in Fig. 4. The best multi-linear regression showed a significant increase in R2 when the number of the descriptors was no more than 5. However, there was negligible change in R2 when the number of descriptors increased from 5 to 6. Descriptors with high t values were accepted and those with low t values were rejected. A “breaking point” indicates that the improvement of the regression model has become insignificant (ΔR2 < 0.02–0.04).37 In addition, the number of the descriptors complies to the linear regressions given by eqn (1).| N ≥ 3(K + 1) | (1) |
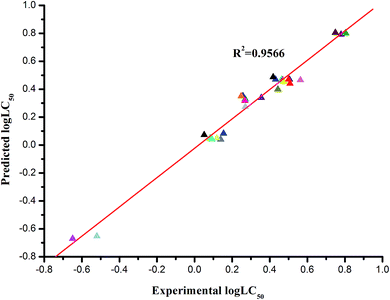
The statistically optimized QSAR equation for the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 data has the following statistical characteristics: R2 = 0.9566, F = 101.46, S2 = 0.0040 (Table 3). This model includes five descriptors in descending order according to their statistical significance (t values), where X and ΔX are the regression coefficients and their standard errors. The comparison between the experimental and predicted log
LC50 data has the following statistical characteristics: R2 = 0.9566, F = 101.46, S2 = 0.0040 (Table 3). This model includes five descriptors in descending order according to their statistical significance (t values), where X and ΔX are the regression coefficients and their standard errors. The comparison between the experimental and predicted log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 is listed in Table 4. Fig. 5 shows the plot of predicted versus experimental activity of the 29 compounds. The five-descriptor QSAR model equation and corresponding statistical criteria are described in the following eqn (2) (Fig. 6).
LC50 is listed in Table 4. Fig. 5 shows the plot of predicted versus experimental activity of the 29 compounds. The five-descriptor QSAR model equation and corresponding statistical criteria are described in the following eqn (2) (Fig. 6).
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 = 2.8005 + 3.4038 × HOMO − 9.2784 × DM + 2.7178 × qOmax − 2.3561 × qNmin + 2.4803 × μh LC50 = 2.8005 + 3.4038 × HOMO − 9.2784 × DM + 2.7178 × qOmax − 2.3561 × qNmin + 2.4803 × μh
| (2) |
| N = 29, R2 = 0.9566, F = 101.46, S2 = 0.0040 |
| Descriptor No. | X | ±ΔX | t-Test | Descriptor |
|---|---|---|---|---|
| a Energy of the highest occupied molecular orbit in atomic units.b Dipole moment.c Max. net atomic charge for a O atom.d Min. net atomic charge for a N atom.e Tot hybridization composite of the molecular dipole. | ||||
| 0 | 2.8005 | 4.9272 × 10−1 | 5.6838 | Intercept |
| 1 | 3.4038 | 3.0849 × 10−1 | 11.0338 | HOMOa |
| 2 | −9.2784 | 9.1786 × 10−1 | −10.1086 | DMb |
| 3 | 2.7178 | 4.0462 × 10−1 | 6.7169 | qOmaxc |
| 4 | −2.3561 | 2.6446 × 10−1 | −8.9090 | qNmind |
| 5 | 2.4803 | 7.6091 × 10−1 | 3.2596 | μhe |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 and predicted log
LC50 and predicted log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50
LC50
| No. | Compd | Calc. log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 LC50 |
Exp. log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50 LC50 |
Difference |
|---|---|---|---|---|
| 1 | 3 | 0.508 | 0.441 | 0.067 |
| 2 | 4a | 0.463 | 0.470 | −0.007 |
| 3 | 4b | 0.431 | 0.470 | −0.039 |
| 4 | 4c | 0.469 | 0.459 | 0.010 |
| 5 | 4d | 0.469 | 0.466 | 0.003 |
| 6 | 4e | 0.474 | 0.455 | 0.019 |
| 7 | 4f | 0.443 | 0.399 | 0.044 |
| 8 | 4g | 0.505 | 0.470 | 0.035 |
| 9 | 4h | 0.779 | 0.792 | −0.013 |
| 10 | 4i | 0.749 | 0.805 | −0.056 |
| 11 | 4j | 0.803 | 0.802 | 0.001 |
| 12 | 4k | 0.269 | 0.329 | −0.060 |
| 13 | 4l | 0.257 | 0.351 | −0.094 |
| 14 | 4m | 0.248 | 0.351 | −0.103 |
| 15 | 4n | 0.355 | 0.339 | 0.016 |
| 16 | 6a | 0.268 | 0.317 | −0.049 |
| 17 | 6b | 0.228 | 0.297 | −0.069 |
| 18 | 6c | 0.270 | 0.274 | −0.004 |
| 19 | 6d | 0.139 | 0.040 | 0.099 |
| 20 | 6e | 0.118 | 0.050 | 0.068 |
| 21 | 6f | −0.619 | −0.652 | 0.033 |
| 22 | 6g | 0.563 | 0.466 | 0.097 |
| 23 | 6h | 0.051 | 0.073 | −0.022 |
| 24 | 6i | 0.418 | 0.487 | −0.069 |
| 25 | 6j | 0.500 | 0.470 | 0.030 |
| 26 | 6k | 0.083 | 0.050 | 0.033 |
| 27 | 6l | 0.154 | 0.083 | 0.071 |
| 28 | 6m | 0.094 | 0.040 | 0.053 |
| 29 | 6n | −0.766 | −0.670 | −0.096 |
 | ||
| Fig. 6 HOMO energy maps for compounds 6f and 6n from DFT calculation of Gaussian 03W. The green parts represent positive molecular orbitals, and the red parts represent negative molecular orbitals. | ||
The internal validation and “leave-one-out” cross-validation methods were used to validate the developed QSAR model.37,38 The internal validation was carried out by dividing the compound data into three subsets A, B and C, with 10, 10 and 9 compounds respectively. The compounds 1, 4, 7, 10, etc., went into subset (A); 2, 5, 8, 11, etc., went into subset (B); and 3, 6, 9, 12, etc., went into the third subset (C). Two of the three subsets, (A and B), (A and C), and (B and C), made up the training set while the remaining subset was treated as a test set. The correlation equations were derived from each of the training sets using the same descriptors and then applied to predict values for the corresponding test set. Internal validation results are presented in Table 5. The RTraining2 and RTest2 are within 5% for all three sets, and the average values of RTraining2 = 0.9575 and RTest2 = 0.9530 were close to the overall R2 value. Therefore, the QSAR model obtained demonstrated the predictive power of 3-fold cross-validation. The “leave-one-out” method was completed in a similar manner to the internal validation. In the “leave-one-out” method, a set of seven compounds (4, 8, 12, etc.) was used as the external test set and the remaining compounds were left in the training subset. The QSAR model containing the same five descriptors was obtained with R2 = 0.9868 from the training set. When the same QSAR model was applied on the test set, R2 = 0.9793 was observed. Therefore, the “leave-one-out” cross-validation results were also satisfactory.
| Training set | N | R2 | F | S2 | Test set | N | R2 | F | S2 |
|---|---|---|---|---|---|---|---|---|---|
| a Compds A: 1, 4, 7, 10, 13, 16, 19, 22, 25, 28. Compds B: 2, 5, 8, 11, 14, 17, 20, 23, 26, 29. Compds C: 3, 6, 9, 12, 15, 18, 21, 24, 27. | |||||||||
| A + B | 20 | 0.9600 | 100.26 | 0.0053 | C | 9 | 0.9644 | 112.34 | 0.0037 |
| B + C | 19 | 0.9599 | 98.44 | 0.0031 | A | 10 | 0.9433 | 100.52 | 0.0039 |
| A + C | 19 | 0.9526 | 118.20 | 0.0048 | B | 10 | 0.9515 | 108.75 | 0.0048 |
| Average | 0.9575 | 105.63 | 0.0044 | Average | 0.9530 | 107.20 | 0.0041 | ||
By interpreting the descriptors involved in the model, we gained some insight into the structural features influencing insecticidal activity. The foremost important descriptor was the HOMO energy (the energy of the highest occupied molecular orbital). This descriptor has a significant effect on the activity as the energy of the HOMO is directly related to the ionization potential of the compounds and characterizes the susceptibility of the molecule to electrophilic attack.39,40 The negative contribution of HOMO energy also suggested that the electron withdrawing substitution groups of rosin-based diamides are favorable for the insecticidal activity against P. xylostella.
The second important descriptor was the dipole moment. This descriptor was important in modulating insecticidal activity against P. xylostella because of the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, N–H, and thiadiazole groups, which exhibited permanent polarization due to an electronegativity difference between the atoms.37,39 The O (C
O, N–H, and thiadiazole groups, which exhibited permanent polarization due to an electronegativity difference between the atoms.37,39 The O (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), N (N–H/thiadiazole group), and S (thiadiazole group) atoms may be involved in binding interactions with insect cells present at the target site. The dipole moment thus played a critical role in modulating the insecticidal activity of the test compounds.41,42
O), N (N–H/thiadiazole group), and S (thiadiazole group) atoms may be involved in binding interactions with insect cells present at the target site. The dipole moment thus played a critical role in modulating the insecticidal activity of the test compounds.41,42
The 3rd and 4th descriptors obtained in the model were max. net atomic charge for a O atom and min. net atomic charge for a N atom. These two descriptors belonged to electrostatic descriptors, and reflected characteristics of the charge distribution of the molecules.39,41–43 The 5th descriptor obtained in the model was tot hybridization composite of the molecular dipole, which belonged to quantum-chemical descriptors. This descriptor reflected the quantitative measure of the lipophilic and hydrophobic properties of the compounds, and was essential for the penetration and distribution of the compounds as well as the interaction of compounds with receptors.44,45 In eqn (2), appearance with a negative sign in the model indicates that a molecule with a lower descriptor value has a higher log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50, and contrary to that a positive sign in the model indicates that a molecule with a higher descriptor value has a higher log
LC50, and contrary to that a positive sign in the model indicates that a molecule with a higher descriptor value has a higher log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50.
LC50.
4. Conclusions
In summary, 29 rosin-based diamides 3, 4a–n and 6a–n were synthesized and their insecticidal activities against P. xylostella and M. separata were evaluated. Thiadiazole-containing diamides 6a–n displayed better activity than others, especially compounds 6f and 6n which exhibited excellent insecticidal activities against P. xylostella. A QSAR study indicated that the involved descriptors for rosin-based diamides may account for their structural features responsible for the insecticidal activity. These promising results are of significant importance to the development of insecticides from common, inexpensive, and non-toxic natural products.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Project no. 31400509 and Project no. 31401783), Forestry Industry Research Special Funds for Public Welfare Projects (Project no. 201404701) and Research Fund for the Doctoral Program of Higher Education of Northwest A&F University (Project no. 2013BSJJ048).Notes and references
- G. P. Lahm, T. M. Stevenson, T. P. Selby, J. H. Freudenberger, D. Cordova, L. Flexner, C. A. Bellin, C. M. Dubas, B. K. Smith, K. A. Hughes, J. G. Hollingshaus, C. E. Clark and E. A. Benner, Bioorg. Med. Chem. Lett., 2007, 17, 6274–6279 CrossRef CAS PubMed.
- G. P. Lahm, D. Cordova and J. D. Barry, Bioorg. Med. Chem., 2009, 17, 4127–4133 CrossRef CAS PubMed.
- G. P. Lahm, D. Cordova, J. D. Barry, T. F. Pahutski, B. K. Smith, J. K. Long, E. A. Benner, C. W. Holyoke, K. Joraski, M. Xu, M. E. Schroeder, T. Wagerle, M. J. Mahaffey, R. M. Smith and M. Tong, Bioorg. Med. Chem. Lett., 2013, 23, 3001–3006 CrossRef CAS PubMed.
- L. A. Teixeira and J. T. Andaloro, Pestic. Biochem. Physiol., 2013, 106, 76–78 CrossRef CAS PubMed.
- T. P. Selby, G. P. Lahm, T. M. Stevenson, K. A. Hughes, D. Cordova, I. B. Annan, J. D. Barry, E. A. Benner, M. J. Currie and T. F. Pahutski, Bioorg. Med. Chem. Lett., 2013, 23, 6341–6345 CrossRef CAS PubMed.
- A. Jeanguenat, Pest Manage. Sci., 2013, 69, 7–14 CrossRef CAS PubMed.
- M. Liu, Y. Wang, W. Wangyang, F. Liu, Y. Cui, Y. Duan, M. Wang, S. Liu and C. Rui, J. Agric. Food Chem., 2010, 58, 6858–6863 CrossRef CAS PubMed.
- B. Troczka, C. T. Zimmer, J. Elias, C. Schorn, C. Bass, T. G. E. Davies, L. M. Field, M. S. Williamson, R. Slater and R. Nauen, Insect Biochem. Mol. Biol., 2012, 42, 873–880 CrossRef CAS PubMed.
- C. Gnamm, A. Jeanguenat, A. C. Dutton, C. Grimm, D. P. Kloer and A. J. Crossthwaite, Bioorg. Med. Chem. Lett., 2012, 22, 3800–3806 CrossRef CAS PubMed.
- B. Wang, H. Zhu, Y. Ma, L. Xiong, Y. Li, Y. Zhao, J. Zhang, Y. Chen, S. Zhou and Z. Li, J. Agric. Food Chem., 2013, 61, 5483–5493 CrossRef CAS PubMed.
- M. Feng, Y. Li, H. Zhu, L. Zhao, B. Xi and J. Ni, J. Agric. Food Chem., 2010, 58, 10999–11006 CrossRef CAS PubMed.
- A. K. Isaacs, S. Qi, R. Sarpong and J. E. Casida, Chem. Res. Toxicol., 2012, 25, 1571–1573 CrossRef CAS PubMed.
- Y. Wang, Y. Shao, Y. Wang, L. Fan, X. Yu, X. Zhi, C. Yang, H. Qu, X. Yao and H. Xu, J. Agric. Food Chem., 2012, 60, 8435–8443 CrossRef CAS PubMed.
- C. L. Cantrell, F. E. Dayan and S. O. Duke, J. Nat. Prod., 2012, 75, 1231–1242 CrossRef CAS PubMed.
- J. Li, J. Song, S. Shang, X. Rao and Y. Gao, Nat. Prod. Res., 2012, 27, 702–710 CrossRef PubMed.
- E. J. Alvarez-Manzaneda, R. Chahboun, J. J. Guardia, M. Lachkar, A. Dahdouh, A. Lara and I. Messouri, Tetrahedron Lett., 2006, 47, 2577–2580 CrossRef CAS PubMed.
- G. Liu, X. Ju, J. Cheng and Z. Liu, Chemosphere, 2010, 78, 300–306 CrossRef CAS PubMed.
- A. Speck-Planche, M. Natalia Dias Soeiro Cordeiro, L. Guilarte-Montero and R. Yera-Bueno, Curr. Comput.–Aided Drug Des., 2011, 7, 304–314 CrossRef CAS.
- A. Speck-Planche, V. V. Kleandrova and M. T. Scotti, Chemom. Intell. Lab. Syst., 2012, 111, 39–45 CrossRef CAS PubMed.
- C. Hansch and R. P. Verma, Eur. J. Med. Chem., 2009, 44, 260–273 CrossRef CAS PubMed.
- P. K. Naik, S. T. Singh and H. Singh, SAR QSAR Environ. Res., 2009, 20, 551–566 CrossRef CAS PubMed.
- H. Xu, J. Wang, H. Sun, M. Lv, X. Tian, X. Yao and X. Zhang, J. Agric. Food Chem., 2009, 57, 7919–7923 CrossRef CAS PubMed.
- J. Li, X. Rao, S. Shang, Y. Gao and J. Song, BioResources, 2012, 17, 1961–1971 Search PubMed.
- H. Wang, S. Shang, Y. Yin, X. Rao and X. Xu, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2009, 65, o1521 CAS.
- Y. Zhou, B. Wang, F. Di, L. Xiong, N. Yang, Y. Li, Y. Li and Z. Li, Bioorg. Med. Chem. Lett., 2014, 24, 2295–2299 CrossRef CAS PubMed.
- Q. Feng, Z. Liu, L. Xiong, M. Wang, Y. Li and Z. Li, J. Agric. Food Chem., 2010, 58, 12327–12336 CrossRef CAS PubMed.
- M. Feng, Y. Li, H. Zhu, J. Ni, B. Xi and L. Zhao, Pest Manage. Sci., 2012, 68, 986–994 CrossRef CAS PubMed.
- J. Shang, W. Wang, Y. Li, H. Song, Z. Li and J. Wang, J. Agric. Food Chem., 2012, 60, 8286–8293 CrossRef CAS PubMed.
- F. Huang, M. Zhao, X. Zhang, C. Wang, K. Qian, R. Kuo, S. Morris-Natschke, K. Lee and S. Peng, Bioorg. Med. Chem., 2009, 17, 6085–6095 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. A. Montgomery, T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez and J. A. Pople, Gaussian 03, Gaussian, Inc., Wallingford CT, 2004 Search PubMed.
- J. Zhang, J. Xu, B. Wang, Y. Li, L. Xiong, Y. Li, Y. Ma and Z. Li, J. Agric. Food Chem., 2012, 60, 7565–7572 CrossRef CAS PubMed.
- M. Mao, Y. Li, Y. Zhou, X. Zhang, Q. Liu, F. Di, H. Song, L. Xiong, Y. Li and Z. Li, J. Agric. Food Chem., 2014, 62, 1536–1542 CrossRef CAS PubMed.
- A. Katritsky, M. Karelson, V. S. Lobanov, R. Dennington, T. A. Keith and R. D. A. T. Keith, Codessa 2.7.15, Semichem, Inc., Shawnee, KS, 2004 Search PubMed.
- M. J. S. Dewar, A. J. Holder, I. Roy, D. Dennington, D. A. Liotard, D. G. Truhlar, T. A. Keith, J. M. Millam and C. D. Harris, AMPAC 9.3.1, Semichem, Inc., Shawnee, KS, 2004 Search PubMed.
- S. Kang, B. Song, J. Wu, M. He, D. Hu, L. Jin, S. Zeng, W. Xue and S. Yang, Eur. J. Med. Chem., 2013, 67, 14–18 CrossRef CAS PubMed.
- J. Song, Z. Wang, A. Findlater, Z. Han, Z. Jiang, J. Chen, W. Zheng and S. Hyde, Bioorg. Med. Chem. Lett., 2013, 23, 1245–1248 CrossRef CAS PubMed.
- Z. Wang, J. Song, J. Chen, Z. Song, S. Shang, Z. Jiang and Z. Han, Bioorg. Med. Chem. Lett., 2008, 18, 2854–2859 CrossRef CAS PubMed.
- S. Liao, J. Song, Z. Wang, J. Chen, G. Fan, Z. Song, S. Shang, S. Chen and P. Wang, Bioorg. Med. Chem. Lett., 2014, 24, 773–779 CrossRef CAS PubMed.
- P. Sharma, A. Kumar, S. Upadhyay, V. Sahu and J. Singh, Eur. J. Med. Chem., 2009, 44, 251–259 CrossRef CAS PubMed.
- D. Sharma, B. Narasimhan, P. Kumar and A. Jalbout, Eur. J. Med. Chem., 2009, 44, 1119–1127 CrossRef CAS PubMed.
- A. D. Pillai, S. Rani, P. D. Rathod, F. P. Xavier, K. K. Vasu, H. Padh and V. Sudarsanam, Bioorg. Med. Chem., 2005, 13, 1275–1283 CrossRef CAS PubMed.
- K. Kalani, D. Yadav, F. Khan, S. Srivastava and N. Suri, J. Mol. Model., 2012, 18, 3389–3413 CrossRef CAS PubMed.
- P. P. Roy, S. Kovarich and P. Gramatica, J. Comput. Chem., 2011, 32, 2386–2396 CrossRef CAS PubMed.
- R. Kumar, A. Kumar, S. Jain and D. Kaushik, Eur. J. Med. Chem., 2011, 46, 3543–3550 CrossRef CAS PubMed.
- M. Witschel, Bioorg. Med. Chem., 2009, 17, 4221–4229 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: IR, 1H NMR, MS, and elemental analysis data for the target compounds. The values of all descriptors of rosin-based diamides with insecticidal activity against P. xylostella are also available. See DOI: 10.1039/c4ra10125a |
| This journal is © The Royal Society of Chemistry 2014 |