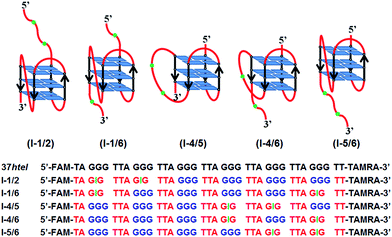
Folding and structural polymorphism of G-quadruplex formed from a long telomeric sequence containing six GGG tracts†
Atsushi Tanaka,
Jungkweon Choi‡
* and
Tetsuro Majima*
The Institute of Scientific and Industrial Research (SANKEN), Osaka University, Mihogaoka 8-1, Ibaraki, Osaka 567-0047, Japan. E-mail: jkchoi@ibs.re.kr; majima@sanken.osaka-u.ac.jp
First published on 20th October 2014
Abstract
The structure and stability of a G-quadruplex formed by a long human telomeric sequence containing five or more TTAGGG repeats are not clear yet. Using the guanine-to-inosine (G-to-I) substitution, we conducted thermodynamic studies on the structural polymorphisms of G-quadruplexes formed by the long telomeric sequences, 37htel and five G-to-I substituted sequences (I-1/2, I-1/6, I-4/5, I-4/6, and I-5/6), and investigated their folding dynamics at single-molecule level. The thermodynamic study reveals that a G-quadruplex formed from I-1/2 has a higher Tm and a larger ΔG298K than those formed by other G-to-I substituted sequences, suggesting that a long telomeric sequence preferentially forms a thermodynamically stable G-quadruplex at the 3′ end. In addition, from changes in the hydrodynamic radius by the formation of a G-quadruplex at single-molecule level, we found that the folding reaction of 37htel may proceed through a two-state mechanism without any detectable intermediate and that the global structure, which leads to the change in molecular size, is still occurring even after the formation of a secondary structure (G-quadruplex).
Introduction
A G-quadruplex is a four-stranded DNA formed by a guanine (G)-rich sequence in the presence of monovalent cations such as Na+ and K+ ions.1–6 Practically, the repetitive (TTAGGG)n overhang of the human telomeric sequence forms a G-quadruplex in vitro. The structure of a G-quadruplex consists of π–π stacking of planar G-tetrads, cyclically bound to each other through eight hydrogen bonds via Hoogsteen base pairs.1–8 Several research groups have revealed experimental evidence on the presence and functions of G-quadruplexes in genomes.2,9–11 However, the structure of G-quadruplex in living cells is not clear yet, and its presence in living cells is the subject of continuing debate.None the less, G-quadruplex has received great attention as a target for a therapeutic agent in anticancer treatments because of its biological ability to regulate the telomere elongation by telomerase in vivo.5,12 Furthermore, G-quadruplex is also considered as a promising material for nanotechnology, including nano-electronic devices because of its reversible conformational change and excellent hole trapping ability.13–15 In particular, the reversible conformational switching between single-stranded DNA (ssDNA) and G-quadruplex can serve as an electronic switch in a nano-electronic device. On the other hand, G-quadruplex-forming sequences such as human telomeric sequences have been utilized as a biosensor for K+ ions in vitro and in vivo because the G-quadruplex structure cannot be conserved in the absence of a monovalent ion, especially K+ ions.16–19 From these perspectives, theoretical and experimental studies on the structures and stabilities of human telomeric G-quadruplexes have been extensively conducted in vivo as well as in vitro, and their structures and biological functions have been highlighted.
Practically, numerous human telomeric sequences containing five or more TTAGGG repeats exist in human chromosomes. Nevertheless, most studies on the folding and structure of the G-quadruplex in the presence of K+ ions have been focused on the human telomeric sequence containing only four TTAGGG repeats (or four GGG tracts). Although the folding and structure of G-quadruplex formed by long telomeric sequences with five and more TTAGGG repeats have recently investigated by many research groups,20,21 their folding dynamics and structural polymorphisms are still unclear. In addition, theoretically a G-quadruplex can be formed anywhere along the long human telomeric sequence. However, information on the position where G-quadruplex is formed along the long telomeric sequence has been lacking. Furthermore, little is known about the structural polymorphism of G-quadruplexes formed by long telomeric sequences. To address this issue, we thoroughly investigated the folding process of a long G-rich sequence as well as the structural polymorphism of G-quadruplex formed by a long telomeric sequence with six GGG tracts using thermodynamic analysis and various spectroscopic techniques. In this study, we synthesized and used the dye-labeled human telomeric sequence (5′-FAM-TAGGG(TTAGGG)5-TT-TAMRA-3′, 37htel) and five guanine-to-inosine (G-to-I) substituted sequences (I-1/2, I-1/6, I-4/5, I-4/6, and I-5/6). (see Scheme 1) FAM (fluorescein) and TAMRA (tetramethylrhodamine) are attached to 5′- and 3′-end, respectively, as an energy donor (D) and an energy acceptor (A) for the fluorescence resonance energy transfer (FRET) experiment. (see Scheme 1) It is known that the substitution of inosine (I) at the middle G of an individual GGG tract can selectively induce the exclusion of GIG tract from the formation of the G-quartet core, resulting in the reduction of the multiplicity of G-quadruplex conformations.20 From the thermodynamic studies for six G-rich sequences, we clearly characterized the structures and stabilities of G-quadruplexes formed by long G-rich sequences in the presence of K+ ions. In addition, we demonstrated the folding process of a long telomeric sequence with six GGG tracts in terms of the change of the hydrodynamic radius of DNA at the single-molecule level.
Results and discussion
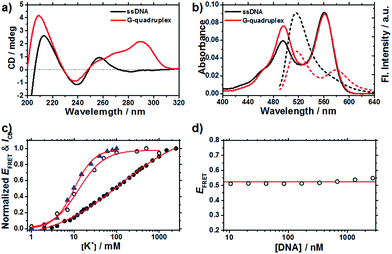
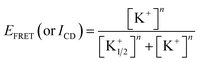
Fig. 1a shows the CD spectra of 37htel in the absence and presence of 100 mM K+ ions in 10 mM Tris-HCl buffer solutions (pH 7.4). The formation of G-quadruplex in the presence of 100 mM K+ ion was confirmed by its characteristic CD spectrum, producing a positive band at 290 nm with a shoulder at 265 nm and a negative band at 236 nm (Fig. 1a), suggesting that the G-quadruplex has a mainly antiparallel/parallel hybrid (3 + 1) structure. This result, which is consistent with previous reports,22,23 also implies that the formation and structure of G-quadruplex upon the addition of K+ ions is unaffected by the covalent attachment of dyes. To further understand the formation of the G-quadruplex structure for 37htel, we measured the absorption and emission spectra of 37htel in the absence and presence of K+ ions under the same experimental conditions. (Fig. 1b) Although the absorbance of FAM at around 490 nm is slightly enhanced by the increase of the ionic strength upon addition of 100 mM K+ ions,24 the absorption spectrum of dyes (D and A) attached to the G-quadruplex structure is similar to that in single-stranded DNA (ssDNA). On the other hand, the fluorescence intensity (ID) of the donor (FAM) attached to 37htel is significantly quenched by the addition of K+ ions, whereas the fluorescence intensity (IA) of the acceptor (TAMRA) is enhanced. (see Fig. 1b and S2†) The changes in ID and IA with the addition of K+ ions are due to the conformational change from ssDNA to G-quadruplex structure, resulting in the efficient energy transfer from D to A due to close proximity.To elucidate the folding reaction of a long telomeric G-quadruplex-forming sequence, we investigated the formation of the G-quadruplex structure for 37htel using the variation of the FRET efficiency (EFRET = IA/(ID + IA)) as a function of [K+] and then compared with the formation of the G-quadruplex structure for 25htel, which has a four TTAGGG repeats (5′-FAM-TAGGG(TTAGGG)3-TT-TAMRA-3′). As shown in Fig. 1c, the transition curve of 37htel upon the addition of K+ ions reveals a broad feature compared to that for 25htel. The transition curves monitored by EFRET (or ICD) are fitted by eqn (1) (classical Hill equation):
 | (1) |
The n is the Hill coefficient and K+1/2 is the concentration of K+ ions at which half the G-quadruplex structures were formed. From the transition curve depicted in Fig. 1c, K+1/2 and n values for 37htel were determined to be 117.0 ± 21.7 mM and 0.7, respectively, while K+1/2 and n values for 25htel were determined as 10.1 ± 0.4 mM and 1.8, respectively. (Fig. S1, Table S1†) In contrast to the transition curve of 37htel monitored by EFRET, however, the transition curve of 37htel monitored by CD intensities is very similar to that of 25htel (K+1/2 = 14.4 ± 0.8 mM and n = 1.5). (see Fig. 2 and 3 and Table S1†) The EFRET has been frequently used to measure the distance between domains of a single biomolecule and to obtain the information about the conformational change of a biomolecule. Meanwhile, the CD signal is used to identify the secondary structure of G-quadruplex. Thus, differences in the K+1/2 and n values of 37htel determined by two parameters (ICD and EFRET) suggest that the secondary structure of the G-quadruplex is rapidly formed in the low concentration of K+ ions and then the additional conformational change involving the change in D-A distance takes place in the presence of a high concentration of K+ ions. However, we cannot rule out the possibility that the broad-response of 37htel for K+ ions monitored by EFRET is attributed to the G-quadruplex intermolecular interaction at high concentrations of K+ ions. To test this possibility, we measured the concentration dependence on EFRET of 37htel in the presence of 1 M K+ ions. As shown in Fig. 1d, no concentration dependence on EFRET of 37htel was observed, indicating that the broad-response of 37htel for K+ ions is probably due to the formation of G-quadruplex and then following an additional conformational change rather than the G-quadruplex intermolecular interaction. The broad-response of 37htel for K+ ions will be further discussed later.
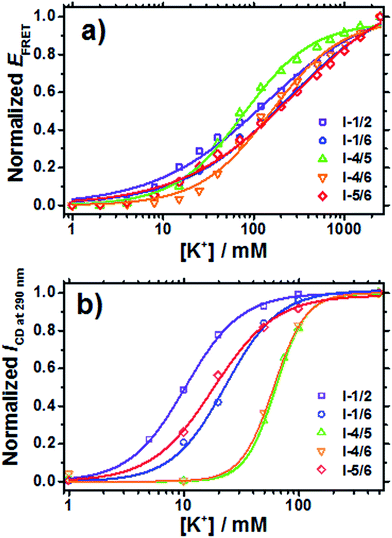 | ||
| Fig. 2 Changes in EFRET (a) and ICD (b) for 37htel, I-1/2, I-1/6, I-4/5, I-4/6, and I-5/6 as a function of [K+]. Theoretical curves obtained from the fitting analysis using eqn (1) are shown in solid lines. | ||
 | ||
| Fig. 3 (a) K+1/2 and (b) n values for 27htel, 37htel and five G-to-I substituted sequences (I-1/2, I-1/6, I-4/5, I-4/6, and I-5/6) determined by EFRET and CD intensities (ICD). | ||
As mentioned above, G-quadruplexes can theoretically be formed anywhere along a long G-rich sequence such as 37htel, resulting in the existence of a multi-species with a different G-quadruplex structure as depicted in Scheme 1. The existence of a multi-species can induce significant difficulty in the study on the structure and stability of G-quadruplexes formed by a long human telomeric sequence. To deal with this difficulty, recently Yue et al. used the G-to-I substituted sequence such as I-4/5.20 As a result, they clearly showed that the GIG tract replaced by inosine (I) at the middle G of individual GGG tract is excluded from the formation of G-quartet core.20 Substantially, their results showed that the single inosine substitution for d[TA-GGG(TTAGGG)nTT] (n = 5–7), which shows multiple G-quadruplex conformations in the presence of K+ ions, reduces the multiplicity of its conformation, and revealed that simultaneous inosine substitution in the fourth and fifth G-tracts of 37htel (I-4/5) led to the emergence of a predominant species (∼70%) with a long propeller loop.20 (see Scheme 1) To elucidate the stabilities and structures of several G-quadruplex structures formed by 37htel in the presence of K+ ions, we synthesized five G-to-I substituted sequences (I-1/2, I-1/6, I-4/5, I-4/6, and I-5/6) and investigated the stability and structure of G-quadruplex formed by each oligonucleotide. As depicted in Fig. S3,† G-quadruplexes formed by five G-to-I substituted sequences show similar CD signals; two positive bands at 270 and 290 nm with a negative band at 240 nm. These results are consistent with that reported by Yue et al.,20 indicating that all G-quadruplexes formed from five G-to-I substituted sequences have a mainly antiparallel/parallel hybrid (3 + 1) structure depicted in Scheme 1. However, I-1/2, I-1/6 and I-5/6 form G-quadruplex with the shortest TTA loop, while I-4/6 and I-4/5 form G-quadruplex harboring one or two GIG tracts within a single loop.
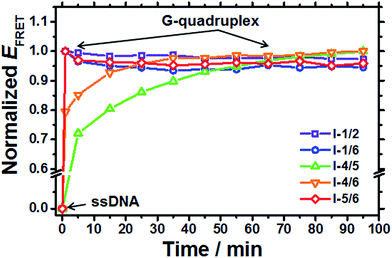
The variation in EFRET and ICD accompanied by the formation of G-quadruplexes for five G-to-I substituted sequences with various [K+] are measured. (see Fig. 2) From the quantitative analysis of transition curves obtained from the variation of the EFRET as a function of [K+], K+1/2 values for I-1/2, I-1/6, I-4/5, I-4/6, and I-5/6 were determined to be 132 mM (n = 0.7), 239 mM (n = 1.0), 77.5 mM (n = 1.1), 154 mM (n = 1.1), and 272 mM (n = 0.7), respectively (Order for K+1/2 value: I-4/5 < I-4/6 < I-1/2 < I-1/6 < I-5/6). (Fig. 3 and Table S1†) Meanwhile, the transition curves of five G-to-I substituted sequences monitored by CD intensities (ICD) showed contrasting results as shown in Fig. 2b; that is, lower K+1/2 values and the higher n values. (Fig. 3 and Table S1†) It is interesting to note that the K+1/2 values determined from ICD were in the order I-1/2 < I-5/6 < I-1/6 < I-4/6 < I-4/5. These results imply that the formation of G-quadruplexes for I-1/2, I-1/6 and I-5/6 is faster than that for I-4/6 and I-4/5. As depicted in Scheme 1, G-quadruplexes formed by I-1/2, I-1/6 and I-5/6 have a short TTA loop compared to those formed by I-4/6 and I-4/5, implying that the loop length affects the formation of the G-quadruplex structure. Here, we measured the kinetic aspect for the formation of G-quadruplexes using changes in the EFRET monitored at different times after the addition of K+ ions. As shown in Fig. 4, the kinetic aspect for the formation of G-quadruplex triggered by the addition of K+ ions showed that the formation of G-quadruplex for I-1/2, I-1/6 and I-5/6 was faster than that for I-4/6 and I-4/5. The kinetic traces for I-4/6 and I-4/5 were reproduced with a single-exponential function and the rate constants for G-quadruplex formation for I-4/6 and I-4/5 were determined to be 1.3 ± 0.1 × 10−3 and 0.5 ± 0.02 × 10−3 s−1, respectively. Although we could not determine the rate constant for I-1/2, I-1/6 and I-5/6 with a short TTA loop, the data presented herein clearly reveals that the long G-rich sequence with five or more TTAGGG repeats preferentially forms G-quadruplexes with a short TTA loop.
To further elucidate the stability of G-quadruplex formed by each sequence, we measured Tm values of all the sequences at various [K+]. (Fig. 5 and S4†) As summarized in Table S2† and Fig. 5, Tm values of I-1/2, I-1/6 and I-5/6 determined at various [K+] are higher than those of I-4/5 and I-4/6, indicating that G-quadruplexes with a short TTA loop are more thermodynamically stable than those with a long loop. This result coincides with that reported by Koirala et al.21 Furthermore, the Gibbs free energy change (ΔG298K) for I-1/2, I-1/6 and I-5/6 at 25 °C in the presence of 120 mM K+ ions are larger than those measured for I-4/5 and I-4/6 (Fig. 5b), implying that the structural stability of G-quadruplex is decreased by the presence of a long loop. Indeed, this result agrees with the previous study that suggested the decrease in the stability of G-quadruplex with increasing a total loop length.25
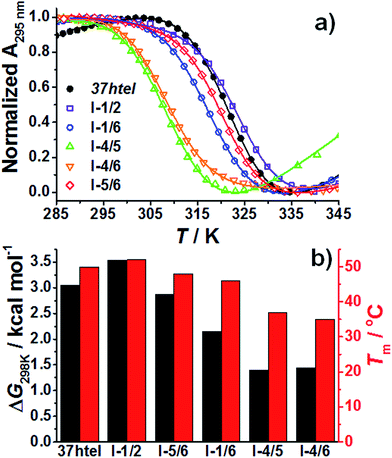 | ||
| Fig. 5 (a) Thermal denaturation curves monitored at 295 nm measured for 37htel, I-1/2, I-1/6, I-4/5, I-4/6, and I-5/6 in the presence of 120 mM K+ ions. ([DNA] = 5 μM). Theoretical curves obtained from the fitting analysis using eqn (S1)† are shown in solid lines. (b) Melting temperatures (Tm) and ΔG298K of 37htel, I-1/6, I-4/5, I-4/6, and I-5/6 determined in the presence of 120 mM K+ ions. | ||
Interestingly, as shown in Fig. 5b, Tm and ΔG values for I-1/2 determined from the melting curve are significantly higher and larger than other G-to-I sequences, implying that G-quadruplexes formed from I-1/2 is much more stable than those formed from other G-to-I sequences. This means that the thermodynamically most stable G-quadruplex structure formed from 37htel in the presence of K+ ions is very similar to that formed from I-1/2. Furthermore, as shown in Fig. 5b, the Tm and ΔG values for 37htel determined from the melting curve are close to those for I-1/2. Considering the G-quadruplex structure formed from I-1/2 (see Scheme 1), we suggest that in vitro, a long telomeric sequence containing five or more TTAGGG repeats mainly forms G-quadruplex structures at the 3′ end rather than the 5′ end or at internal positions. This coincides with the result reported by Tan and coworkers.26 They suggested that when an open 5′ end is not present, which mimics the telomere G-rich strand in vivo, the probability at the 3′ end is higher than at the 5′ end using DMA footprinting and exonuclease hydrolysis.26 In this study, moreover, we clearly show that from the thermodynamic analysis on G-quadruplex structure, the long telomeric sequence containing five or more TTAGGG repeats with both open 5′ and 3′ ends can form G-quadruplexes preferentially at the 3′ end rather than at 5′ end or at internal positions.
To further understand the folding dynamics of 37htel at single-molecule level, we measured the molecular diffusion time of 37htel as a function of the concentration of K+ ions using the fluorescence correlation spectroscopy (FCS). FCS is a very useful tool to measure the molecular diffusion time that can provide information on the change in molecular size associated with the conformational change of biomolecules such as DNA, protein, and so on.27–30 As depicted in Fig. 6a, all FCS curves show two dynamics; one is due to the fast relaxation process corresponding to singlet–triplet (S–T) relaxation of fluorescein and the other is attributed to the translational diffusion of each chemical species. Thus, the autocorrelation function can be expressed by.
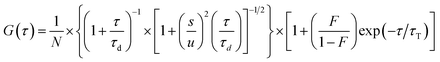 | (2) |
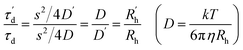 | (3) |
 | ||
| Fig. 6 (a) Representative FCS curves of 37htel measured with 0 (red), 0.01 (orange), 0.1 (yellow), 1 (green), 10 (olive), 100 (blue) and 1000 mM (purple) mM K+ ions. (b) Plot of changes in the molecular diffusion times (τd) of 37htel as a function of [K+]. The τd and τ′d are the molecular diffusion times in the absence and presence of K+ ions, respectively. The τd values are corrected for the increase of the solution viscosity upon addition of K+ ions. The inset is the log scale plot. Theoretical curves obtained from the fitting analysis using eqn (2) are shown in solid red lines. | ||
Fig. 6b shows changes in the molecular diffusion times (τd) of 37htel determined by the quantitative analysis of FCS curves measured at various [K+] with eqn (2). It has been suggested that a partially folded G-triplex is an intermediate in the folding reaction of the G-quadruplex, and can stably exist in the low concentration of K+ ions.21,31–36 The formation of a G-triplex in the low concentration of K+ ions, should induce a significant change in the τd of 37htel because the molecular size of G-triplex is significantly different to that of single-stranded DNA. As shown in Fig. 6b, however, 37htel shows the constant τd values in the range of 0–10 mM, suggesting that the G-triplex is not formed at low concentrations of K+ ions. As shown in Fig. 6b, the τd of 37htel is significantly decreased by the addition of ≥10 mM K+ ions and then shows constant τd values in the presence of a high concentration of K+ ions. As expressed in eqn (3), the τd is proportional to the Rh of a molecule. Thus, the decrease of τd upon the addition of K+ ions means the decrease of the Rh due to the formation of G-quadruplex. Our result is in contrast to the results reported by several previous studies, suggesting that a partially folded G-triplex is an intermediate in the folding reaction of G-quadruplex, and can stably exist in the low concentration of K+ ions.21,31–34,37 Recently, Koirala et al. revealed that human telomeric sequences containing four to seven TTAGGG repeats showed the conformational transition into G-quadruplex through a partially folded triplex as well as the direct conformational change from ssDNA to G-quadruplex.21 However, the folding mechanism of G-quadruplexes is still the subject of debate. Indeed, Koirala et al. could not observe the population of a triplex for hTelo-6, 5′-(TTAGGG)6TTA-3′,21 which is consistent with our result. The result reported by Koirala et al. indicates that not all human telomeric sequences containing four or more TTAGGG repeats are folded to G-quadruplex via a G-triplex.21 In addition, the folding (or unfolding) reaction of G-quadruplex studied by single-molecule techniques did not show the multi-step folding process that occurs with an intermediate such as a hairpin or triplex.38–40 The presented data herein is consistent with those obtained by several previous single-molecule experiments. Therefore, we suggest that in terms of DNA size change, the folding reaction of 37htel at single-molecule level occurs without any detectable intermediates, indicating two-state transition.
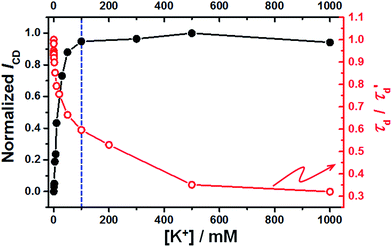
On the other hand, the K+1/2 value of 99 ± 9 mM for 37htel obtained from changes of τd is close to that determined by variation of the EFRET, but is larger than that determined by the variation of ICD. (Fig. 7) In addition, the transition region of the transition curve is wider than that obtained from the ICD. (see Fig. 7). As mentioned above, the change of τd reflects the change in molecular size associated with the global structural of a molecule, whereas the CD signal is sensitive to the secondary structure of G-quadruplex. Thus, the difference in K+1/2 values determined by τd and ICD can be interpreted in terms of the change in the kinetics for the secondary and the global structural change of a DNA sequence upon addition of K+ ions. In this respect, the smaller K+1/2 value for 37htel obtained from changes of ICD is probably due to the fast formation of a secondary structure such as G-quadruplex. The unexpected higher K+1/2 values for 37htel (or five G-to-I substituted sequences) determined by variation of the EFRET may stem from the slow global structural change occurring even after the formation of a secondary structure such as G-quadruplex. The global structural change, which results in the additional change of a molecular size, is due to the existence of a long loop as well as a long overhang at 5′ or 3′ ends.
Conclusions
In contrast to human telomeric sequences containing four TTAGGG repeats, relatively few studies have been done on the structure and stability of G-quadruplex formed from a long human telomeric sequence containing five or more TTAGGG repeats. Lots of human telomeric sequences containing five or more TTAGGG repeats exist in human chromosomes, and G-quadruplexes can be formed at different positions along the long telomeric sequence. Thus, it is very important to obtain information on the structure and stability of G-quadruplexes formed from a long human telomeric G-rich sequence. From this respect, we have thoroughly investigated the folding process of a long G-rich sequence as well as the structural polymorphism of G-quadruplex formed from long telomeric sequences with six GGG tracts using the thermodynamic analysis and various spectroscopic techniques. The results provided herein revealed that a long telomeric sequence containing five or more TTAGGG repeats forms G-quadruplex dominantly at the 3′ end rather than at the 5′ end or at internal positions. Furthermore, we show that from changes in the hydrodynamic radius associated by the formation of G-quadruplex at single-molecule level, the folding reaction of 37htel with six GGG tracts can be explained by two-state mechanism without any detectable intermediate. After the formation of a secondary structure (G-quadruplex), a long telomeric sequence containing five or more TTAGGG repeats shows a global structural change, resulting in the additional change in molecular size. We believe that the results provided herein will certainly contribute to understanding the structure and stability of G-quadruplexes.Experimental sections
Full experimental details and characterization of compounds can be found in the Supporting Information.FAM phosphoramidite, TAMRA CPG, and nucleotide phosphoramidite reagents were purchased from GLEN RESEARCH, Ltd. All oligonucleotides (25htel, 37htel, I-1/2, I-1/6, I-4/5, I-4/6 and I-5/6) studied here were synthesized using Applied Biosystems 3400 DNA synthesizer with standard solid-phase techniques and purified on a JASCO HPLC with a reversed phase C-18 column with an acetonitrile/50 mM ammonium formate gradient. The purified oligonucleotides were lyophilized for three times. Then, all DNA sequences were characterized by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra (Fig. S1†), and their concentrations were determined by the absorption of FAM labeled to each oligonucleotides.
The steady-state UV-visible absorption, fluorescence, and CD spectra were measured using Shimadzu UV-3100, Horiba FluoroMax-4 and JASCO CD-J720, respectively. To obtain the kinetic aspect for the formation of G-quadruplex at room temperature, the fluorescence spectra were measured at different times following addition of K+ ions using Horiba FluoroMax-4. The EFRET was determined by ID and IA obtained from each spectrum. (EFRET = IA/(ID + IA))
The melting temperature (Tm) was measured using a JASCO V-530. Fluorescence spectra were measured with an excitation wavelength of 485 nm. All sample solutions for the measurement of UV, fluorescence, CD spectrum were prepared with 10 mM Tris-HCl buffer (pH 7.4). The Tm was measured in 10 mM K+ phosphate buffer (pH 7.4). The thermodynamic parameters and Tm values were determined from the fitting analysis of melting curves using eqn (S1–S3†).
FCS experiments carried out with a time-resolved fluorescence microscope using confocal optics (MicroTime 200; PicoQuant, Berlin-Adlershof, Germany). We used 2 nM sample solutions for measuring FCS curves with various concentrations of K+ ion.
Acknowledgements
We thank the members of the Research Laboratory for Quantum Beam Science of ISIR, Osaka University for running the linear accelerator. This work has been partly supported by a Grant-in-Aid for Scientific Research (Project 24550188, 25220806, and 25288035) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japanese Government.Notes and references
- A. N. Lane, J. B. Chaires, R. D. Gray and J. O. Trent, Nucleic Acids Res., 2008, 36, 5482–5515 CrossRef CAS PubMed.
- H. J. Lipps and D. Rhodes, Trends Cell Biol., 2009, 19, 414–422 CrossRef CAS PubMed.
- J. L. Huppert, FEBS J., 2010, 277, 3452–3458 CrossRef CAS PubMed.
- J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893–5909 RSC.
- Y. Xu, Chem. Soc. Rev., 2011, 40, 2719–2740 RSC.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed.
- A. Gonnelli, M. G. Ortore, E. J. Baldassarri, G. P. Spada, S. Pieraccini, R. C. Perone, S. S. Funari and P. Mariani, J. Phys. Chem. B, 2013, 117, 1095–1103 CrossRef CAS PubMed.
- S. Lena, G. Brancolini, G. Gottarelli, P. Mariani, S. Masiero, A. Venturini, V. Palermo, O. Pandoli, S. Pieraccini, P. Samorì and G. P. Spada, Chem.–Eur. J., 2007, 13, 3757–3764 CrossRef CAS PubMed.
- A. Verma, V. K. Yadav, R. Basundra, A. Kumar and S. Chowdhury, Nucleic Acids Res., 2009, 37, 4194–4204 CrossRef CAS PubMed.
- D. Sun, K. Guo and Y. J. Shin, Nucleic Acids Res., 2011, 39, 1256–1265 CrossRef CAS PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- G. W. Collie and G. N. Parkinson, Chem. Soc. Rev., 2011, 40, 5867–5892 RSC.
- Y. C. Huang and D. Sen, J. Am. Chem. Soc., 2010, 132, 2663–2671 CrossRef CAS PubMed.
- S. Liu, X. Zhang, W. Luo, Z. Wang, X. Guo, M. L. Steigerwald and X. Fang, Angew. Chem., Int. Ed., 2011, 50, 2496–2502 CrossRef CAS PubMed.
- J. Choi, J. Park, A. Tanaka, M. J. Park, Y. J. Jang, M. Fujitsuka, S. K. Kim and T. Majima, Angew. Chem., Int. Ed., 2013, 52, 1134–1138 CrossRef CAS PubMed.
- Y. Hong, M. Haussler, J. W. Lam, Z. Li, K. K. Sin, Y. Dong, H. Tong, J. Liu, A. Qin, R. Renneberg and B. Z. Tang, Chem.–Eur. J., 2008, 14, 6428–6437 CrossRef CAS PubMed.
- C. K. Kwok, M. E. Sherlock and P. C. Bevilacqua, Angew. Chem., Int. Ed., 2013, 52, 683–686 CrossRef CAS PubMed.
- C. K. Kwok, M. E. Sherlock and P. C. Bevilacqua, Biochemistry, 2013, 52, 3019–3021 CrossRef CAS PubMed.
- Z. Chen, L. Chen, H. Ma, T. Zhou and X. Li, Biosens. Bioelectron., 2013, 48, 108–112 CrossRef CAS PubMed.
- D. J. Yue, K. W. Lim and A. T. Phan, J. Am. Chem. Soc., 2011, 133, 11462–11465 CrossRef CAS PubMed.
- D. Koirala, C. Ghimire, C. Bohrer, Y. Sannohe, H. Sugiyama and H. Mao, J. Am. Chem. Soc., 2013, 135, 2235–2241 CrossRef CAS PubMed.
- H. Q. Yu, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 15461–15468 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, K. N. Luu and D. J. Patel, Nucleic Acids Res., 2007, 35, 6517–6525 CrossRef CAS PubMed.
- R. Sjöback, J. Nygren and M. Kubista, Spectrochim. Acta, Part A, 1995, 51, L7–L21 CrossRef.
- A. Guedin, J. Gros, P. Alberti and J. L. Mergny, Nucleic Acids Res., 2010, 38, 7858–7868 CrossRef CAS PubMed.
- J. Tang, Z. Y. Kan, Y. Yao, Q. Wang, Y. H. Hao and Z. Tan, Nucleic Acids Res., 2008, 36, 1200–1208 CrossRef CAS PubMed.
- H. D. Kim, G. U. Nienhaus, T. Ha, J. W. Orr, J. R. Williamson and S. Chu, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4284–4289 CrossRef CAS PubMed.
- J. Choi, S. Kim, T. Tachikawa, M. Fujitsuka and T. Majima, J. Am. Chem. Soc., 2011, 133, 16146–16153 CrossRef CAS PubMed.
- A. P. Fields and A. E. Cohen, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 8937–8942 CrossRef CAS PubMed.
- A. Tanaka, J. Choi, S. K. Kim and T. Majima, J. Phys. Chem. B, 2013, 117, 6711–6717 CrossRef CAS PubMed.
- M. Bončina, J. Lah, I. Prislan and G. Vesnaver, J. Am. Chem. Soc., 2012, 134, 9657–9663 CrossRef PubMed.
- V. Limongelli, S. De Tito, L. Cerofolini, M. Fragai, B. Pagano, R. Trotta, S. Cosconati, L. Marinelli, E. Novellino, I. Bertini, A. Randazzo, C. Luchinat and M. Parrinello, Angew. Chem., Int. Ed., 2013, 52, 2269–2273 CrossRef CAS PubMed.
- W. Li, X. M. Hou, P. Y. Wang, X. G. Xi and M. Li, J. Am. Chem. Soc., 2013, 135, 6423–6426 CrossRef CAS PubMed.
- J. J. Green, L. Ying, D. Klenerman and S. Balasubramanian, J. Am. Chem. Soc., 2003, 125, 3763–3767 CrossRef CAS PubMed.
- M. Boncina, J. Lah, I. Prislan and G. Vesnaver, J. Am. Chem. Soc., 2012, 134, 9657–9663 CrossRef CAS PubMed.
- W. Li, X.-M. Hou, P.-Y. Wang, X.-G. Xi and M. Li, J. Am. Chem. Soc., 2013, 135, 6423–6426 CrossRef CAS PubMed.
- R. D. Gray, R. Buscaglia and J. B. Chaires, J. Am. Chem. Soc., 2012, 134, 16834–16844 CrossRef CAS PubMed.
- R. D. Gray and J. B. Chaires, Nucleic Acids Res., 2008, 36, 4191–4203 CrossRef CAS PubMed.
- M. H. Qureshi, S. Ray, A. L. Sewell, S. Basu and H. Balci, J. Phys. Chem. B, 2012, 116, 5588–5594 CrossRef CAS PubMed.
- X. Long, J. W. Parks, C. R. Bagshaw and M. D. Stone, Nucleic Acids Res., 2013, 41, 2746–2755 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and extra supporting data (Table S1 and S2 and Fig. S1–S4). See DOI: 10.1039/c4ra08053j |
| ‡ Present address: Center for Nanomaterials and Chemical Reactions, Institute for Basic Science (IBS), Daejeon 305-701, Republic of Korea. |
| This journal is © The Royal Society of Chemistry 2014 |