DOI:
10.1039/C4RA08730E
(Paper)
RSC Adv., 2015,
5, 1893-1901
Structure, dynamics and thermodynamics of single-file water under confinement: effects of polarizability of water molecules†
Received
15th August 2014
, Accepted 20th November 2014
First published on 24th November 2014
Abstract
Various structural, dynamic and thermodynamic properties of water molecules confined in single-wall carbon nanotubes (CNTs) are investigated using both polarizable and non-polarizable water models. The inclusion of polarizability quantitatively affects the nature of hydrogen bonding, which governs many properties of confined water molecules. Polarizable water leads to tighter hydrogen bonding and makes the distance between neighboring water molecules shorter than that for non-polarizable water. Stronger hydrogen bonding also decreases the rotational entropy and makes the diffusion constant smaller than in TIP3P and TIP3PM water models. The reorientational dynamics of the water molecules is governed by a jump mechanism, the barrier for the jump being highest for the polarizable water model. Our results highlight the role of polarizability in governing the dynamics of confined water and demonstrate that the inclusion of polarizability is necessary to obtain agreement with the results of ab initio simulations for the distributions of waiting and jump times. The SPC/E water model is found to predict various water properties in close agreement with the results of polarizable water models with much lower computational costs.
1 Introduction
The structure, dynamics and thermodynamics of water confined in the narrow channels of CNTs have been studied extensively in recent years.1–4 Most of the interest in CNT–water systems arises from their potential nano-technological applications,5,6 such as water desalination7 and nano-sensor.8 This system also provides a simple model to study the properties of water in various biological channels and pores. Classical Molecular Dynamics (MD) simulations have provided a great deal of information about the structure and dynamics of confined water molecules. The outcome of these simulations depends to a large extent on the quality of the force field used in the simulation. Due to the anomalous properties of water, no single water model is able to reproduce the physical properties of water over the whole range of thermodynamic states of interest. Many water models have hence been developed in recent years. These include models with three (TIP3P,9 SPC,10 SPC/E11), four (TIP4P,12 TIP4P-Ice,13 TIP4P-2005),14 and five (TIP5P,15 TIP5P-Ew16) coulombic interaction sites with rigid geometry and other models that incorporate the effects of polarizability (POL3,17 AMOEBA,18 POL5),19 fluctuating charges (TIP4P-Fq,20 QTPIE21) and flexible bond lengths and angles (F3C,22 SPCFw,23 TIP4PFw,24 BK325and GCPM26). Each of these water models has been optimized to reproduce only a few of the thermodynamic properties of water, usually at ambient temperature and pressure. To reduce the computational cost and for the sake of simplicity, most of the classical force fields are based on non-polarizable pair potentials. These additive force fields take care of polarizability in a mean-field way by scaling up the partial charges on the constituent atoms. In a further approximation, most of the water potentials involve orientation-independent Lennard-Jones (LJ) interactions and the classical force fields do not include LJ-type interactions for the hydrogen atoms. This is done in order to make the water potential independent of the combination rule, which helps in making the interaction potential transferable across various force fields. These approximations work quite well for homogenous systems such as bulk water. However, in inhomogeneous systems, such as those with interfaces, cavities and pores, the polarizability may play an important role, as shown by Dang et al.27 Pertsin et al.28 have demonstrated that the thermodynamic and structural properties of confined water molecules near a graphene surface are very sensitive to the range and orientation-dependence of the potential used in the simulation.
Most of the existing simulations of confined water molecules have employed non-polarizable and orientation-independent water models, such as TIP3P, SPC/E and TIP4P.1,29,30 Although there have been a few studies of water molecules confined in single-wall CNTs using flexible water models,31,32 polarizable water models33–35 and ab initio methods,36–38 most of these studies are limited to investigations of vibrational spectra, hydrogen bond dynamics, dipole moment of the water molecules and diffusion constants. There have been no systematic studies of how the polarizability and flexibility of water molecules and the anisotropic nature of the water potential affect the thermodynamic, dynamic and structural properties of confined water molecules. This situation demands a systematic comparison of the results for the dynamics and structure of confined water molecules that are obtained using different models of water including polarizability, as well as flexibility. In this study, we have computed various structural, dynamic and thermodynamic quantities for water molecules confined in narrow single-wall CNT channels using five different water models: TIP3P,9 modified TIP3P39 (hereafter referred to as TIP3PM), SPC/E,11 SPCFw,23 and POL3.17 TIP3P and SPC/E are three-site rigid water models; SPCFw is a flexible water model; POL3 is a polarizable water model, which includes the induced dipole moment along with the permanent one, while TIP3PM is a modified TIP3P model that includes LJ potential interaction sites for all three atoms. We have also considered the TIP3P water model in combination with a polarizable force field for the carbon atoms in the nanotube (hereafter referred to as TIP3PP). Studying this set of water models allows us to compare different approaches used in previous studies to model bulk water and also to assess the effects of polarizability and flexibility on the dynamics and thermodynamics of confined water molecules.
Entry of water molecules into carbon nanotube has been confirmed by experiments3 and simulations.1 When water molecules are confined in (6,6) CNTs of diameter 8.1 Å, they cannot pass each other, resulting in a single file arrangement. Due to this single file arrangement, each water molecule makes hydrogen bonds with only its two nearest-neighbor water molecules: oxygen acts as an acceptor and one of the hydrogen atoms of each of the neighboring molecules acts as a donor. This leads to a highly oriented hydrogen-bonded network2 inside the CNT. The position of the donor hydrogen is repeatedly exchanged during reorientational relaxation via angular jumps.4 This relaxation dynamics of confined water molecules is similar to that of bulk water molecules,40 except for the fact that the hydrogen atoms involved in the exchange belong to the same water molecule in the case of single-file water and to different neighboring water molecules in bulk water. In addition to this reorientational relaxation, confined water molecules exhibit another relaxation mode in which the dipole moment of the entire chain of molecules flips collectively on a timescale of nanoseconds, depending on the length of the CNT.2,41 This collective flipping is mediated by the propagation of a defect along the chain of water molecules. A defect is created at one end of the CNT and, as it moves along the CNT, the dipole moment of the water molecules it encounters gets flipped. The translational dynamics of confined water molecules is also significantly affected by confinement. The translational diffusion constant of water in (6,6) CNTs has been found to be roughly half of the bulk value42 in the TIP3P model. Interestingly, in spite of their single file arrangement, Fickian diffusion has been observed42 for the confined water molecules. Due to changes in the hydrogen bond network, the structural properties of the confined water molecules differ significantly from those in the bulk. Confined water molecules exhibit solid-like ordering even at room temperature. This ordering arises from the strong hydrogen bonding between neighboring water molecules inside the nanotube. Alexiadis et al.43 have systematically studied the density of water molecules inside CNTs of various diameters and shown that the density of water inside narrow CNTs is very different from that of bulk water. The thermodynamics of confined water has been investigated recently,44,45 using the two-phase thermodynamics (2PT) method.46,47 It has been demonstrated that water molecules have higher energy inside narrow CNTs, and the spontaneous entry of water molecules into CNTs has been attributed to the gain in the rotational entropy of the molecules inside CNTs. There is, however, a contradictory report48 in which the driving force for water entry was found to be a favorable energy transfer.
An important question that naturally arises in this context is whether the above mentioned properties of water molecules under confinement would be affected if the simulations were performed using different water models that include the polarizability and flexibility of water molecules, as well as the polarizability of the carbon atoms in the CNT. In this paper, we systematically study the effects of the inclusion of polarizability and flexibility on various properties of confined water, e.g. the diffusion constant, hydrogen bond lifetime and the free energy barrier for jump reorientation. We find that the inclusion of polarizability of both CNT and water is necessary in order to make accurate predictions of water properties – the inclusion of nanotube polarizability alone is not sufficient for capturing all the effects of polarizability.
2 Method
An armchair CNT with chirality index (6,6) and length of 24 unit cells (56 Å) was solvated in a bath in accordance with different water models (TIP3P, TIP3PM, SPC/E, SPCFw, POL3, approximately 7000 water molecules) and simulated using AMBER12 (ref. 49) with periodic boundary conditions applied along all three axes. Carbon atoms in the CNT were modeled as uncharged LJ particles (εC = 0.086 kcal mol−1, σ = 3.4 Å) and parameters were taken from the AMBER ff10 force-field (atom type CA). Equilibrium bond length between CA–CA atoms was 1.4 Å, equilibrium angle between CA–CA–CA was 2π/3 radians, while the corresponding spring constants were 938 kcal mol−1 Å−2 and 128 kcal mol−1 rad−2, respectively. To model the polarizable nanotube and POL3 water, the ff02·pol50 force field was used. This polarizable force field takes into account the effects of polarization by explicitly including a polarizable center for each atom to compute the non-additive polarization energy in the Hamiltonian. In the case of POL3, both carbon and water molecules were assigned polarizability, while for TIP3PP only carbon atoms were assigned polarizability, and this was used to compute the induced dipole moment and hence the modified energies. The non-additive potential energy is given by:| |
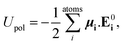 | (1) |
where E0i, the electric field at site i produced by the fixed charges in the system, is given by| |
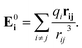 | (2) |
In eqn (1), μi is the induced dipole moment at site i, given by
where
Ei is the total electric field at atom
i and
αi is the atomic polarizability. To compute the total electric field
Ei, the following equation is used:
| |
 | (4) |
where
Tij is the dipole tensor, defined as
| |
 | (5) |
where
I is the unity tensor. The induced dipole moment is computed using a self-consistent iterative method from
eqn (3)–(5). Initial induced dipole moment for a given time step to start the iterative loop was interpolated from the previous step using a 2
nd order polynomial, and convergence criteria to stop self-consistent calculation was an induced dipole tolerance of 10
−5 Debye. This induced dipole moment was further used to compute the non-additive potential energy term from
eqn (1).
As TIP3PM uses a switching function that is different from that used in AMBER, we have used LAMMPS51 to simulate TIP3PM which allows us to use the same switching function as in CHARMM (pair style: lj/charmm/coul/long). Note that TIP3PM has non-zero LJ parameters for hydrogen as well, in contrast to all the other water models; this makes the van der Waals energy dependent on the orientation of the water molecules relative to the CNT. To make the comparison more exhaustive, we have also studied the SPCFw water model, a flexible water model in which the dipole moment can change due to fluctuating OH bond length and HOH angle. The properties of these water models have been compared with those of the most commonly studied non-polarizable water models, such as TIP3P and SPC/E. After an equilibration period of 2 ns in the NPT ensemble at T = 300 K and P = 1 atm, the coordinates and velocities of the water molecules obtained from a simulation in the NVT ensemble were used to analyze various structural and dynamic properties of the water molecules inside the CNT. Equations of motion were integrated using leap-frog algorithm, while temperature was kept constant using Berendsen weak coupling with a coupling constant of 2.0 ps. To study the reorientational dynamics of confined water molecules, since a typical angular jump time is of the order of femtoseconds, we used an integration time step of 0.5 fs and stored the coordinates after each integration step for a simulation time of 40 ps. Throughout the simulation, the nanotube was held fixed with its long axis along the z-axis. For statistical accuracy, 10 different sets of simulations were performed for each water model and averaged physical quantities were obtained from these runs.
3 Results
3.1 Structure
To examine the differences in the positional ordering of confined water molecules in the different water models studied here, we have computed the time-averaged pair correlation function along the nanotube axis, defined as:| |
 | (6) |
where zij is the distance between the centers of mass of the ith and jth water molecules along the nanotube axis and N is the total number of confined water molecules used to compute the pair correlation function.
As shown in Fig. 1, g(z) shows signatures of solid-like ordering of the water molecules inside the CNT, even at room temperature (300 K). Distinct peaks appear with a periodicity of 2.5 Å for all water models, except TIP3P for which the periodicity is 2.6 Å. This result shows that the effective intermolecular oxygen interaction between neighboring water molecules is relatively weak for TIP3P in comparison to the other water models studied here. Wu et al.23 have shown that the length of the O–H bonds affects the strength of the effective interaction between neighboring water molecules in the bulk: longer O–H bonds lead to stronger hydrogen bonding. This fact can be used to explain the increased intermolecular distance for confined water molecules observed here for TIP3P water, as it has a shorter O–H bond length (0.9572 Å) compared to the other water models, which have O–H bond lengths of 1.00 Å (Table 1). Interesting structural information emerges from the OH bond orientation with respect to the oxygen–oxygen bond of nearest-neighbor water molecules. The distribution of the OH bond tilt angle p(θ) has been plotted in Fig. 2. It is clear from this plot that each water molecule has two preferred orientations for the O–H bond: one at around 10° and the other at the angle complementary to the HOH angle. The OH bond in the SPC family of water models and POL3 is more inclined to the oxygen–oxygen axis, which is a reflection of tighter hydrogen bonding with adjacent water molecules as compared to the TIP3P family of water models. It is interesting to note that the structural arrangement of water molecules inside a polarizable CNT (TIP3PP model) is very similar to that of TIP3P water molecules, indicating that the inclusion of CNT polarizability does not alter the structural properties of the confined water molecules.
 |
| | Fig. 1 Axially projected pair correlation function g(z) of the confined water molecules inside a CNT. Distinct peaks represent solid-like arrangement of the water molecules. TIP3P shows the weakest hydrogen bonding among these water models. POL3 shows the highest degree of ordering followed by SPCFw. | |
 |
| | Fig. 2 Distribution of the angle that an OH bond makes with the vector joining the oxygen of a water molecule with the oxygen of the nearest water molecule. POL3, SPC/E and SPCFw models have more tilted OH bonds than the TIP3P family of water models. | |
Table 1 Force field (ff) parameters for the different water models studied in this work
| Model |
SPC/E |
SPCFw |
TIP3P |
TIP3PM |
POL3 |
| εOO (kJ mol−1) |
0.6491 |
0.6500 |
0.6358 |
0.6358 |
0.6521 |
| σOO (Å) |
3.1657 |
3.1660 |
3.1507 |
3.1507 |
3.2037 |
| εHH (kJ mol−1) |
0.0000 |
0.0000 |
0.0000 |
0.1923 |
0.0000 |
| σHH (Å) |
0.0000 |
0.0000 |
0.0000 |
0.4000 |
0.0000 |
| qO |
−0.8476 |
−0.8400 |
−0.8340 |
−0.8340 |
−0.7300 |
| qH |
0.4238 |
0.4100 |
0.4170 |
0.4170 |
0.3650 |
| μ (Debye) |
2.35 |
2.39 |
2.35 |
2.35 |
2.87 |
| OH (Å) |
1.00 |
1.00 |
0.9572 |
0.9572 |
1.01 |
| HOH (θ°) |
109.47 |
113.24 |
104.52 |
104.52 |
104.52 |
3.2 Dynamic properties
3.2.1 Translational diffusion. To compare the translational dynamics of the confined water molecules in different water models, we have computed the time-origin averaged mean-square displacement (MSD),| |
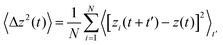 | (7) |
Here 〈…〉t represents an average over different time origins. As pointed out by Mukherjee et al.,42 MSD computation for confined water requires extensive averaging. 50 ps of MSD data was computed by averaging 20 separate 2.5 ns long blocks from a 50 ns long simulation trajectory and performing multiple time-origin averaging within each block. Note that the MSD of the confined water cannot be computed for arbitrarily long times, due to the limited duration of stay of the water molecules inside the nanotube – since the ends of the nanotube are open, the confined water molecules exchange their positions with the water molecules in the outside reservoir. The dependence of the MSD on time for different water models has been plotted in Fig. 3. In an earlier detailed study by Mukherjee et al.,42 it was demonstrated that water molecules inside short, open-ended (6,6) SWCNT exhibit normal Fickian diffusion. Assuming Fickian diffusion for all water models (see Fig. S1 in the ESI†), we compute the diffusion constant by linear fitting of MSD data. Although the nature of diffusion is the same for all water models studied here, the value of the diffusion constant differs substantially for different water models. Table 2 lists the diffusion constants for all the water models studied here, along with the diffusion constant values for bulk water. TIP3PP shows the highest value for the diffusion constant, followed by TIP3P and TIP3PM. Models in which the water molecules are flexible or polarizable show relatively low diffusion constants. It should be noted that the trend observed for the diffusion constants of confined water molecules is not the same as that observed for their bulk counterparts. These results show that polarizability has a significant effect on the translational motion of water in the presence of strong confinement. The dynamic properties of the water molecule may be affected by the choice of thermostat.52 To check the effect of the thermostat, we have calculated the diffusion constants of water molecules for the TIP3P model in both NVE and NVT ensembles. The diffusion constant of TIP3P water molecules inside a (6,6) SWCNT using the NVT ensemble is 2.38 × 10−5 cm2 s−1, which is quantitatively very similar to the value of 2.31 × 10−5 cm2 s−1 obtained in the micro-canonical (NVE) ensemble (see Fig. S2 in the ESI†).
 |
| | Fig. 3 MSD of water molecules along the nanotube axis. Water models in the TIP3P family have higher diffusion constants. Anisotropic interaction of water with the nanotube does not have a significant effect on the translational dynamics of water molecules, as TIP3P and TIP3PM have almost the same diffusion constant. SPC/E has the lowest diffusion constant. In the figure, points represent simulation data and straight lines are linear fits to the simulation data. For clarity, every 100th simulation data point has been shown. | |
Table 2 Translational diffusion constants for different water models. The diffusion constant was computed from the slope of the MSD vs. time plot
| Model |
Dconf (10−5 cm2 s−1) |
Dbulk (10−5 cm2 s−1) |
| SPCE |
0.78 |
2.49 |
| SPCFw |
1.19 |
2.32 |
| POL3 |
1.41 |
3.10 |
| TIP3P |
2.38 |
5.19 |
| TIP3PM |
2.18 |
4.20 |
| TIP3PP |
2.83 |
5.19 |
3.2.2 Hydrogen bond dynamics. Since many dynamic and thermodynamic properties of water depend on the nature of the hydrogen bonding, we have compared hydrogen bond dynamics of confined water molecules for different water models. Hydrogen bond dynamics can be characterized by the autocorrelation of the hydrogen bond population, given by:53| |
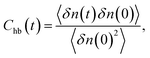 | (8) |
where δn(t) = n(t) − 〈n〉, and 〈n〉 is the average number of hydrogen bonds. Here, n(t) is the hydrogen bond descriptor, n(t) = 1 if a tagged water molecule is hydrogen bonded and n(t) = 0 otherwise. A pair is assumed to be hydrogen bonded if the distance between the two oxygen atoms is less than or equal to 3.5 Å and the angle between the vector joining the two oxygen atoms and one of the OH vectors of the donor is less than 30°. This correlation function has been plotted in Fig. 4. The hydrogen bond correlation decays exponentially with time for all the water models considered here. The time scale for this exponential decay is different for different models.
 |
| | Fig. 4 Temporal evolution of the hydrogen bond population autocorrelation function. The lines represent exponential fits. POL3 has the slowest decay, indicating the strongest hydrogen bond. | |
A measure of the decay time can be obtained by fitting Chb(t) to an exponential function. Fig. 4 suggests that the hydrogen bonding is very strong for POL3 water – the correlation function in this model decays over a longer time scale compared to the other models. Water models in the SPC family show a faster rearrangement of hydrogen bonds, but the rearrangement is slower than that in the TIP3P family. The values of the decay time τh, computed from single exponential fits of the hydrogen bond population autocorrelation function, are given in Table 3. We observe that the hydrogen bond life time for confined water is much longer compared to that for bulk water molecules, which is close to 2 ps. This slower decay can be attributed to stronger hydrogen bonding between confined water molecules. Polarization of only the CNT, in the case of TIP3PP, leads to faster decay of the autocorrelation function in comparison to TIP3P.
Table 3 Waiting time (τw), jump time (τj), jump angle θ(°), free-energy barrier (ΔF) for a typical hydrogen bond exchange, hydrogen bond lifetime τh, OH bond relaxation time τ2 and rotational entropy of confined water molecules for different water models. All calculations were performed at 300 K
| Model |
τw (fs) |
τj (fs) |
θ (°) |
ΔF (kcal mol−1) |
τh (fs) |
τ2 (fs) |
TSrot (kcal mol−1) |
| SPCE |
4068 |
71 |
94 |
2.27 |
5377 |
3946 |
1.55 |
| SPCFw |
3621 |
65 |
94 |
2.20 |
5217 |
3792 |
1.62 |
| POL3 |
4310 |
67 |
94 |
2.64 |
7127 |
5558 |
1.52 |
| TIP3P |
2347 |
128 |
84 |
1.35 |
2738 |
1416 |
1.69 |
| TIP3PM |
3101 |
126 |
86 |
1.65 |
2639 |
2019 |
1.65 |
| TIP3PP |
2250 |
128 |
85 |
1.42 |
2227 |
1314 |
1.73 |
3.2.3 Spectral density. The power spectrum of the velocity autocorrelation function computed in MD simulations (also called spectral density) is an important quantity that can be directly related to experimentally observable quantities, such as IR spectra and Raman spectra. The peaks in the spectral density can be associated with various hindered translational and vibrational modes. From MD simulation trajectories, we have computed the power spectrum using the Fourier transform of the velocity autocorrelation function. The spectral density of single-file water molecules confined in CNTs has been studied by Marti et al.32 using SPCFw water models. They observed a shift of low-frequency peaks and the emergence of an additional peak in the high frequency domain near 3400 cm−1. It was proposed that this additional peak appears due to splitting of the bulk water stretching mode into symmetric and asymmetric modes as an effect of confinement. Later, we44 studied the power spectrum of confined water molecules for the TIP3P rigid-water model. The power spectrum was decomposed into contributions coming from translational and rotational motion. The low-frequency modes were attributed to librational motion and hindered translational motion of the water molecules inside the nanotube. The presence of additional librational modes and restricted translational motion were further confirmed from the enhanced rotational entropy and suppressed translational entropy.Fig. 5 presents the results for the translational and rotational power spectrum for different water models. The position of the peak in the translational power spectrum is almost the same for all the water models, as shown in Table 4. The emergence of this peak is associated with the hindered translation of water molecules in the direction perpendicular to the nanotube axis. Since this motion is governed by the confinement potential produced by the nanotube, the translational power spectrum is very similar for different water models. The rotational power spectrum shows the emergence of distinct peaks near 150 cm−1 and 450 cm−1. The emergence of these peaks is associated with the librational motion of confined water molecules due to the presence of hydrogen atoms that do not participate in hydrogen bonding. The rotational spectrum of POL3 water has peaks at 156.4 cm−1 and 442.5 cm−1. These are blue shifted compared to the corresponding peaks in all the other water models studied here, again reflecting the presence of stronger hydrogen bonding for POL3 water.
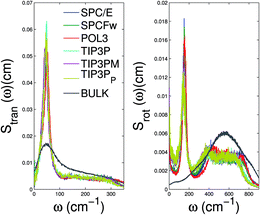 |
| | Fig. 5 (a) Translational and (b) rotational power spectra of confined water molecules. The translational power spectra are nearly the same for all the water models, while the rotational power spectra show the same trend as the hydrogen bond lifetime. The power spectra of bulk POL3 water are also shown for reference. | |
Table 4 Peak positions in translational and rotational spectra of single-file water confined in a (6,6) CNT, computed from the power spectrum of velocity autocorrelation functions. The rotational power spectrum has two peaks
| Model |
Translational (cm−1) |
Rotational (cm−1) |
| SPCE |
46.6 |
152.2 |
429.5 |
| SPCFw |
50.0 |
150.1 |
415.2 |
| POL3 |
53.5 |
156.4 |
442.7 |
| TIP3P |
49.0 |
148.2 |
392.4 |
| TIP3PM |
42.3 |
144.5 |
398.6 |
| TIP3PP |
49.1 |
147.4 |
396.7 |
3.2.4 Rotational dynamics. The hydrogen bond network of water molecules is known to govern many of their properties. For bulk water, it is known that the dynamics of this hydrogen bond network is governed by the rotation of water molecules. Mukherjee et al.4 have shown using classical MD simulations that the orientational relaxation of single-file water inside CNTs involve large amplitude angular jumps. It was shown that, during a successful jump, a bonded hydrogen atom exchanges its position with the other non-bonded hydrogen atom of the same water molecule (see Fig. 6). A typical reorientation event consists of a jump over a free energy barrier of a few kBT, separated by a jump angle of almost 90°. A similar analysis was performed here to investigate the reorientational dynamics of confined water molecules and all jumps were detected using the same procedure as that followed by Mukherjee et al.4
 |
| | Fig. 6 Illustration of the jump-mediated reorientational relaxation of single-file water molecules inside a CNT. The water molecule at the centre exhibits an angular jump: H1* is initially hydrogen bonded to O′ and at a later time, H2* is hydrogen bonded to O′. Figure adopted from ref. 4. | |
To characterize the reorientational dynamics of water in different water models, we have calculated the waiting time (the average time between two successive jumps), the jump time (the average time to complete a jump) and the free energy barrier for all the different water models. To compute these quantities, first we define two binary functions fH1(t) and fH2(t) that each take the value 1 or 0. The value of fH1(t) is equal to 1 if the hydrogen atom H1* (see Fig. 6) is hydrogen bonded with the nearest oxygen O′ and to 0 if it is free. fH2(t) is defined in a similar way for H2*. These functions were computed for each water molecule inside the CNT. A jump is considered to be successful when fH1(t) changes from 1 to 0 at some point of time and fH2(t) changes from 0 to 1 at a later time and vice versa. From this analysis, we find the starting time and the end time for each successful jump. These data are then used to compute the distribution of the waiting time τw between two successful jumps and the distribution of the time taken in completing a successful jump (jump time τj). The free energy barrier to complete a jump was computed from the logarithm of the probability distribution of the angles O′O*H1* and O′O*H2* (see Fig. 6):
| |
ΔF = −kBT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln[p(θO′O*H*)] ln[p(θO′O*H*)]
| (9) |
As shown in Fig. 7, the height of the free energy barrier (2.64 kcal mol−1) for POL3 is larger than that in the other water models studied here. The SPC/E and SPCFw water models have very similar barrier heights in the range of 2.27–2.20 kcal mol−1. TIP3P and TIP3PM have much lower free energy barriers of 1.35 kcal mol−1 and 1.65 kcal mol−1, respectively. The effect of this free energy barrier is also reflected in the waiting time between two jumps. POL3 has the highest barrier height and hence the longest waiting time of 4.3 ps, as shown in Table 3, and SPC/E and SPCFw have slightly shorter waiting times, as these models have lower free energy barriers. This enhanced barrier height can be attributed to the stronger hydrogen bonding for POL3 and SPC family of water models. This observation is consistent with the hydrogen bond population correlation analysis performed in the section on hydrogen bond dynamics. Larger OH bond lengths in SPC and POL3 models lead to stronger hydrogen bonding, and hence slower hydrogen bond population dynamics. From these observations, it can be concluded that the inclusion of the polarizability of the water molecules increases the barrier height for the jumps. Bankura et al.38 have studied the reorientational dynamics of single-file water confined in CNTs using ab initio dynamics and found a much longer waiting time compared to that obtained from classical MD simulations. This is consistent with our finding that the hydrogen bond dynamics become slower if polarizability is included. Our present work demonstrates that the inclusion of polarizability is necessary for an accurate description of the reorientational dynamics. The slowing down of the hydrogen bond dynamics in the presence of polarizability can be understood in terms of dipole–dipole interactions. Inside the nanotube, the dipoles of all water molecules point roughly in the same direction. They have the lowest interaction energy in the perfectly aligned state. The inclusion of polarizability leads to energetically more favorable configurations and higher barriers for reorientation. In the case of TIP3PP, where only the polarization of CNT is included, the height of the free-energy barrier for a jump remains almost the same as that for TIP3P, but the jump time decreases, leading to a shorter H-bond lifetime.
 |
| | Fig. 7 Free energy profile for the reorientation of water molecules in a single-file arrangement inside a (6,6) CNT. The quantity ΔF is computed as ΔF(θ) = −kBT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln[p(θ)], where p(θ) is the probability distribution of the angle θ between the OH vector and the vector joining the two oxygen atoms of adjacent water molecules that form a hydrogen bond (the angle O′O*H2* in Fig. 6). ln[p(θ)], where p(θ) is the probability distribution of the angle θ between the OH vector and the vector joining the two oxygen atoms of adjacent water molecules that form a hydrogen bond (the angle O′O*H2* in Fig. 6). | |
To further investigate the rotational dynamics of confined water, we have computed the more familiar time-dependent reorientational correlation function for the water molecules. This correlation function is defined as:
| |
 | (10) |
where
eiα can be any vector joining two points in the
ith molecule and
Pl is the
lth order Legendre polynomial. The reorientation time of a molecule can be obtained by defining an appropriate vector and fitting the corresponding correlation function to an exponential decay. The correlation function of most experimental interest for this system is obtained when we take
eiα to be the O–H vector of the
ith water molecule and compute the correlation function for
l = 2. This correlation function characterizes the reorientational relaxation of the O–H vector and hence the dynamics of the hydrogen bonds. This function has been plotted for various water models in
Fig. 8 and the relaxation time
τ2 has been computed by fitting its long-time decay to an exponential function. The values of the relaxation time have been listed in
Table 3. The observed behavior of the orientational correlation is consistent with the hydrogen bond dynamics and the free-energy barrier obtained earlier. TIP3P water exhibits faster relaxation, because reorientational jumps can occur more frequently, due to a lower free-energy barrier. In contrast, POL3 water exhibits the slowest relaxation, because it has the highest free-energy barrier for hydrogen bond dynamics.
 |
| | Fig. 8 Orientational correlation function C2 for the OH vector of confined water molecules. | |
3.3 Thermodynamics of confined water
Water molecules are found to spontaneously enter CNTs in both experiments and simulations. To understand the thermodynamics of water entry, it is necessary to evaluate the free energy of transfer which involves both the energy and the entropy of transfer. Using 2PT,46,47 we have shown in an earlier study44 that the rotational entropy of a water molecule is higher inside a CNT. The 2PT method computes the thermodynamic quantities of a fluid by decomposing its vibrational density of states, which can be obtained from the Fourier transformation of the velocity autocorrelation function, into solid-like and gas-like densities of states. Details of this method can be found in a paper by Lin et al.46 This method has been shown to yield entropy values for bulk water in excellent agreement with those from experiments over a large range of thermodynamic states.46 Similar agreement has also been found for several organic liquids.54–56 Using the same method, we have computed the entropy of transfer (ΔS) from bulk to confined water for various water models at T = 300 K. Absolute values of the rotational entropy of the confined water molecules are listed in Table 3. All the water models consistently show that the water molecules gain entropy inside the hydrophobic cavity of a CNT. Due to weaker hydrogen bonding, the TIP3P family of water models shows higher values of entropy while POL3 has the lowest rotational entropy. The absolute values of the rotational entropy are thus again consistent with the results for the reorientational dynamics studied in the previous section, where we found that TIP3P shows faster reorientational relaxation, while POL3 shows the slowest reorientational dynamics.
The energy of transfer (ΔE = Econf − Ebulk) can also be estimated from the difference between the average energy per water molecule inside the nanotube cavity and that in the bulk. We were not able to make a direct computation of the transfer of energy for POL3 water molecules, as the estimate of the polarization term for individual water molecules was not available. In the TIP3P family of models, the energy of water molecules increases by about 0.6 kcal mol−1 upon entering the CNT (see Table 5). However, for TIP3PP (TIP3P water inside a polarizable nanotube), the energy of transfer is 0.3 kcal mol−1 higher than that for TIP3P water inside a non-polarizable nanotube. This is consistent with the results of Schyman et al.,35 who found that water molecules have higher energy near a polarizable carbon surface, as compared to a non-polarizable one. The SPC/E family of water models shows a higher energy transfer, as well as a higher entropy transfer, as compared to the TIP3P family of models. In the calculation of the energy transfer, the contribution of the interaction of the water molecules with the carbon atoms of the CNT was divided equally between the water molecules and the CNT. Therefore, the change in the energy of the CNT has to be included in a calculation of the change in the total energy, as a water molecule is transferred from the bulk to the cavity inside a CNT. The free energy of transfer, ΔF = ΔE − TΔS, is found to be negative (−1.7 kcal mol−1 for the TIP3P model), if the change in the energy of the CNT is included in ΔE.
Table 5 The energy ΔE and entropy ΔS of transfer of water molecules, computed at T = 300 K. For the POL3 water model, we were not able to compute the value of the energy transfer due to lack of information about the polarization term in the energy of a water molecule
| Model |
ΔE (kcal mol−1) |
TΔS (kcal mol−1) |
| SPCE |
1.154 ± 0.18 |
1.031 ± 0.25 |
| SPCFw |
1.205 ± 0.18 |
1.066 ± 0.23 |
| POL3 |
na |
0.918 ± 0.30 |
| TIP3P |
0.561 ± 0.20 |
0.560 ± 0.30 |
| TIP3PM |
0.578 ± 0.20 |
0.504 ± 0.30 |
| TIP3PP |
0.884 ± 0.16 |
0.649 ± 0.30 |
4 Conclusions
Several structural, dynamic and thermodynamic properties of single-file water molecules confined in narrow CNTs have been analyzed using various water models. It has been shown that POL3 reproduces the hydrogen bond dynamics more accurately (as compared to ab initio simulations) than the other water models studied here. Hydrogen bond dynamics governs most of the structural and dynamic properties of the confined water molecules. These properties hence also follow the same trend in the accuracy of the results obtained for different water models. The POL3 water model leads to stronger hydrogen bonding, which is reflected in stronger translational order and shorter nearest-neighbor distance, longer hydrogen bond lifetime, larger values for the waiting time for angular jumps and the orientational relaxation time of the OH bond, and smaller rotational entropy of the confined water molecules. Both SPC and TIP3P families of models exhibit dynamic behavior characteristic of weaker hydrogen bonding (faster relaxation) compared to that in the POL3 model. The results obtained for the SPC family of models are closer to those for POL3 than the results for the TIP3P family of models. The inclusion of flexible bonds and angles does not improve the results for the dynamic behavior and sometime leads to results that are worse than those obtained for the SPC/E model. This is consistent with the findings of previous studies57,58 carried out for bulk water. The inclusion of anisotropic van der Waals interaction does not have any significant effect on the dynamics of the TIP3P water model: most of the features of the dynamics remain the same for TIP3P and TIP3PM. The inclusion of polarizability only for the CNT does not improve the accuracy of the calculation. The SPC/E model leads to results for dynamics that are similar to those obtained for the POL3 water model, but with much lower computational costs. Considering both accuracy and computational cost, based on our observations in this study, the SPC/E water model appears to be the optimum choice for the study of confined water molecules.
Acknowledgements
We acknowledge financial support from DST, India. HK would like to thank the University Grants commission, India, for a Senior Research Fellowship.
References
- G. Hummer, J. C. Rasaiah and J. P. Noworyta, Nature, 2001, 414, 188–190 CrossRef CAS PubMed.
- D. J. Mann and M. D. Halls, Phys. Rev. Lett., 2003, 90, 195503 CrossRef.
- A. I. Kolesnikov, J.-M. Zanotti, C.-K. Loong, P. Thiyagarajan, A. P. Moravsky, R. O. Loutfy and C. J. Burnham, Phys. Rev. Lett., 2004, 93, 035503 CrossRef.
- B. Mukherjee, P. K. Maiti, C. Dasgupta and A. K. Sood, J. Phys. Chem. B, 2009, 113, 10322–10330 CrossRef CAS PubMed.
- F. Fornasiero, H. G. Park, J. K. Holt, M. Stadermann, C. P. Grigoropoulos, A. Noy and O. Bakajin, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17250–17255 CrossRef CAS PubMed.
- M. Whitby, L. Cagnon, M. Thanou and N. Quirke, Nano Lett., 2008, 8, 2632–2637 CrossRef CAS PubMed.
- B. Corry, Energy Environ. Sci., 2011, 4, 751–759 CAS.
- S. Ghosh, A. K. Sood and N. Kumar, Science, 2003, 299, 1042–1044 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS PubMed.
- H. J. C. Berendsen, J. R. Grigera and T. P. Straatsma, J. Phys. Chem., 1987, 91, 6269–6271 CrossRef CAS.
- P. Mark and L. Nilsson, J. Phys. Chem. A, 2001, 105, 9954–9960 CrossRef CAS.
- W. L. Jorgensen and J. D. Madura, Mol. Phys., 1985, 56, 1381–1392 CrossRef CAS.
- J. L. F. Abascal, E. Sanz, R. G. Fernandez and C. Vega, J. Chem. Phys., 2005, 122, 234511–234519 CrossRef CAS PubMed.
- J. L. F. Abascal and C. Vega, J. Chem. Phys., 2005, 123, 234505–234512 CrossRef CAS PubMed.
- M. W. Mahoney and W. L. Jorgensen, J. Chem. Phys., 2000, 112, 8910–8922 CrossRef CAS PubMed.
- S. W. Rick, J. Chem. Phys., 2004, 120, 6085–6093 CrossRef CAS PubMed.
- J. W. Caldwell and P. A. Kollman, J. Phys. Chem., 1995, 99, 6208–6219 CrossRef CAS.
- P. Ren and J. W. Ponder, J. Phys. Chem. B, 2004, 108, 13427–13437 CrossRef CAS.
- H. A. Stern, F. Rittner, B. J. Berne and R. A. Friesner, J. Chem. Phys., 2001, 115, 2237–2251 CrossRef CAS PubMed.
- S. W. Rick, J. Chem. Phys., 2001, 114, 2276–2283 CrossRef CAS PubMed.
- J. Chen and T. J. Martinez, Chem. Phys. Lett., 2007, 438, 315–320 CrossRef CAS PubMed.
- M. Levitt, M. Hirshberg, R. Sharon, K. E. Laidig and V. Daggett, J. Phys. Chem. B, 1997, 101, 5051–5061 CrossRef CAS.
- Y. Wu, H. L. Tepper and G. A. Voth, J. Chem. Phys., 2006, 124, 024503–024512 CrossRef PubMed.
- C. P. Lawrence and J. L. Skinner, Chem. Phys. Lett., 2003, 372, 842–847 CrossRef CAS.
- P. T. Kiss and A. Baranyai, J. Chem. Phys., 2013, 138, 204507 CrossRef PubMed.
- P. Paricaud, M. Predota, A. A. Chialvo and P. T. Cummings, J. Chem. Phys., 2005, 122, 244511 CrossRef PubMed.
- L. X. Dang, H. V. R. Annapureddy, X. Q. Sun, P. K. Thallapally and B. P. McGrail, Chem. Phys. Lett., 2012, 551, 115–120 CrossRef CAS PubMed.
- A. Pertsin and M. Grunze, J. Phys. Chem. B, 2003, 108, 1357–1364 CrossRef.
- N. Giovambattista, P. J. Rossky and P. G. Debenedetti, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2006, 73, 041604 CrossRef.
- A. Alexiadis and S. Kassinos, Chem. Rev., 2008, 108, 5014–5034 CrossRef CAS PubMed.
- I. Gladich and M. Roeselova, Phys. Chem. Chem. Phys., 2012, 14, 11371–11385 RSC.
- J. Marti and M. C. Gordillo, J. Chem. Phys., 2001, 114, 10486–10492 CrossRef CAS PubMed.
- A. Alexiadis and S. Kassinos, Mol. Simul., 2008, 34, 671–678 CrossRef CAS.
- Y. Nakamura and T. Ohno, Mater. Chem. Phys., 2012, 132, 682–687 CrossRef CAS PubMed.
- P. Schyman and W. L. Jorgensen, J. Phys. Chem. Lett., 2013, 4, 468–474 CrossRef CAS PubMed.
- I. Hanasaki, A. Nakamura, T. Yonebayashi and S. Kawano, J. Phys.: Condens. Matter, 2008, 20, 015213 CrossRef.
- L. Wang, J. Zhao, F. Li, H. Fang and J. P. Lu, J. Phys. Chem. C, 2009, 113, 5368–5375 CAS.
- A. Bankura and A. Chandra, J. Phys. Chem. B, 2012, 116, 9744–9757 CrossRef CAS PubMed.
- A. D. MacKerell, D. Bashford, M. Bellott, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin and M. Karplus, J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef CAS PubMed.
- D. Laage and J. T. Hynes, Science, 2006, 311, 832–835 CrossRef CAS PubMed.
- B. Mukherjee, P. K. Maiti, C. Dasgupta and A. K. Sood, ACS Nano, 2008, 2, 1189–1196 CrossRef CAS PubMed.
- B. Mukherjee, P. K. Maiti, C. Dasgupta and A. K. Sood, J. Chem. Phys., 2007, 126, 124704–124708 CrossRef PubMed.
- A. Alexiadis and S. Kassinos, Chem. Eng. Sci., 2008, 63, 2047–2056 CrossRef CAS PubMed.
- H. Kumar, B. Mukherjee, S.-T. Lin, C. Dasgupta, A. K. Sood and P. K. Maiti, J. Chem. Phys., 2011, 134, 124105–124108 CrossRef PubMed.
- T. A. Pascal, W. A. Goddard and Y. Jung, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 11794–11798 CrossRef CAS PubMed.
- S.-T. Lin, P. K. Maiti and W. A. Goddard, J. Phys. Chem. B, 2010, 114, 8191–8198 CrossRef CAS PubMed.
- S.-T. Lin, M. Blanco and W. A. Goddard, J. Chem. Phys., 2003, 119, 11792–11805 CrossRef CAS PubMed.
- A. Waghe, J. C. Rasaiah and G. Hummer, J. Chem. Phys., 2012, 137, 044709 CrossRef PubMed.
- D. A. Case, T. E. Cheatham, T. Darden, H. Gohlke, R. Luo, K. M. Merz, A. Onufriev, C. Simmerling, B. Wang and R. J. Woods, J. Comput. Chem., 2005, 26, 1668–1688 CrossRef CAS PubMed.
- Z.-X. Wang, W. Zhang, C. Wu, H. Lei, P. Cieplak and Y. Duan, J. Comput. Chem., 2006, 27, 781–790 CrossRef CAS PubMed.
- S. Plimpton, J. Comput. Phys., 1995, 117, 1–19 CrossRef CAS.
- K. E. Gubbins, Y.-C. Liu, J. D. Moore and J. C. Palmer, Phys. Chem. Chem. Phys., 2011, 13, 58–85 RSC.
- A. Luzar and D. Chandler, Nature, 1996, 379, 55–57 CrossRef CAS.
- S. Mogurampelly and P. K. Maiti, J. Chem. Phys., 2013, 138, 034901 CrossRef PubMed.
- A. Debnath, K. G. Ayappa and P. K. Maiti, Phys. Rev. Lett., 2013, 110, 018303 CrossRef.
- S. G. Lee, T. A. Pascal, W. Koh, G. F. Brunello, W. A. Goddard and S. S. Jang, J. Phys. Chem. C, 2012, 116, 15974–15985 CAS.
- J.-L. Barrat and I. R. McDonald, Mol. Phys., 1990, 70, 535–539 CrossRef CAS.
- A. A. Chialvo and P. T. Cummings, Advances in Chemical Physics, John Wiley & Sons, Inc., 2007, pp. 115–205 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra08730e |
|
| This journal is © The Royal Society of Chemistry 2015 |
Click here to see how this site uses Cookies. View our privacy policy here. 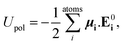
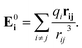



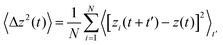
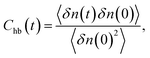


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln[p(θO′O*H*)]
ln[p(θO′O*H*)]

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln[p(θ)], where p(θ) is the probability distribution of the angle θ between the OH vector and the vector joining the two oxygen atoms of adjacent water molecules that form a hydrogen bond (the angle O′O*H2* in Fig. 6).
ln[p(θ)], where p(θ) is the probability distribution of the angle θ between the OH vector and the vector joining the two oxygen atoms of adjacent water molecules that form a hydrogen bond (the angle O′O*H2* in Fig. 6).





