Complete degradation of dimethyl phthalate by biochemical cooperation of the Bacillus thuringiensis strain isolated from cotton field soil†
Muhammad Ali Surhioa,
Farah N. Talpur*a,
Shafi M. Nizamania,
Farah Amina,
Chui Wei Bongb,
Choon Weng Leeb,
M. A. Ashrafc and
Muhammad Raza Shahd
aNational Centre of Excellence in Analytical Chemistry, University of Sindh, Jamshoro-76080, Pakistan. E-mail: farahtalpur@hotmail.com; Tel: +92-222-772065
bInstitute of Biological Sciences, Faculty of Science Building, University of Malaya, 50603, Kuala Lumpur, Malaysia
cDepartment of Geology, Faculty of Science, University of Malaya, 50603, Kuala Lumpur, Malaysia
dH.E.J. Research Institute of Chemistry, International Center for Chemical and Biological Sciences, University of Karachi, Karachi 75270, Pakistan
First published on 17th October 2014
Abstract
Dimethyl phthalate (DMP), a phthalate ester, is widely used in cosmetics, perfumes, and plasticizers. It has been classified as a suspected endocrine disruptor by many countries. The present study describes the biodegradation of DMP by a new aerobic bacterium, isolated from soil samples of a cotton field by an enrichment culture technique utilizing DMP as the sole source of carbon and energy. The isolate was identified as Bacillus thuringiensis based on the morphological and biochemical characteristics as well as gene sequence analysis. Bacillus thuringiensis grows best in a mineral salt medium of pH 7.0 at 30 °C incubation for 48 hours. The effects of temperature, inoculum size, substrate concentration and incubation time on DMP degradation were also studied. Bacillus thuringiensis is able to biodegrade 400 mg L−1 of DMP under aerobic conditions with 99% degradation potential. A combination of GC and GC-MS analysis revealed a complete DMP biodegradation pathway. The results indicate that Bacillus thuringiensis may prove a promising source for DMP bioremediation at a commercial scale.
Introduction
Dimethyl phthalate (DMP) is widely used as a plasticizer in the plastic industry during the manufacturing of compounds such as latex, cellulose acetate films and plastic. DMP leaches into the environment from tubing, dishes, paper, and containers via the general use of plastic.1 DMP molecules are bound physically to the plastic structure; they are easily released from plastic products and leached into the environment.2 Moreover it is a relatively stable compound in the natural environment with a half-life of ≈20 years.3 Therefore, DMP has been frequently identified in diverse environmental samples including groundwater, river water, drinking water, open ocean water, soil humates, lake and marine sediments.4 DMP promotes chromosome injuries in human leucocytes and known to be an endocrine disrupting chemical (EDC) that interferes with the reproductive system and normal development of animals and humans.5Natural processes such as hydrolysis and photo decomposition have been previously reported for degradation of phthalates from environment.6–8 Unfortunately degradation of DMP under abiotic conditions has been reported to be slow and insignificant process.3
Breakdown by microorganisms plays a major role in DMP degradation and bacterial activities bring about actual assimilation of various compounds under eco-friendly conditions. Several investigators have demonstrated successful degradation of DMP by microbes under aerobic conditions in soil, natural water and wastewater.9 Different bacterial strains have been isolated with the ability to degrade DMP and their isomers from activated sludge, mangrove sediment, wastewater, etc.9–11 The microbial strains such as Sphingomonas paucimobilis, Pseudomonas fluorescens, Pseudomonas aureofaciens, Xanthomonas maltophilia,12 Rhodococcus rubber,13 Pseudomonas fluorenscens FS1,14 Flavobacterium sp.,15Candida rugosa16 and Arthrobacter sp.17 have reported as to degrade DMP. Studies have shown that bioremediation can offer a potential solution for the conversion of phthalic acid esters into harmless end products such as CO2 and H2O.18 However; many authors have not described the degradation pathways, although few have reported the conversion of DMP into monomethyl phthalate (MMP) and phthalic acid (PA).
Hence in the present study, an aerobic bacterial strain Bacillus thuringiensis was enriched from cotton field soil. Bacillus thuringiensis is a soil dwelling bacterium and is deadly to insects but it is non-pathogenic to human beings. The biodegradation potential of this strain is investigated first time in literature for DMP degradation and its end product in present study. The influences of environmental factors on degradation ability were also examined. Gene sequence and GC-MS were employed to analyze the microbial strain structure and DMP degradation intermediate respectively.
Experimental
Chemicals
Dimethyl phthalate (DMP) 99% purity was purchased from Scharlau (Barcelona, Spain). Luria–Bertani (LB) medium was procured from Caisson laboratories inc (USA). n-Hexane and chloroform was procured from Sigma-Aldrich (Steinheim, Germany). All other chemical reagents used were of analytical grade.Source of microorganisms
The bacteria used in the degradation experiments were isolated from cotton field soil collected from village of district Mirpurkhas, Sindh, Pakistan. The soil samples were stored in plastic bags and kept at 4 °C in refrigerator until start of experiments. The soil was air dried at room temperature (28 °C). Next the soil ground gently in mortar & pestle and passed through a 2 mm-mesh sieve to remove stones and plant debris before the physico-chemical testing. The physical and chemical properties of the soil are given as ESI.†Enrichment and isolation of bacteria
The mineral salt medium (MSM) used in all experiments including enrichment experiments contained (L−1): (NH4)2SO4 1 g, KH2PO4 0.8 g, K2HPO4 0.2 g, MgSO4·7H2O 0.5 g, FeSO4 0.01 g, CaCl2 0.05 g, NiSO4 0.032 g, Na2B4O7·10H2O 0.021 g, (NH4)6Mo7O24·H2O 0.0144 g, ZnCl2 0.023 g, CoCl2·H2O 0.021 g, CuCl2·2H2O 0.01 g and MnCl2·4H2O 0.03 g.19 The pH of MSM was adjusted to 7.0 with 0.1 M HCl or NaOH and then sterilized by autoclaving for 20 min at 121 °C.The agar plates were prepared by adding 20 g of Luria–Bertani (LB) medium, 15 g Agar and the minimal salt medium consisting of (L−1): K2HPO4 4.5648 g, KH2PO4 3.4023 g, Na2HPO4·12H2O 5.3721 g, NH4Cl 0.8024 g, CaCl2 0.0368 g, MgSO4·7H2O 0.1232 g and FeCl3·6H2O 0.0014 g, at pH 7.0.20 The enrichment procedure was similar to that described by Chao et al.21 with some modifications. Initially, 2.0 g of soil was added to a 250 mL Erlenmeyer flask containing 100 mL of MSM solution amended with DMP (1 mg L−1). The suspension was incubated for 7 days in the dark at 30 °C in an orbital incubator shaker operated at 150 rpm. Subsequently, 2 mL of the enrichment culture was serially transferred four times to fresh medium, each time containing a higher concentration of DMP (3–10 mg L−1) and incubated under the same conditions. Then the final enrichment was streaked onto the agar plates and incubated one week at 30 °C. Presumptive colonies were picked on the basis of differences in colony, morphology and coloration and re-streaked onto agar plates amended with the DMP. The bacterial isolates were further purified by streaking on LB Nutrient agar plates and then re-streaked onto agar plates with and without DMP to confirm their degradation abilities. Isolates showing growth in the presence of DMP were selected for further degradation study.
Identification and characterization of bacteria
Gram reaction and cell morphology were determined by observing stained cells under a light microscope. Biochemical tests for the identification of bacteria were performed using standard methods.22 The bacterial isolates of pure culture were further characterized using 16S rRNA gene sequencing methods. To extract the genomic DNA of bacterial strains for 16S rRNA gene amplification, we used the phenol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :chloroform
:chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) isoamyl alcohol (25
isoamyl alcohol (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) extraction and ice-cold ethanol precipitation method. DNA was resuspended in Tris–EDTA (TE) buffer (pH 8.0) and stored at −20 °C. For isolated strain, one pair of universal primers was used for the amplification of the 16S rDNA gene: the forward primer, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′), and the reverse primer, 1525R (5′-AAGGAGGTGWTCCARCC-3′) were used to produce an amplicon of approximately 1498 bp. Polymerase chain reaction (PCR) was performed in a 15 μL reaction mixture containing 10 × PCR buffer, 1.5 mM of MgCl2, 0.4 μM of each primer, 3 μL of DNA template (approximately 200 ng μL−1), 0.2 mM of dNTPs, and 0.5 U of Taq polymerase (FinnzymesDyNAzyme™ II, Finland). PCR amplification was carried out according to the following conditions: initial denaturation at 94 °C for 2 min; 35 cycles of denaturation for 30 s at 94 °C, annealing for 40 s at 55 °C, extension for 90 s at 72 °C; and a final extension at 72 °C for 7 min. PCR products were then purified using the QIAquick® PCR Purification Kit (QIAGEN, Germany) as reported earlier by Lee et al.23 The 16S rDNA sequences of each isolates were compared to GenBank entries using Basic Local Alignment Tool (BLAST) in order to obtain a preliminary affiliation.
1) extraction and ice-cold ethanol precipitation method. DNA was resuspended in Tris–EDTA (TE) buffer (pH 8.0) and stored at −20 °C. For isolated strain, one pair of universal primers was used for the amplification of the 16S rDNA gene: the forward primer, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′), and the reverse primer, 1525R (5′-AAGGAGGTGWTCCARCC-3′) were used to produce an amplicon of approximately 1498 bp. Polymerase chain reaction (PCR) was performed in a 15 μL reaction mixture containing 10 × PCR buffer, 1.5 mM of MgCl2, 0.4 μM of each primer, 3 μL of DNA template (approximately 200 ng μL−1), 0.2 mM of dNTPs, and 0.5 U of Taq polymerase (FinnzymesDyNAzyme™ II, Finland). PCR amplification was carried out according to the following conditions: initial denaturation at 94 °C for 2 min; 35 cycles of denaturation for 30 s at 94 °C, annealing for 40 s at 55 °C, extension for 90 s at 72 °C; and a final extension at 72 °C for 7 min. PCR products were then purified using the QIAquick® PCR Purification Kit (QIAGEN, Germany) as reported earlier by Lee et al.23 The 16S rDNA sequences of each isolates were compared to GenBank entries using Basic Local Alignment Tool (BLAST) in order to obtain a preliminary affiliation.
Sample preparation for AFM imaging
The bacterial culture was grown in 100 mL MSM at 30 °C. Bacteria were harvested at mid-exponential growth phase and washed three times in phosphate-buffered saline (PBS) (contained 5 mM KH2PO4, 25 mM Na2HPO4, and 120 mM NaCl, pH adjusted to 7.4 by using NaOH) by low-speed centrifugation, so as to avoid shear, and resuspended in PBS. The obtained solution was vortexed for 1 min and sonicated (KQ 500-DE) for 30 min. About 30 μL solution was taken out and deposited on a freshly cleaved mica surface, and then the sample was analyzed through atomic force microscopy (AFM). The images for MSM culture was also taken directly without washing with PBS buffer after appropriate dilution.Topographic images at room temperature were recorded with AFM 5500 (Agilent, USA). Silicon nitride probes with a triangular soft cantilever (Veeco, model MLCT-AUHW) with a nominal value of the spring constant of 0.01 N m−1 and 0.1 N m−1 being used in the non-contact topography measurements.
DMP degradation experiment
Bacterial cultures were grown in the MSM on 30 °C at 150 rpm, growing cells were then harvested after 48 hours. For degradation experiment 100 mL MSM was prepared in 250 mL Erlenmeyer flask sterilized and supplemented with DMP at various concentrations (100–1000 mg L−1) and then inoculated with freshly harvested culture and incubated at 30 °C. After incubation period, 30 mL of media containing degraded DMP and n-hexane was added directly to separating funnel and shaken vigorously for few minutes. The resulting heavy emulsion was centrifuged (5000 rpm, 5 min) and the aqueous phase was extracted. The hexane phase was evaporated to dryness and reconstitute for further analysis. All the degradation experiments were performed in triplicate.GC-FID conditions for substrate analysis
The substrate analysis was done by using RT-2560 (0.25 mm ID) 100 m long polar capillary column, with the gas chromatograph (GC-8700 PERKIN ELMER). Nitrogen was used as a carrier gas, oven temperature was increased from 150 to 200 °C at a ramp rate of 11 °C min−1 and then final temperature 220 °C at a ramp rate of 2 °C min−1 held for 20 min, injection volume was 2 μL, injector and detector temperature were kept 240 °C and 260 °C respectively.Spectrophotometric determinations
The microbial biomass in the culture flasks was determined spectrophotometrically by measuring optical density at 600 nm (OD600) using UV-visible spectrophotometer (Biochrom Libra S22).Analysis of DMP-derived degradation products by GC-MS
To detect end products after the course of incubation, the samples extracted above were concentrated to 1 mL and then subjected to GC-MS analysis using a Agilent 6890 N gas chromatograph, Injector auto sampler 7683-B, equipped with a MS-5975 inert XL mass selective mass spectrometer (GC-MS, Agilent Technologies, Santa Clara, USA). Gas chromatography was performed on a Capillary column HP-5MS (30 m × 0.25 mm × 0.25 μm); temperature maintained at 190 °C for 1 min, increased from 15 °C min−1 to 300 °C for 15 min, then cooled rapidly to 60 °C, using ultra high purity helium as carrier (flow rate, 0.8 mL min−1), injector and detector temperature were kept 310 °C and 350 °C respectively. For MS spectra collected in the positive electron ionization (EI) mode, the source temperature was 250 °C with an electron ionization energy of 70 eV, the mass range was 50–550 m/z, and 1 μL of the extracted sample was injected in split mode (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The NIST Mass Spectral Search Program (National Institutes of Standard and Technology, Gaithersburg, MD) was used to identify signals by comparison to the retention time and mass spectra of authentic compounds.
1). The NIST Mass Spectral Search Program (National Institutes of Standard and Technology, Gaithersburg, MD) was used to identify signals by comparison to the retention time and mass spectra of authentic compounds.
Statistical analysis
Excel XL State (Microsoft Corp., Redmond, WA) was used to perform tests for statistical significance. Student t-test was performed to assess the significance of the differences between percent degradation and experimental variables at probability level of 0.05.Results and discussion
Identification and characterization
The strain was Gram-positive, rod-shaped and was a facultative anaerobe. In addition, this strain exhibited positive reaction for spore formation and negative reaction for mannitol test. The 16S rDNA partial gene sequence of this bacterium were aligned and compared with the 16S rDNA bacterial gene sequences in GenBank and the results indicated that it bore the closest relationship to Bacillus thuringiensis with 97% of sequence similarity (GenBank accession no. KF218168). The strain was also identified as Bacillus thuringiensis through its morphology, physiochemical characteristics and 16S rDNA sequence. However, there have been no comprehensive studies of DMP biodegradation by this bacterium. Only one paper is available on concurrent degradation of dimethyl phthalate (DMP) during production of Bacillus thuringiensis based biopesticides.24Bacterial cell morphology by AFM analysis
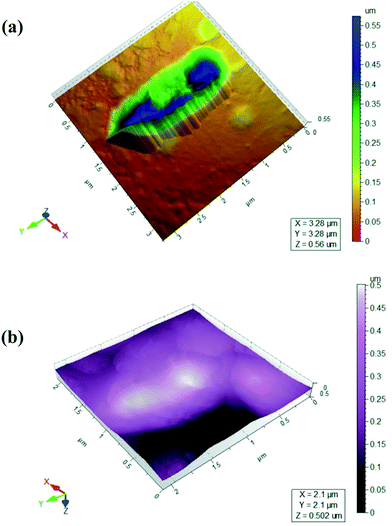
The observation of the morphology of bacterial cell and their ultrastructures is fundamental for understanding the structure and behaviour of bacteria, since morphology is one way for bacteria to cope with their environment and gain a competitive advantage.25 The surface morphology of isolated strain in various environmental conditions (buffer and media) was observed. Freshly cleaved mica is an appropriate substrate for visualization of bacteria. The images of isolated bacterial strain were taken from bacterial culture harvested from growing medium and imaged in PBS and MSM (Fig. 1a and b). Although bacteria imaged in PBS gave universally good results in the qualitative tests (Fig. 1a), bacterial images comparable to those derived from mineral media were obtained by AFM. In all cases, the bacteria maintained a hydrated appearance with no evidence of collapse of the cell wall. Isolated bacterial strain have different sizes, cells are longer and thicker as seen in Fig. 1b. The obtained parameters characterizing the bacteria dimensions for isolated strain are summarized in as: length 2.75 μm, width 0.75–1.75 μm, height 0.55 μm. The preparation technique used gives a monolayer and bacterium is clearly seen in AFM images (Fig. 1a). Most of bacteria are packed close to one another, forming compact coverings due to surrounding medium (Fig. 1b). Packing of bacteria leads to a reduced contact area and may contribute to greater collective stability. In all cases examined attachment was robust enough to allow individual bacterium to be repeatedly imaged.Biodegradation of DMP
A microbial strain capable of mineralizing DMP under aerobic conditions was enriched and isolated from cotton field soil. Bacterial strain could utilize DMP as the sole carbon and energy source. Results have demonstrated that DMP was completely degraded by B. thuringiensis as evident from GC-FID chromatogram for control of DMP (Fig. 2a) as compared to reaction with B. thuringiensis (Fig. 2b). It can be clearly seen that the DMP was degraded very quickly, with more than 99% removal in less than 72 h. Control experiment was also carried out by autoclaving and not inoculating the flask, the results showed that no obvious degradation was observed. This suggested that abiotic loss of phthalates can be neglected during this study.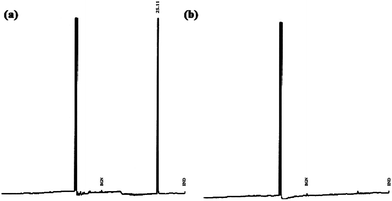 | ||
| Fig. 2 GC-FID chromatogram of (a) control and (b) reaction with B. thuringiensis containing DMP at 400 mg L−1. | ||
Similarly Jianlong et al.4 has also reported complete degradation of DMP in soil bioaugmented with acclimated activated sludge at much slower rate i.e. within 15 days. Authors suggested that the mixed microbial consortium can enhance the degradation rates of DMP in the presence of Pseudomonas fluorescens, Pseudomonas aureofaciens and Sphingomonas paucimobilis, possibly due to the coordinated metabolic interaction of different bacterial strains.19 However in present study single strain B. thuringiensis was capable of degrading DMP in 72 h incubation time. Table 1 shows the comparison of degradation potential of DMP by B. thuringiensis with other species which are reported in literature.
Influence of pH and time on growth of DMP degrading bacteria
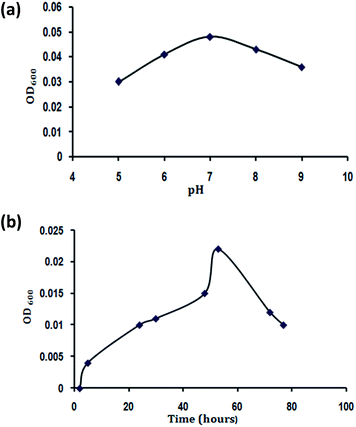
The culture medium hydrogen ion concentration greatly influenced DMP degrading bacterial growth as pH limited enzyme activities is shown in Fig. 3a. The growth of DMP degrading bacteria increased rapidly when pH was increased from 5.0 to 7.0. The highest growth was achieved at pH 7.0. When the pH exceeded 9.0, the growth decreased dramatically. Therefore, from this study it was found that pH 7.0 was optimal for the growth of B. thuringiensis sp.Fig. 3b displays the effects of the time on the growth of B. thuringiensis. The OD600 increased significantly (P < 0.05) with the increase of time from 2 to 48 hours, with a slight increase in the growth rate when the time was increased to 53 hours. However, when the time exceeded 53 hours, the microbial biomass decreased notably. Hence 48 hours were optimal for B. thuringiensis growth.
Effects of temperature on DMP biodegradation
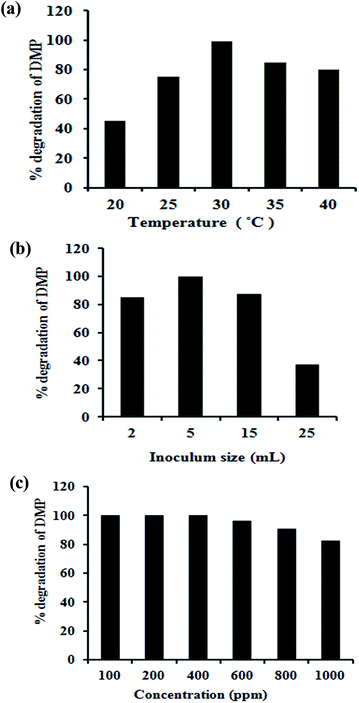
Bacterial growth is temperature sensitive. As shown in Fig. 4a, the DMP degradation rate increased rapidly (P < 0.05) as temperature increased from 25 °C to 30 °C. The highest DMP degradation rate (about 99%) for B. thuringiensis sp. was achieved at 30 °C. Degradation decreased at temperatures above 30 °C, indicating that the optimum temperature was 30 °C. The optimum temperatures for microbes in degrading various organic pollutants have been reported in the range of 30 °C to 38 °C.2,29,30Effects of inoculum size on DMP degradation
Since biodegradation is highly dependent on inoculum concentration,31 the effect of inoculum concentrations on the degradation of DMP was studied (Fig. 4b). Under aerobic conditions, degradation was only 85% when 2 mL inoculums made up to 100 mL medium, degradation increased to 99% when 5 mL inoculums made up to 100 mL medium. However, further increase in the inoculums concentration did not increase the degradation efficiency (P > 0.05). On the contrary inoculums concentrations greater than 5 mL to 100 mL medium resulted in a decrease in degradation efficiency to 87%. Thus, considering the degradation efficiency and treatment costs, an inoculums concentration of 5 mL in 100 mL medium was the most appropriate for aerobic degradation of DMP.The decrease in degradation efficiency with inoculums concentrations was observed previously under both anaerobic and aerobic conditions.32 Increasing inoculums concentration did not enhance the DMP degradation, because increasing population of bacteria also increases the bacterial competition for the resources (e.g., food), and therefore limits bacterial growth when the food in the limited environment becomes depleted.33
Effects of DMP concentration on degradation efficiency
The effect of various initial concentrations of DMP on biodegradation efficiency was subsequently determined under optimal conditions. When DMP was initially added in a range of 100–400 mg L−1, degradation profiles were approximately similar (Fig. 4c). On the other hand, when the substrate concentration was increased to the range of 400–1000 mg L−1, the degradation significantly decreased (P < 0.05).The results (Fig. 4c) also showed that B. thuringiensis can completely degrade DMP within 72 h when the initial concentration is not higher than 400 mg L−1. When the initial concentration of DMP increased to 600 mg L−1, the degradation rate may reach to 96% within 72 h.
Luo et al.34 reported that Trichosporon sp. could degrade DMP at much lower concentration i.e. 21.5 mg L−1 and its degradation rate just reached to about 70% after 24 days incubation. Jianlong et al.4 also reported that the degradation rate of DMP (100 mg L−1) in the soil augmented with acclimated sludge reached to 90% after 15 days incubation time. Therefore, B. thuringiensis is much efficient in degrading DMP compared with previous reports. Furthermore, the results of our study are evident that B. thuringiensis can tolerate high concentration i.e., 1000 mg L−1. Similarly Wang et al.,35 has reported that the free cells of several bacillus sp. can tolerate 400 mg L−1. In addition Hai and Yun-xia36 has reported that the green alga Dunaliella tertiolecta can hydrolyze DMP to the product PA with the maximum tolerance level reaching up to 300 mg L−1.
Kinetics of DMP biodegradation
DMP biodegradability by B. thuringiensis sp. was investigated at initial DMP concentration 400 mg L−1, pH 7.0 and 30 °C. First-order kinetics has been frequently used to describe the biodegradation kinetics of organic pollutants1.4,28,37 Here, DMP biodegradation by B. thuringiensis sp. was assumed to fit to the first-order kinetic equation, with the form:ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C = −Kt + A C = −Kt + A |
t1/2 = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/K 2/K |
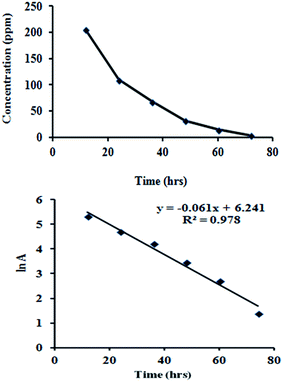
The fitting of DMP biodegradation to experimental data using the first-order kinetic model and the kinetic equation is shown in Fig. 5.
Here, the calculated results indicated that the first-order model was a good fit. When the initial concentration of DMP was 400 mg L−1, the rate constant (K) was independent of initial concentration and the first-order equation could be expressed as ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C = −0.0616t + A with 0.9783 R2 value, the biodegradation half-life was 11.25 h. Jianlong et al.4 reported that in soil augmented with acclimated sludge the biodegradation half-life for DMP (100 mg L−1) was 2.29 days On other hand Wang et al.38 reported that biodegradation half-life for DMP alone (10 mg L−1) by using digested sludge was 1 day. Donglei, et al.39 reported that half-life for DMP (100 mg L−1) biodegradation by using Arthrobacter sp. half-life was 2.25 days. Hence in present study DMP was degraded at much higher concentration and rate.
C = −0.0616t + A with 0.9783 R2 value, the biodegradation half-life was 11.25 h. Jianlong et al.4 reported that in soil augmented with acclimated sludge the biodegradation half-life for DMP (100 mg L−1) was 2.29 days On other hand Wang et al.38 reported that biodegradation half-life for DMP alone (10 mg L−1) by using digested sludge was 1 day. Donglei, et al.39 reported that half-life for DMP (100 mg L−1) biodegradation by using Arthrobacter sp. half-life was 2.25 days. Hence in present study DMP was degraded at much higher concentration and rate.
Biochemical degradation pathway
To explore the pathways of DMP degradation by B. thuringiensis sp., metabolites of DMP were identified by GC-MS. Two major metabolites besides the parent compound DMP, MMP and PA were detected and identified during the course of degradation by comparing the mass spectrum at a particular retention time with the published mass spectra from National Institute of Standards and Technology (NIST) Database (Fig. 6a–c). Ester hydrolysis of DMP resulting in MMP and PA was carried out in two steps before aromatic ring cleavage and full mineralization. In the first step, MMP was formed and followed by hydrolysis of MMP to PA presumably by the same hydrolytic enzyme. The above results are similar to those previously reported.9,40 Based on the above results, a tentative metabolic pathway for the degradation of DMP by B. thuringiensis sp. is proposed in Fig. 6d.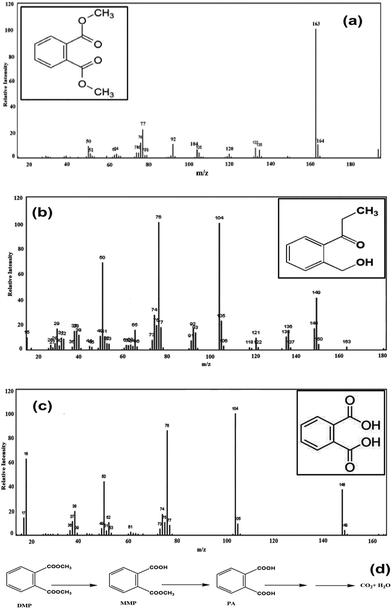 | ||
| Fig. 6 Mass spectrum of (a) dimethyl phthalate (b) monomethyl phthalate (c) phthalic acid and (d) proposed degradation pathway. | ||
Similarly, the proposed degradation biochemical pathway of PAEs involves sequential cleavage of the ester bond to yield the phthalate monoester and then PA which can be further metabolized to produce carbon dioxide and water.36,41–44
Conclusions
DMP degrading bacterium was isolated from cotton field soil. Based on 16S rDNA partial gene sequencing analysis, the dominating bacterial strain B. thuringiensis sp. was identified. This study investigated the optimal pH and time for growth of DMP degrading microbes in MSM and the effects of inoculums size, substrate concentration and time on DMP degradation. MMP and PA could be monitored in metabolic DMP intermediates. The B. thuringiensis sp. metabolized DMP completely at higher concentrations, suggesting that B. thuringiensis sp. represents a novel tool for removing DMP from contaminated waste.Notes and references
- B. Xu, N.-Y. Gao, H. Cheng, S.-J. Xia, M. Rui and D.-D. Zhao, J. Hazard. Mater., 2009, 162, 954–959 CrossRef CAS PubMed.
- C.-R. Fang, J. Yao, Y. G. Zheng, C. J. Jiang, L. F. Hu, Y. Y. Wu and D. S. Shen, Int. Biodeterior. Biodegrad., 2010, 64, 442–446 CrossRef CAS PubMed.
- C. A. Staples, D. R. Peterson, T. F. Parkerton and W. J. Adams, Chemosphere, 1997, 35, 667–749 CrossRef CAS.
- W. Jianlong, Z. Xuan and W. Weizhong, Process Biochem., 2004, 39, 1837–1841 CrossRef PubMed.
- B. L. Yuan, X. Z. Li and N. Graham, Chemosphere, 2008, 72, 197–204 CrossRef CAS PubMed.
- Y. H. Chen, L. L. Chen and N. C. Shang, J. Hazard. Mater., 2009, 172, 20–29 CrossRef CAS PubMed.
- S. Jonsson, V. A. Vavilin and B. H. Svensson, Water Sci. Technol., 2006, 53, 119–127 CrossRef CAS.
- T. K. Lau, W. Chu and N. Graham, Chemosphere, 2005, 60, 1045–1053 CrossRef CAS PubMed.
- D. Liang, T. Zhang, H. Fang and J. He, Appl. Microbiol. Biotechnol., 2008, 80, 183–198 CrossRef CAS PubMed.
- P. Roslev, K. Vorkamp, J. Aarup, K. Frederiksen and P. Nielsen, Water Res., 2007, 41, 969–976 CrossRef CAS PubMed.
- X. Xu, H. Li, J. Gu and X. Li, J. Hazard. Mater., 2007, 140, 194–199 CrossRef CAS PubMed.
- Y. Y. Wang, Y. Z. Fan and J.-D. Gu, World J. Microbiol. Biotechnol., 2003, 19, 811–815 CrossRef CAS.
- J. Li, J.-D. Gu and L. Pan, Int. Biodeterior. Biodegrad., 2005, 55, 223–232 CrossRef CAS PubMed.
- F. Zeng, K. Y. Cui, X. D. Li, J. M. Fu and G. Y. Sheng, Process Biochem., 2004, 39, 1125–1129 CrossRef CAS.
- Y. Kido, T. Tanaka, K. Yamada, H. Hachiyanagi, H. Baba, T. Iriguchi and M. Uyeda, J. Health Sci., 2007, 53, 740–744 CrossRef CAS.
- L. Mita, V. Sica, M. Guida, C. Nicolucci, T. Grimaldi, L. Caputo, M. Bianco, S. Rossi, U. Bencivenga, M. S. M. Eldin, M. A. Tufano, D. G. Mita and N. Diano, J. Mol. Catal. B: Enzym., 2010, 62, 133–141 CrossRef CAS PubMed.
- D. Vega and J. Bastide, Chemosphere, 2003, 51, 663–668 CrossRef CAS.
- W. Donglei, M. Qaisar, W. Lili and Z. Ping, J. Environ. Sci., 2008, 20, 922–926 CrossRef.
- Y. Wang, Y. Fan and J.-D. Gu, Int. Biodeterior. Biodegrad., 2004, 53, 93–101 CrossRef CAS PubMed.
- B. V. Chang, C. M. Yang, C. H. Cheng and S. Y. Yuan, Chemosphere, 2004, 55, 533–538 CrossRef CAS PubMed.
- W. L. Chao, C. M. Lin, I. I. Shiung and Y. Kuo, Chemosphere, 2006, 63, 1377–1383 CrossRef CAS PubMed.
- R. E. Buchanna and N. E. Gibbons, Bergey's Manual of Determinative Bacteriology, ed. J. G. Holt, Lippincott Williams and Wilkins, 530 Walnut St. Philadelphia, USA, 9th edn, 1994 Search PubMed.
- C. W. Lee, A. Y. F. Ng, K. Narayanan, E. U. H. Sim and C. C. Ng, Cienc. Mar., 2009, 35, 153–167 CAS.
- S. K. Brar, M. Verma, R. D. Tyagi, J. R. Valéro and R. Y. Surampalli, J. Hazard. Mater., 2009, 171, 1016–1023 CrossRef CAS PubMed.
- K. D. Young, Curr. Opin. Microbiol., 2007, 10, 596–600 CrossRef PubMed.
- E. W. Merrill and A. Sagar, US Pat., 203952, 1995.
- J. K. H. Cheung, R. K. W. Lam, M. Y. Shi and J.-D. Gu, Sci. Total Environ., 2007, 381, 126–133 CrossRef CAS PubMed.
- C. V. Netten, K. V. Teschke, Y. C. Leung and K. Bartlett, Sci. Total Environ., 2000, 263, 155–160 CrossRef.
- Y. Lu, F. Tang, Y. Wang, J. H. Zhao, X. Zeng, Q. F. Luo and L. Wang, J. Hazard. Mater., 2009, 168, 938–943 CrossRef CAS PubMed.
- K. Khleifat, Process Biochem., 2006, 41, 2010–2016 CrossRef CAS PubMed.
- S. F. Pesce and D. A. Wunderlin, Water Res., 1997, 31, 1601–1608 CrossRef CAS.
- P. D. Jensen, M. T. Hardin and W. P. Clarke, Bioresour. Technol., 2009, 100, 5219–5225 CrossRef CAS PubMed.
- L. A. Mellefont, T. A. McMeekin and T. Ross, Int. J. Food Microbiol., 2008, 121, 157–168 CrossRef CAS PubMed.
- Z.-H. Luo, K.-L. Pang, Y.-R. Wu, J.-D. Gu, R. K. K. Chow and L. L. P. Vrijmoed, in Biology of marine fungi, ed. C. Raghukumar, Springer-Verlag, Berlin Heidelberg, 2012, pp. 299–328 Search PubMed.
- J. L. Wang, Y.-C. Ye and W.-Z. Wu, Biomed. Environ. Sci., 2003, 16, 126–132 Search PubMed.
- Y. Hai and L. Yun-xia, J. Environ. Sci., 1998, 10, 296–301 Search PubMed.
- X. R. Xu, H. B. Li and J. D. Gu, Int. Biodeterior. Biodegrad., 2005, 55, 9–15 CrossRef CAS PubMed.
- J. L. Wang, L. J. Chen, H. C. Shi and Y. Qian, Chemosphere, 2000, 41, 1245–1248 CrossRef CAS.
- W. Donglei, Z. Ping, M. Qaisar and Y. X-sheng, J. Zhejiang Univ., Sci., A, 2007, 8, 1469–1474 CrossRef.
- Y. Wang, B. Yin, Y. Hong, Y. Yan and J. Gu, Ecotoxicology, 2008, 17, 845–852 CrossRef CAS PubMed.
- J. L. Wang, P. Liu and Y. Qian, Process Biochem., 1999, 34, 745–749 CrossRef CAS.
- X. R. Xu, H. B. Li and J. D. Gu, J. Microbiol. Biotechnol., 2005, 15, 946–951 CAS.
- X. R. Xu, H. B. Li and J. D. Gu, Environ. Technol., 2007, 28, 1055–1061 CrossRef CAS PubMed.
- X. C. Duan, X. Y. Yi, X. W. Yang, Z. Dang, G. N. Lu and C. Yang, J. Agro-Environ. Sci., 2007, 26, 1937–1941 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra09465d |
| This journal is © The Royal Society of Chemistry 2014 |