Shyamaprosad Goswami, Rinku Chakrabarty, Swapan Dey and Hoong-Kun Fun
RSC Adv., 2014,4, 49663-49671
DOI:
10.1039/C4RA06994C,
Paper
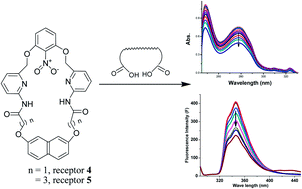
Five receptors (1–5) including two macrocyclic receptors have been designed and synthesized for the recognition of dicarboxylic acids. The receptors having an NO2 group show inhibition in hydrogen bonding molecular recognition towards the dicarboxylic acid and this effect becomes more prominent in the case of macrocyclic receptors 4 and 5 in CHCl3. The binding behavior of these receptors has been studied by UV-vis, fluorescence and 1 : 1 1H NMR binding studies.