A hydrazone crosslinked zwitterionic polypeptide nanogel as a platform for controlled drug delivery
Caicai Lua,
Bingqiang Lia,
Na Liua,
Guolin Wu*ab,
Hui Gaoc and
Jianbiao Ma*c
aKey Laboratory of Functional Polymer Materials, Institute of Polymer Chemistry, Nankai University, Tianjin 300071, PR China
bCollaborative Innovation Center of Chemical Science and Engineering, Tianjin, PR China. E-mail: guolinwu@hotmail.com
cSchool of Chemistry and Chemical Engineering, Tianjin University of Technology, Tianjin 300191, PR China
First published on 1st October 2014
Abstract
A pH-responsive polypeptide nanogel was prepared via hydrazone self-crosslinking under mild conditions. Zwitterionic sulfobetaine was introduced to the polypeptide by aminolysis reaction of polysuccinimide with N,N-dimethylaminopropylamine, and the subsequent ring-opening reaction between tertiary amine and 1,3-propanesultone. The hydrazine and aldehyde modified poly(α,β-L-aspartic acid) precursors, poly(aspartamide sulfobetaine-co-aspartylhydrazide) and poly(aspartamide sulfobetaine-co-2-oxoethyl aspartamide) were prepared by the successive aminolysis reaction of polysuccinimide. The obtained zwitterionic poly(aspartamide) nanogel exhibited excellent anti-protein adsorption ability and stability in protein solutions. An anticancer drug, doxorubicin (DOX), was entrapped into the nanogel, and showed accelerated release under acidic conditions. Additionally, the nanogel entrapped DOX could be successfully released into the cancer cells and showed high cytotoxicity. This easily prepared nanogel, with features of biocompatibility, anti-protein adsorption ability, and pH-responsiveness, is a promising controlled drug delivery system.
Introduction
It is known that the conventional polymer nanoparticles prepared by self-assembled amphiphilic copolymers would disintegrate after dilution below the critical micelle concentration (CMC). However, nanogels could retain their structure in the dilute solutions because of the cross-linked networks between the polymers, which are useful for the controlled delivery of bioactive compounds in vivo. Nanogels are nanometer-sized hydrogel nanoparticles with three-dimensional networks of cross-linked polymer chains. The porosity between the cross-linked networks of nanogels provides an ideal reservoir for loading drugs,1–3 oligonucleotides,4 and imaging agents.5 What's more, nanogels show rapid responses to micro-environmental factors due to their nanoscale dimensions. Therefore, polymeric nanogels have attracted significant interest over the last several years owing to their potential applications in biomedical fields.Currently, one of the major challenges of nanocarriers used in the drug delivery system is the limited blood circulation time after intravenous systemic administration.6 The prevention of surface protein adsorption is becoming increasingly important for achieving a prolonged blood circulation time for drug delivery systems. Poly(ethylene glycol) (PEG) is the most widely used anti-protein adsorption surface modification material for its high hydrophilicity, flexibility, non-toxicity and non-immunoreaction. However, PEG has relatively low stability in the presence of oxygen and transition-metal ions.7–11 Moreover, the previous studies have shown that the PEG surfaces limit the interaction between carriers and the target tissues, and hard to be functionalized thus compromising the effective cellular uptake of the loaded cargo.12,13 Over the past few years, it has been demonstrated that zwitterionic materials and surfaces are highly resistant to protein adsorption from blood plasma.14 Results showed that due to their strong electrostatically induced hydration, zwitterionic polymers form a tightly bound and structured water layer around polymer chains to repel nonspecific protein adsorption. Betaines represent a special class of zwitterionic materials that bear both cationic moiety, such as quaternary ammonium group, and anionic functionalities, such as sulfo-, carboxy-, and phosphobetaines, on the same repeat unit.15 They have a good resistance to protein adsorption capacity and have been used to modify nanoparticles. Jiang et al. reported that polycarboxybetaine methacrylate coated Fe3O4 nanoparticles would be highly stable in biological media and could evade the innate immune system.16 They also developed a series of zwitterionic particles based on the zwitterionic polymer poly(carboxybetaine methacrylate).17–20 These particles exhibited excellent biophysical stability in fetal bovine serum. However, the biomedical applications of these polymeric nanogels are limited because of their difficult biodegradability and elimination in vivo. Until now, zwitterionic-modified nanogels based on biodegradable polymeric materials have been rarely studied.
Polypeptides have been extensively studied as carriers for drug delivery because of their non-antigenicity, biocompatibility, biodegradability, and various side groups for further functionalization. Unlike most other polypeptides prepared by the complicated ring-opening polymerization of amino acid, N-carboxyanhydrides (NCAs), poly(α,β-L-aspartic acid) (PAsp) can be easily prepared by the hydrolysis of polysuccinimide (PSI),21,22 which can be easily obtained by the thermal polycondensation of L-aspartic acid. Depending on the ring opening reagents, various functional groups can be easily introduced into the side chain of PAsp.22–26 Therefore, PAsp is a good candidate for preparing polypeptide derivatives. Recently, a mixed-charge polypeptide derivative has been developed in our group by amidation of PAsp with L-histidine methyl ester, which exhibited distinct anti-fibrinogen-fouling property.23 However, for this kind of polymers the positive and negative charges distribute randomly and separately, which cannot ensure the surface contains a nanometer-scale homogenous mixture of balanced charged groups as the zwitterionic polymers. The latter is crucial for them to achieve a highly resistant to nonspecific protein adsorption.27 Herein, zwitterionic sulfobetaine was introduced to the polypeptide by aminolysis reaction of polysuccinimide with N,N-dimethylaminopropylamine, and the subsequent ring-opening reaction between tertiary amine and 1,3-propanesultone. The obtained polypeptide derivative bears both cationic moiety like quaternary ammonium and anionic functionality like sulfo- on the same repeat side group, which provides a nanometer-scale homogenous mixture of balanced charges.
Hydrazone has been extensively studied in biomedical researches because of two aspects. First, it can be prepared under mild conditions, and second, it is an acid-cleavable linkage that is stable at the physiological pH and hydrolyzes under acidic conditions.28 Therefore, hydrazone has been widely used in medical biotechnology for developing pH-triggered controlled release system because of the slightly acidic pH of cancer cells.25,29 Recently, an injectable hydrogel by hydrazone cross-linking under the physiological condition from hydrazine and aldehyde modified PAsp was developed in our group without adding any cross-linkers or catalysts.30 In this paper, a polypeptide nanogel with zwitterionic side groups was prepared via hydrazone crosslinking by the reaction of poly(asparamide sulfobetaine-co-aspartylhydrazide) (PASB-Hy) and poly(asparamide sulfobetaine-co-2-oxoethyl asparamide) (PASB-Al) under mild conditions. Hydrazine and aldehyde modified PAsp were used as two nanogel precursors. Both of them are water-soluble and biodegradable polymers with a protein-like structure, and obtained by aminolysis reaction of polysuccinimide. The chemical structure, morphology, stability and anti-protein adsorption ability of the zwitterionic nanogel were studied. Moreover, the potential of the nanogel as pH-sensitive drug delivery system was evaluated by using doxorubicin (DOX) as a model drug.
Experimental
Materials
PSI (Mn = 2.1 × 104, Mw/Mn = 1.30, determined by gel permeation chromatography (GPC) using DMF as eluent) was synthesized in our laboratory, according to the reference.23,31 Sodium periodate (NaIO4) and hydrazine hydrate aqueous solution (80%) were purchased from Tianjin Chemical Reagent. 3-Amino-1,2-propanediol was provided by J&K chemical Ltd. DOX·HCl was obtained from Beijing Huafeng United Technology Co., Ltd and was used as received. 1,3-Propane sultone and N,N-dimethylaminopropylamine were purchased from Aladdin Chemistry Co. Ltd. Fetal blood serum (FBS), fibrinogen (FIB) and albumin (Alb) were provided by Lianxing Biotechnology Co. Ltd.Synthesis of partially sulfobetaine grafted PSI (PASB-SI)
N,N-Dimethylaminopropylamine (510 mg, 5 mmol) was added to the DMF (10 mL) solution of PSI (970 mg, 10 mmol), and reacted at 40 °C for 6 h. After that, 1,3-propane sultone (732 mg, 6 mmol) was added into the solution and reacted for another 12 h. PASB-SI was obtained by precipitation in diethyl ether.Synthesis of PASB-Hy
PASB-SI (2.09 g) was dissolved in H2O–DMSO (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10 mL). Hydrazine hydrate aqueous solution (4.06 g, 62.5 mmol) was added drop-wise to the continuously stirred solution and reacted at 25 °C for 4 h. The mixture was precipitated with excess diethyl ether and washed with acetone. The rough product was then dissolved in deionized water, transferred to a dialysis and purified by exhaustive dialysis against distilled water. The purified product was freeze-dried and kept at 4 °C.
1, 10 mL). Hydrazine hydrate aqueous solution (4.06 g, 62.5 mmol) was added drop-wise to the continuously stirred solution and reacted at 25 °C for 4 h. The mixture was precipitated with excess diethyl ether and washed with acetone. The rough product was then dissolved in deionized water, transferred to a dialysis and purified by exhaustive dialysis against distilled water. The purified product was freeze-dried and kept at 4 °C.
Synthesis of PASB-Al
PASB-SI (2.09 g) was dissolved in H2O–DMSO (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10 mL), and then 3-amino-1,2-propanediol (0.546 g, 6 mmol) was added drop-wise to the continuously stirred solution and reacted at 40 °C for 24 h. The sulfobetaine and 3-amino-1,2-propanediol modified PAsp (PASB-Di) was obtained by precipitation in diethyl ether. After that, PASB-Di (2.55 g) was dissolved in 10 mL water, and cooled to 0 °C. An excess amount of aqueous solution of NaIO4 (0.33 M, 12.5 equiv. per 1,2-diol group) was added and stirred for 30 min at 0 °C in the dark. The mixture was then dialyzed against deionized water overnight using dialysis membrane with a molecular weight cut-off of 7 kDa. The dialyzed solution was freeze-dried to give a pale yellow powder.
1, 10 mL), and then 3-amino-1,2-propanediol (0.546 g, 6 mmol) was added drop-wise to the continuously stirred solution and reacted at 40 °C for 24 h. The sulfobetaine and 3-amino-1,2-propanediol modified PAsp (PASB-Di) was obtained by precipitation in diethyl ether. After that, PASB-Di (2.55 g) was dissolved in 10 mL water, and cooled to 0 °C. An excess amount of aqueous solution of NaIO4 (0.33 M, 12.5 equiv. per 1,2-diol group) was added and stirred for 30 min at 0 °C in the dark. The mixture was then dialyzed against deionized water overnight using dialysis membrane with a molecular weight cut-off of 7 kDa. The dialyzed solution was freeze-dried to give a pale yellow powder.
Preparation of PAsp nanogel
Two polymer precursor solutions were prepared by dissolving PASB-Hy or PASB-Al in PBS solution (pH 7.4), with concentration of 2.5, 5, 10 mg mL−1. The PAsp nanogels were obtained by mixing the two zwitterionic polymer precursor solutions with the same concentration in stirring for 24 h. So, the polymer concentrations of PAsp nanogels is 2.5, 5, 10 mg mL−1, respectively. Then the obtained solutions were dialyzed in 12–14 kDa cut off membrane against water for 48 h.1H NMR characterization
1H NMR spectra were recorded on a Varian UNITY-plus 400 spectrometer using D2O as the solvent. Chemical shifts were reported in ppm. For the characterization of the PAsp nanogels, the obtained PAsp nanogel solution was first dialyzed overnight in 12–14 kDa cut off membrane and then freeze-dried for further measurements such as 1H NMR and FT-IR.Fourier transformed infrared (FT-IR)
FT-IR spectra of PSI, PASB-Al, PASB-Hy and PAsp nanogel were recorded with FT-IR spectrometer model FTS 6000 (BIO-RAD, USA) against a blank KBr pellet background. A total of 64 scans were performed at a resolution of 4 cm−1.Molecular weight measurement
The molecular weight and distribution of PASB-Hy and PASB-Al were determined by GPC (Waters 2414 system Milford, MA) at 35 °C. The average weights were calibrated with standard PEG samples.Dynamic light scattering (DLS)
The hydrodynamic size of the nanogels was analyzed with DLS using a Zetasizer Nano ZS90 (Malvern Instruments, Southborough, MA). The scattering angle was fixed at 90°, and the measurements were recorded at a constant temperature (25 °C). Every measurement was repeated three times.Zeta potential measurement
The zeta potentials of PAsp nanogel to pH values were recorded using a Zetasizer Nano ZS90 (Malvern Instruments, Southborough, MA). Before measurement, the pH of PAsp nanogel solution was adjusted with 1 M HCl and then kept stirring for 1 h. The zeta potential was calculated using the Helmholtz–Smoluchowski equation.Transmission Electron Microscopy (TEM)
The morphology of nanogels was characterized by TEM. The digital images were recorded on a JEOL microscope (Japan). A specimen suspension under different conditions was prepared as below: a drop of the suspension was placed onto a 200 mesh copper grid, and then dried at the corresponding temperature in a vacuum drying chamber for preparing TEM samples.Drug loading and in vitro drug release
DOX·HCl solution (10 mg mL−1, 1 mL) was added into the PBS (pH 7.4) solution of PASB-Hy (6 mL, 2.5 mg mL−1). Then, the PBS (pH 7.4) solution of PASB-Al (6 mL, 2.5 mg mL−1) was added and vigorous stirred for 24 h. The mixed solution was dialyzed in 7 kDa cut off dialyzing membrane against PBS (pH 7.4) solution to remove the free DOX. The dialyzed solution was freeze-dried to obtain the DOX-loaded nanogel particles. The DOX contents in the nanogels were measured using a UV-visible spectrophotometer by the absorbance at 485 nm. The drug loading efficiency (LE%) and entrapment efficiency (EE%) were calculated by the following equations:| LE% = We/Po × 100% |
| EE% = We/Wo × 100% |
In which Wo, We, and Po are the amount of DOX in feed, capsulated drug and weight of the nanogels with drug, respectively.
To evaluate the drug release of the obtained DOX-loaded nanogels at different pH values, the DOX-loaded nanogel particles were first dispersed in PBS solution with a concentration of 2 mg mL−1 and then sealed in dialysis membrane tubes with a molecular weight cut-off of 7 kDa and were immersed in 10 mL of buffers (PBS buffer at pH 7.4 and acetate buffer at pH 5.0). The mixtures were shaking at 37 °C in the dark. At predetermined intervals, 4 mL of release medium was collected for testing and replaced by an equal volume of fresh buffer. The concentration of released DOX was quantified by a UV spectrophotometer at 485 nm. The cumulative DOX release was calculated as:
Cytotoxicity test
The cytotoxicities of PASB-Al, PASB-Hy and PAsp nanogel were assessed to NIH-3T3 and cells by MTT assay. NIH-3T3 cells were seeded in a 96-well plate at an initial density of 2 × 104 cells per well in DMEM complete medium. After 24 h of culture, the original medium was replaced with fresh DMEM medium. Then the copolymer and the PAsp nanogel solutions, were added to the medium at concentrations ranging from 0 to 500 μg mL−1 (0, 50, 100, 250, and 500 μg mL−1). Each dosage was replicated in 6 wells. After 48 h of further incubation, the DMEM was removed and 100 μL of MTT solution were added to each well and then replaced with 200 μL DMSO after 4 h. The absorbance value was measured using a microplate reader (Labsystem, Multiskan, Ascent, Model 354 Finland) at a wavelength of 570 nm, and the percentage of cell viability was measured relative to the negative control (media alone). Six replicates were counted for each sample. The mean values were used as the final data.MTT assay was also used to evaluate the cytotoxicity of the PAsp nanogel and the DOX-loaded nanogel to HepG2 cells. The process was the same as above. The cytotoxicity of PAsp nanogels were assessed to HepG2 without loading DOX with concentrations ranging from 0 to 500 μg mL−1. The cytotoxicity of DOX-loaded nanogel and free DOX were assessed to HepG2 with DOX concentrations ranging from 0 to 20 μg mL−1.
Intracellular drug release tests
Inverted fluorescence microscopy was employed to examine the intracellular drug release from the micelles. HepG2 cells were seeded in a culture plate at a density of 2 × 104 cells per well and cultured for 48 h. Then the cells were incubated with DOX-loaded nanoparticles. After 8 h, 24 h of incubation, the medium was removed and washed with PBS three times. Then cells were fixed by 4% formaldehyde solution. For nuclear staining, the cells were incubated with Hoechst for 10 min at room temperature, followed by washing with PBS. The intracellular localization of DOX from different samples was observed using an IX81-ZDC focus drift compensating microscope (Olympus, Tokyo, Japan).Results and discussion
Synthesis and characterization of copolymer precursors
PSI, the thermal polycondensation product of L-aspartic acid, has been used for preparing various polyaspartamide derivatives. The synthesis routes of PASB-Hy (Mn = 2.5 kDa, Mw/Mn = 1.17, determined by GPC using phosphate buffered saline (0.1 M, pH 7.0) as the eluent) and PASB-Al (Mn = 2.6 kDa, Mw/Mn = 1.21, determined by GPC using H2O as eluent) are shown in Scheme 1. Sulfobetaine was introduced to the PSI by using N,N-dimethylaminopropylamine as the PSI ring-opening reagent and the subsequent ring-opening reacted with 1,3-propanesultone, giving a quaternary ammonium and sulfobetaine structure. Hydrazine and aldehyde groups were introduced to the PAsp by ring-opening reaction of PSI as reported before.30The structures of PASB-Hy and PASB-Al were confirmed by FT-IR (Fig. 1) and 1H NMR (Fig. 2). Fig. 1 shows the FT-IR spectra of PSI, PASB-Hy, PASB-Al and PAsp nanogel. The spectrum of PSI (Fig.1A) showed the characteristic absorption bands of an imide ring at 1795 and 1720 cm−1. In Fig. 1B and C, the disappearance of bands at 1795 and 1720 cm−1, and appearance of bands around 1675 cm−1 (amide I) and 1530 cm−1 (amide II) corresponding to the amide structure confirmed the complete ring opening reaction of PSI.
In Fig. 1B and C, the absorption peaks at 1039, 1194, 601 and 525 cm−1 are ascribed to the symmetric and the asymmetric stretching vibration of SO3−. In Fig. 2, the peaks at 3.67, 3.48, 2.96, 2.81 and 2.21 ppm are assigned to the methylene protons of sulfobetaine units. In addition, in Fig. 2, peak at 3.14 ppm is ascribed to the methyl protons of quaternary ammonium groups (–N(CH3)2). All these suggested that the sulfobetaine groups were successfully introduced into PAsp backbone. By comparing the peaks at 3.14 and 2.5–3.0 ppm in NMR spectra of PASB-Al and PASB-Hy, the degree of sulfobetaine grafting in two polymer precursors was calculated to be 48%.
In the FT-IR spectrum of PASB-Al, the peak at 1732 cm−1 is ascribed to the aldehyde stretch (Fig. 1C). However, in the 1H NMR spectrum of PASB-Al (Fig. 2b), almost no aldehyde resonance is detected. This is because that aldehyde is in equilibrium with its hydrated form, 1,1-diol structure, which shows the peak at 5.2 ppm.32 It is worth to note that sulfobetaine with the negative and positive charged groups has a strong interaction with water, which is much stronger than that between the aldehyde and water. As a result, the aldehyde signal is weaker compared with the protons on sulfobetaine in 1H NMR spectrum.
Preparation and characterization of PAsp nanogel
A series of PAsp nanogels were prepared by mixing PASB-Al and PASB-Hy solutions with the same concentration. In Fig. 1, it can be observed that the aldehyde stretch peak (1732 cm−1) in PASB-Al is disappeared in the FT-IR spectrum of PAsp nanogel (Fig. 1D). At the same time, compared with PASB-Hy and PASB-Al, the amide absorption peaks shift from 1675 and 1530 cm−1 to 1667 and 1536 cm−1 in the IR spectrum of PAsp nanogel, respectively. The IR spectra confirmed the hydrazide–aldehyde reaction and chemical linkages between PASB-Hy and PASB-Al. Besides, the disappearance of peak at 5.2 ppm (in Fig. 2b) in the 1H NMR spectrum of the nanogel (Fig. 2c) also proved the crossing reaction between the two polymeric precursors. What's more, the peak at 3.3 ppm (Fig. 2b) decreased drastically in Fig. 2c. This is because hydrazone is wrapped inside of the gels during the formation of crosslinking, resulting in limited movement.Fig. 3 shows the size and distribution of PAsp nanogels prepared with different polymer precursor concentrations. As the initial polymer concentration changed, the size of the PAsp nanogel was modulated in the range of 200 to 300 nm. The average sizes of nanogels were 205, 250 and 275 nm, when initial polymer concentrations were 2.5, 5 and 10 mg mL−1, respectively.
 | ||
| Fig. 3 Hydrodynamic diameters and distribution of nanogels prepared from different polymer concentration solutions. | ||
TEM was used to observe the morphology of PAsp nanogels. Take the nanogels prepared with 2.5 mg mL−1 polymer precursor solutions as an example, the TEM images are shown in Fig. 4. Being consistent with the DLS data, TEM images indicated a very good dispersion of nanogels as individual spherical particles. What's more, the size of the nanogel is around 150 nm in diameter, a little smaller than that got by DLS. The slight decrease of the size is caused by the collapse of the hydration layer on the surface of the nanogel particles.
Stability of PAsp nanogel
The disintegrating of nanoparticles caused by the dilution upon intravenous injection into the body is a big challenge of the current nanoparticles drug delivery system. To evaluate the stability of the obtained PAsp nanogel, FT-IR and DLS were used to track the structure and size of the nanogels as dilution. Fig.1E shows the FT-IR of PAsp nanogels by dilution to 0.05 mg mL−1. No absorption peaks were reappeared at 1732 cm−1, and the amide absorption peaks were observed at 1667 and 1536 cm−1. All these proved that the PAsp nanogel can keep the hydrazone crosslink and no particle disintegration during the dilution. Fig. 5a shows the size change as the PAsp nanogel solution dilution. As dilution, the hydrodynamic size increases due to the swelling of the nanogel. When the concentration is lower than 0.1 mg mL−1, as dilution, the hydrodynamic diameter of nanogel is stable at 400 nm instead of increasing.The long term stability of PAsp nanogels was tested in PBS solution at room temperature for three weeks. The hydrodynamic size was test by DLS and the data were shown in Fig.5b. The PAsp nanogels maintained the same mean diameter over three weeks. All these results indicated that the PAsp nanogels were stable in both dilution and in PBS solution for long term.
One of the largest obstacles for the intravenous administration of nanoparticles is nonspecific protein adsorption onto the particles from blood, which can result in nanoparticle aggregation, macrophage cellular uptake, clearance by immune system before the nanoparticles can reach their intended target.6 The stability of nanoparticles in blood is critical to the success of drug delivery. In order to achieve a prolonged blood circulation time, PEG is used to modify the nanoparticles to reduce the protein adsorption from blood.33 Zwitterionic amphiphilic copolymers and conjugates are reported to form much more stable particles than PEG counterparts, and can provide a much longer blood circulation time.34
The stability of zwitterionic PAsp nanogels in protein solutions was tested. DLS was used to track the size changes of the zwitterionic PAsp nanogels in FBS/PBS (10%), FIB/PBS (1 mg mL−1) and Alb/PBS (2 mg mL−1) solutions incubated at room temperature. FIB is the first adsorbed protein during the blood coagulation process. Effectively reducing the adsorption of fibrinogen can greatly inhibit the occurrence of the blood coagulation.35 After the nanogels were added, transparent protein solutions change to light blue because of the light scattering of the nanoparticles. As shown in Fig. 6, PAsp nanogels show excellent stability without an obvious size increase during a 12 h incubation period in FBS, FIB and Alb solutions. Moreover, the size distribution of the nanoparticles after incubation is similar to that before incubation (Fig. 6d–g). The excellent stability of zwitterionic PAsp nanogels in all three protein solutions indicates that the zwitterionic layer is highly effective to protect the nanogels against aggregation in complex physiological conditions.
The pH sensitivity of PAsp nanogel
Hydrazone is a kind of acid-cleavable linkage, which is stable at physiological pH and hydrolyzes under acidic conditions.28 As a result of this advantage, hydrazone has been extensive studied in medical biotechnology to develop pH-triggered controlled release system since cancer tissues and cells have a slightly acidic pH. The pH-sensitivity of the hydrazone cross-linked PAsp nanogels was investigated by monitoring size change of nanogels after pH value changed. The initial pH value of the obtained nanogel solution was 7.4. HCl solution was added to adjust the pH value to 4.0 as soon as possible. Then DLS measurement proceeded immediately. Fig. 7 shows the size change of PAsp nanogels as a function of time. The initial size of nanogels was around 200 nm. Once pH value dropped to 4.0, the size of PAsp nanogel changed rapidly. A new peak more than 600 nm appeared. As time went on, the initial small size peak decreased while the emerging big size peak enhanced and got bigger. After 8 min, only a little of small size particles (<5%) was observed at around 300 nm. After 12 min, no small size particle left, all particles were observed with hydrodynamic size around 1000 nm. Besides, the size distribution of the nanogel at pH 4.0 is broader than the initial nanogels at pH 7.4. In Fig.1F, it showed the FT-IR spectrum of PAsp nanogel dialyzed at pH 5.0 for 24 h. The adsorption peaks at 1732 cm−1 is ascribed to the aldehyde stretch. All these results indicate that the cleavage of hydrazone linkages at low pH values decreases the cross-link density of nanogels, induces the swelling and eventually disintegration of the nanogels. | ||
| Fig. 7 (a) The size change of PAsp nanogels as a function of time as the pH changed from 7.4 to 4.0; (b) zeta potential of PAsp nanogels in different pHs. | ||
To detect the surface charges of the PAsp nanogel, zeta potentials of zwitterionic nanogel in different pHs were measured. As shown in Fig. 7b, the zeta potential at pH 7.4 is −17 mV. As the surface of the cell was negatively charged, it is conducive to the stability of the PAsp nanogel when incubated in the body. As pH increasing, the zeta potential changes from positive to negative. At about 5.2, the net charge was zero. At pH 5.0, the zeta potential is +1.6 mV. The positively charged nanoparticle surface is readily taken up by tumor cells.
DOX loading and in vitro drug release
Nanogels are promising as drug delivery carriers because of their high loading capacity, high stability, and responsiveness to environmental factors, such as ionic strength, pH, and temperature.36 To evaluate the drug loading and release ability of this hydrazone cross-linked zwitterionic PAsp nanogel, DOX was chosen as a model drug to be loaded into the nanogels. The LE% and EE% were calculated to be 18% and 74%, respectively.The evaluation of drug release has been performed in different pH environments. The release profiles are reported in Fig. 8, and show the significant difference between the samples incubated at pH 7.4 and at 5.0. The DOX loaded nanogels showed an efficient release of DOX at pH 5.0, more than 70% of payload was released in 12 h. In contrast, the nanogels incubated at pH 7.4, only less than 25% of the drug was released within the same time. The release of drug from the nanogels is influenced both by the swelling of the matrix and diffusion of drugs. Ritger–Peppas equation37 was used to study the mechanism of the DOX release from the nanogels:
| Mt/M∞ = ktn |
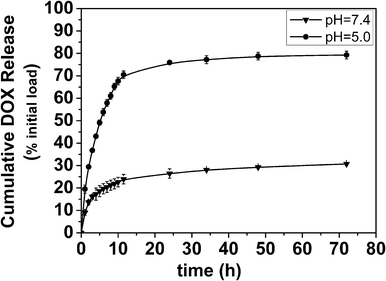 | ||
| Fig. 8 Cumulative drug release of DOX-loaded nanogels at pH 7.4 and 5.0. Data are presented as mean ± SD (n = 3). | ||
In vitro cytotoxicity
The in vitro cytotoxicity tests were performed using MTT assay to evaluate the biocompatibility of PASB-Hy, PASB-Al and the zwitterionic nanogel. The corresponding results are shown in Fig. 9a. The cytotoxicity to cells was evaluated at different concentrations ranging from 0 to 500 μg mL−1. The polymers or PAsp nanogels did not show significant cytotoxicity to NIH-3T3 cells (nanogel-1 in Fig. 9a) or HepG 2 cells (nanogel-2 in Fig. 9a) even at a high concentration (500 μg mL−1) after 48 h incubation. These results suggest that the polymer precursors and the nanogel have good compatibility, and have potential use in biomedical applications.The in vitro cytotoxicity of DOX-loaded nanogel particles against HepG2 cells in comparison with free DOX species determined by MTT assay is shown in Fig. 9b. The drug concentration ranges 0–20 μg mL−1, free DOX exhibited more cytotoxicity against cancer cells compared with the DOX loaded nanoparticles. The cytotoxicity of free DOX and the DOX-loaded nanoparticles to HepG2 cancer cells both increased with the increase of their dose. To verify the cellular uptake and intracellular drug release, fluorescence microscope was used to identify the location of the DOX molecules in cells. The fluorescence microscope images of HepG2 cells treated with the DOX-loaded nanogels after 8 and 24 h incubation are shown in Fig. 9c1 and c2, respectively. In first 8 h, the red fluorescence of DOX was observed in both cytoplasm and nucleus, and the nuclear regions apparently showed much higher fluorescence intensity. After 24 h incubation, the intensity of red fluorescence was much stronger than that after 8 h incubation. Most of the released DOX was found to be located in the nucleus of HepG2 cells, or near the nuclear membranes. The results suggested that DOX loaded in nanogels could be successfully released out and delivered into the nucleus of tumor cells. Different from the free DOX molecules could easily be transported into cancer cells by passive diffusion, for DOX-loaded nanogels, the drug could only be released after the particles are endocytosed into the cells, thereby postponing the nuclear uptake in cancer cells.
Conclusions
In this study, a zwitterionic polypeptide nanogel drug carrier was prepared via hydrazone crosslinking in PBS solution, which exhibited good biocompatibility, stability, and pH-responsive drug release. The nanogel developed in this work could potentially load other types of hydrophilic drugs, such as proteins, DNA, or siRNA, or imaging nanoparticles, such as water soluble quantum dots or gold nanoparticles. Besides, the zwitterionic polypeptide surfaces are easier to immobilize bio-recognition elements for targeting specific disease areas, which is difficult for PEG surfaces.Acknowledgements
This work was funded by NSFC (51203079), the Natural Science Foundation of Tianjin (14JCYBJC18100), PCSIRT (IRT1257), and NFFTBS (J1103306).Notes and references
- T. Zhou, C. Xiao, J. Fan, S. Chen, J. Shen, W. Wu and S. Zhou, Acta Biomater., 2013, 9, 4546–4557 CrossRef CAS PubMed
.
- K. Zhu, T. Ye, J. Liu, Z. Peng, S. Xu, J. Lei, H. Deng and B. Li, Int. J. Pharm., 2013, 441, 721–727 CrossRef CAS PubMed
.
- C. W. Park, H.-M. Yang, H. J. Lee and J.-D. Kim, Soft Matter, 2013, 9, 1781–1788 RSC
.
- C. A. Hong, J. S. Kim, S. H. Lee, W. H. Kong, T. G. Park, H. Mok and Y. S. Nam, Adv. Funct. Mater., 2013, 23, 316–322 CrossRef CAS
.
- L. Yin, C. He, C. Huang, W. Zhu, X. Wang, Y. Xu and X. Qian, Chem. Commun., 2012, 48, 4486–4488 RSC
.
- K. Raemdonck, J. Demeester and S. De Smedt, Soft Matter, 2009, 5, 707–715 RSC
.
- D. A. Herold, K. Keil and D. E. Bruns, Biochem. Pharmacol., 1989, 38, 73–76 CrossRef CAS
.
- L. Y. Li, S. F. Chen, J. Zheng, B. D. Ratner and S. Y. Jiang, J. Phys. Chem. B, 2005, 109, 2934–2941 CrossRef CAS PubMed
.
- E. Ostuni, R. G. Chapman, R. E. Holmlin, S. Takayama and G. M. Whitesides, Langmuir, 2001, 17, 5605–5620 CrossRef CAS
.
- Y.-Y. Luk, M. Kato and M. Mrksich, Langmuir, 2000, 16, 9604–9608 CrossRef CAS
.
- N. V. Efremova, S. R. Sheth and D. E. Leckband, Langmuir, 2001, 17, 7628–7636 CrossRef CAS
.
- H. Hatakeyama, H. Akita, K. Kogure, M. Oishi, Y. Nagasaki, Y. Kihira, M. Ueno, H. Kobayashi, H. Kikuchi and H. Harashima, Gene Ther., 2007, 14, 68–77 CrossRef CAS PubMed
.
- B. Romberg, W. E. Hennink and G. Storm, Pharm. Res., 2008, 25, 55–71 CrossRef CAS PubMed
.
- H. Vaisocherova, W. Yang, Z. Zhang, Z. Cao, G. Cheng, M. Piliarik, J. Homola and S. Jiang, Anal. Chem., 2008, 80, 7894–7901 CrossRef CAS PubMed
.
- Z. Zhang, S. Chen and S. Jiang, Biomacromolecules, 2006, 7, 3311–3315 CrossRef CAS PubMed
.
- L. Zhang, H. Xue, Z. Cao, A. Keefe, J. Wang and S. Jiang, Biomaterials, 2011, 32, 4604–4608 CrossRef CAS PubMed
.
- W. Yang, J.-R. Ella-Menye, S. Liu, T. Bai, D. Wang, Q. Yu, Y. Li and S. Jiang, Langmuir, 2014, 30, 2522–2529 CrossRef CAS PubMed
.
- G. Jia, Z. Cao, H. Xue, Y. Xu and S. Jiang, Langmuir, 2009, 25, 3196–3199 CrossRef CAS PubMed
.
- G. Cheng, L. Mi, Z. Cao, H. Xue, Q. Yu, L. Carr and S. Jiang, Langmuir, 2010, 26, 6883–6886 CrossRef CAS PubMed
.
- Z. Zhang, T. Chao, L. Liu, G. Cheng, B. D. Ratner and S. Jiang, J. Biomater. Sci., Polym. Ed., 2009, 20, 1845–1859 CrossRef CAS PubMed
.
- T.-B. Ren, W.-J. Xia, H.-Q. Dong and Y.-Y. Li, Polymer, 2011, 52, 3580–3586 CrossRef CAS PubMed
.
- Y. Wang, Y. Wang, G. Wu, Y. Fan and J. Ma, Colloids Surf., B, 2009, 68, 13–19 CrossRef CAS PubMed
.
- X. Wang, G. Wu, C. Lu, Y. Wang, Y. Fan, H. Gao and J. Ma, Colloids Surf., B, 2011, 86, 237–241 CrossRef CAS PubMed
.
- X. Gu, J. Wang, X. Liu, D. Zhao, Y. Wang, H. Gao and G. Wu, Soft Matter, 2013, 9, 7267–7273 RSC
.
- X. Wang, G. Wu, C. Lu, W. Zhao, Y. Wang, Y. Fan, H. Gao and J. Ma, Eur. J. Pharm. Sci., 2012, 47, 256–264 CrossRef CAS PubMed
.
- K. Seo and D. Kim, Acta Biomater., 2010, 6, 2157–2164 CrossRef CAS PubMed
.
- S. Chen, Z. Cao and S. Jiang, Biomaterials, 2009, 30, 5892–5896 CrossRef CAS PubMed
.
- Y. Bae and K. Kataoka, Adv. Drug Delivery Rev., 2009, 61, 768–784 CrossRef CAS PubMed
.
- L. Zhou, R. Cheng, H. Tao, S. Ma, W. Guo, F. Meng, H. Liu, Z. Liu and Z. Zhong, Biomacromolecules, 2011, 12, 1460–1467 CrossRef CAS PubMed
.
- C. Lu, X. Wang, G. Wu, J. Wang, Y. Wang, H. Gao and J. Ma, J. Biomed. Mater. Res., Part A, 2014, 102, 628–638 CrossRef PubMed
.
- X. Wang, G. Wu, C. Lu, W. Zhao, Y. Wang, Y. Fan, H. Gao and J. Ma, Eur. J. Pharm. Sci., 2012, 47, 256–264 CrossRef CAS PubMed
.
- D. A. Ossipov, K. Brännvall, K. Forsberg-Nilsson and J. Hilborn, J. Appl. Polym. Sci., 2007, 106, 60–70 CrossRef CAS
.
- H. Otsuka, Y. Nagasaki and K. Kataoka, Adv. Drug Delivery Rev., 2012, 64, 246–255 CrossRef PubMed
.
- A. J. Keefe and S. Jiang, Nat. Chem., 2012, 4, 59–63 CrossRef CAS PubMed
.
- C. Sperling, M. Fischer, M. F. Maitz and C. Werner, Biomaterials, 2009, 30, 4447–4456 CrossRef CAS PubMed
.
- A. V. Kabanov and S. V. Vinogradov, Angew. Chem., Int. Ed., 2009, 48, 5418–5429 CrossRef CAS PubMed
.
- P. L. Ritger and N. A. Peppas, J. Controlled Release, 1987, 5, 37–42 CrossRef CAS
.
| This journal is © The Royal Society of Chemistry 2014 |








