DOI:
10.1039/C4RA06664B
(Paper)
RSC Adv., 2014,
4, 46157-46163
Unraveling the caspase-mediated mechanism for phloroglucinol-encapsulated starch biopolymer against the breast cancer cell line MDA-MB-231†
Received
4th July 2014
, Accepted 27th August 2014
First published on 27th August 2014
Abstract
The main objective of the study is to decipher the mechanism underlying the anticancer activity of phloroglucinol-encapsulated starch biopolymer against the breast cancer cell line MDA-MB-231. An MTT assay confirmed that MDA-MB-231 cells are highly susceptible to treatment with the biopolymer in a dose-dependent manner. Morphological evidence by fluorescence staining revealed chromatin condensation, nuclear beading and loss of mitochondrial membrane potential. DNA fragmentation and cell cycle analysis confirm the progression of apoptosis in the MDA-MB-231 breast cancer cell line. The semi-quantitative RT-PCR showed increased levels of pro-apoptotic genes such as those for caspase-3, 8 and 9. Western blotting analysis was done to substantiate the caspase-3 and 8 expressions in an efficient execution of apoptosis. To conclude, the controlled release of the polyphenolic compound phloroglucinol from starch induces cytotoxicity against the MDA-MB-231 breast cancer cell line.
Introduction
Recent scientific and engineering advances have improved the biological activities of several pharmaceuticals. However, currently used conventional drugs are generally immediately released into the body,1 and such release can have unintended side effects. These issues can be minimized and superior therapeutic efficiency can be achieved by controlled release systems.2 To achieve this, numerous polymeric materials have been employed that control the release of manufactured drugs. The use of starch has in particular become increasingly prevalent due to its non-toxic, eco-friendly and biocompatible properties, unlike those of other, i.e., synthetic, chemicals.3 The presence of numerous hydroxyl groups in starch can anchor a wide variety of naturally found as well as manufactured chemicals.
Polyphenolic compounds are secondary metabolites found in organisms ranging from simple prokaryotes to complex eukaryotes.4 These compounds have several aromatic rings bearing one or more hydroxyl groups that, because of their anchoring properties mentioned above, have made these polyphenols the subjects of many investigations. During extraction from the natural sources, these compounds degrade and require special storage conditions.5 To circumvent these drawbacks, better encapsulation of the polyphenolic compounds could be employed to protect their structural integrity and biological activity. With this in mind, we recently carried out the encapsulation of the polyphenolic compound phloroglucinol by soluble starch in the form of a biopolymer.6
Phloroglucinols are naturally occurring polyphenolic compounds found abundantly in brown seaweeds.7 The enormous biological and pharmaceutical importance of phloroglucinols has led several researchers to synthesize various derivatives of this compound.8 Compared to bulk material, however, the effectiveness of a phloroglucinol compound would be increased by its targeted release into the biological system. It has indeed been shown that, compared to phloroglucinol alone, phloroglucinol-encapsulated starch biopolymer possesses improved antioxidant and cytotoxic effect against the HepG2 hepatocellular carcinoma cancer cell line.6 However, the exact mechanism by which this encapsulated biopolymer is involved in progression to cell death is unclear. Meanwhile, breast cancer is the second leading cancer to cause death in women. Keeping this in mind, the present study was carried out to unravel the molecular mechanisms by which the phloroglucinol-encapsulated starch biopolymer kills cells of the breast cancer cell line MDA-MB-231.
Materials and methods
Chemicals and reagents
Phloroglucinol, methylthiazolyldiphenyl-tetrazolium bromide, acridine orange, ethidium bromide, Hoechst 33258, agarose (low & high), propidium iodide, and JC-1 were purchased from Sigma Aldrich (St. Louis, MO, USA). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and antibiotics (streptomycin & gentamicin) were procured from HiMedia Laboratories, India. Gene specific primers for β-actin and caspases-3, 8 & 9 were obtained from Eurofins, India. Trizol reagent (Invitrogen, USA), Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA), polyvinyl difluoride (PVDF) membrane (Millipore, Billerica, MA, USA), and an enhanced chemiluminescence blotting detection kit (Perkin Elmer Life Sciences, Inc., Boston, MA, USA) were also used in this study. Rabbit monoclonal antibody for caspases-3 & 8 were procured from Abcam, USA and β-actin used in the study was obtained from Santa Cruz (CA, USA).
Preparation of phloroglucinol-encapsulated starch biopolymer
Phloroglucinol-encapsulated starch biopolymer (Fig. 1) was prepared as previously described.6 The loading efficiency of phloroglucinol into the soluble starch was determined spectrophotometrically at 267 nm to be 97%. The Proton Nuclear Magnetic Resonance (1H NMR) data exhibited by the biopolymer was comparable with the literature values obtained earlier.
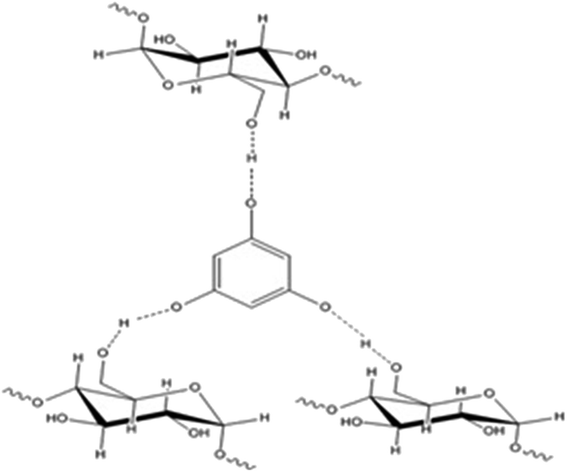 |
| | Fig. 1 Phloroglucinol-encapsulated starch biopolymer. | |
Cell culture and growth
The MDA-MB-231 breast cancer cell line was obtained from the National Centre for Cell Science (NCCS), Pune, India. The cells were grown in Dulbecco's modified Eagle's minimal medium supplemented with 10% fetal bovine serum, 100 U mL−1 streptomycin, 100 mg mL−1 gentamicin. The cells were grown at 37 °C with 5% (v/v) CO2 in a humidified incubator.
MTT assay
To evaluate the cytotoxicity of the biopolymer, an MTT assay was performed against the MDA-MB-231 breast cancer cell line.9 Cells were seeded onto 96-well plates at a density of 1 × 105 cells per well. The MTT assay was performed in triplicate for test compounds such as biopolymer, phloroglucinol and soluble starch at different concentrations (0–250 μg mL−1). The half-maximum inhibitory concentration value (IC50) for the MTT assay was obtained by plotting a bar graph using OriginPro. 8.0. In addition, the free phloroglucinol present in the encapsulated biopolymer was determined by a 2,4-dimethoxybenzaldehyde (DMBA) assay as reported earlier.6
Morphological staining
MDA-MB-231 cells were seeded in a six-well plate at a density of 1 × 105 cells per well. After 24 h, the cells were detached from the surface of the plate and treated with the IC50 concentration of the biopolymer. The cultured plates were incubated for 24, 36 and 48 h. After the desired time interval of incubation, both live and dead cells were collected, incubated and centrifuged at 4000 rpm. The obtained pellet in the form of a cell suspension was washed twice with phosphate buffer saline (PBS), and stained with AO/EB (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) and Hoechst 33258 (5 mg mL−1). The stained cells were visualized under a fluorescence microscope (Olympus BX-51, Tokyo, Japan) and photographed. The untreated cells were used as control.
1 ratio) and Hoechst 33258 (5 mg mL−1). The stained cells were visualized under a fluorescence microscope (Olympus BX-51, Tokyo, Japan) and photographed. The untreated cells were used as control.
JC-1
MDA-MB-231 cells at a confluence of 2 × 105 cells per well were trypsinized, centrifuged and collected for counting using a hemocytometer. Later, in a six-well plate, the cells were incubated (at different time intervals – 24, 36 and 48 h), supplemented with growth media and treated with the IC50 concentration of biopolymer. At predetermined intervals, the cover slips were stained with a working solution of JC-1 at 37 °C for 20 minutes [working solution – 20 μl of stain from stock solution in 1 mL of DMSO; stock solution – 5 mg mL−1]. The adhered cells in cover slip were kept inversely on a glass slide and visualized under a fluorescence microscope (Olympus BX-51, Tokyo, Japan) using a 377-355 nm filter at 400× magnification and photographed.
DNA laddering
The cells were harvested (after 24 and 48 h treatment with biopolymer), centrifuged (at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C) and the pellet was maintained in lysis buffer (10 mM Tris–Cl, pH 7.5, 1 mM EDTA and 0.2% Triton X-1000) for 30 min. The supernatant was collected and incubated with DNA extraction buffer (0.1 mg mL−1 proteinase K & 0.2 mg mL−1 RNase) for 1 h, at 60 °C. The resulting mixture was extracted with phenol/chloroform (1
000 rpm for 10 min at 4 °C) and the pellet was maintained in lysis buffer (10 mM Tris–Cl, pH 7.5, 1 mM EDTA and 0.2% Triton X-1000) for 30 min. The supernatant was collected and incubated with DNA extraction buffer (0.1 mg mL−1 proteinase K & 0.2 mg mL−1 RNase) for 1 h, at 60 °C. The resulting mixture was extracted with phenol/chloroform (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and run on an agarose gel containing 2% ethidium bromide. The DNA fragments were visualized under a UV-transilluminator and photographed.
1) and run on an agarose gel containing 2% ethidium bromide. The DNA fragments were visualized under a UV-transilluminator and photographed.
Cell cycle analysis
The cells treated with biopolymer at different time intervals (24 & 48 h) were trypsinized, collected, centrifuged (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min) and resuspended in 1 mL of ice-cold phosphate buffer solution (free from Mg & Ca). The cells were then fixed with ethanol at 4 °C for 12 h, and stained with propidium iodide before analysis by flow cytometry (FACSCalibur, BD Biosciences).
000 rpm for 10 min) and resuspended in 1 mL of ice-cold phosphate buffer solution (free from Mg & Ca). The cells were then fixed with ethanol at 4 °C for 12 h, and stained with propidium iodide before analysis by flow cytometry (FACSCalibur, BD Biosciences).
Caspase activity assay
The caspase activity assay was determined by a chromogenic assay using caspase-3 (Calbiochem, Merck) and caspase-8 (Chemicon International Inc.) colorimeter activity assay kits. Briefly, after treatment of biopolymer at different time intervals (24, 36 and 48 h), 1.5 × 106 cells were harvested, lysed with cell lysis buffer (50 mM HEPES, 100 mM NaCl, 0.1% CHAPS, 1 mM DTT, 100 mM EDTA) followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 min. About 50 μl of supernatant was incubated with specific substrate (at 37 °C) for 2 h in a water bath. The absorbance of the cleaved substrate was measured at 405 nm using a microtiter plate reader (BioRad, UK).
000 rpm for 1 min. About 50 μl of supernatant was incubated with specific substrate (at 37 °C) for 2 h in a water bath. The absorbance of the cleaved substrate was measured at 405 nm using a microtiter plate reader (BioRad, UK).
Semi-quantitative RT-PCR
The total RNA from the IC50-treated biopolymer at predetermined intervals (24, 36 & 48 h) was prepared by using Trizol reagent (Invitrogen, USA). 1 μg of RNA was reverse transcribed and amplified by one-step semi-quantitative RT-PCR using specific primers (ESI Table 1†). The conditions for PCR are as follows: 1 × (94 °C for 5 min); 35 × (94 °C for 1 min; 55 °C for 1 min & 72 °C for 1 min) & 1 × (72 °C for 10 min). The amplified products were verified by 1% agarose gel electrophoresis under a UV-transilluminator and then photographed. β-actin used in the study served as an internal control.
Western blotting
After treatment with the IC50 concentration of biopolymer (for 24, 36 & 48 h), the adherent and floating cells were harvested and lysed in lysis buffer (50 mM Tris–Cl (pH 8.0), 150 mM NaCl, 0.02% sodium azide, phenyl methane sulfonyl fluoride (PMSF), aprotinin, and 1% Triton-X 100) for 30 min. The lysed cells were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C. The total protein content present in the sample was determined by using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). The obtained protein (30 μg) was subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinyl difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was then blocked and probed with primary antibodies (anti-caspase-3 & 8) overnight at 4 °C. The membrane was again incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Finally, the blots were visualized using enhanced chemiluminescence blotting detection kit (Perkin Elmer Life Sciences, Inc., Boston, MA, USA). β-actin was used as an internal control.
000 rpm for 10 min at 4 °C. The total protein content present in the sample was determined by using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). The obtained protein (30 μg) was subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinyl difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was then blocked and probed with primary antibodies (anti-caspase-3 & 8) overnight at 4 °C. The membrane was again incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Finally, the blots were visualized using enhanced chemiluminescence blotting detection kit (Perkin Elmer Life Sciences, Inc., Boston, MA, USA). β-actin was used as an internal control.
Results and discussion
In vitro cytotoxic assay
The in vitro cytotoxicities of soluble starch, phloroglucinol and phloroglucinol-encapsulated starch biopolymer were investigated against the MDA-MB-231 breast cancer cell line using the MTT assay. The cytotoxicity of the biopolymer is pronounced; it has a high inhibitory effect on the MDA-MB-231 cell line in a dose-dependent manner with an IC50 value of 187.5 μg mL−1 (Fig. 2). Phloroglucinol (255.54 μg mL−1) and soluble starch (854.67 μg mL−1) showed reduced cytotoxic effects compared to the biopolymer. The increased cytotoxic effect of the biopolymer may well be compromised by the loss of cell membrane integrity. Altogether, the controlled release of phloroglucinol within the cancer cell at physiological pH (7.4) induces cell death leading to apoptosis.6 Meanwhile, the actual phloroglucinol present in the biopolymer was found to be 45.49 μg mL−1 according to the DMBA assay.
 |
| | Fig. 2 Cytotoxic effects of biopolymer, phloroglucinol and soluble starch against cells of the breast cancer cell line MDA-MB-231. | |
AO/EB staining
The morphological changes within the cells were examined by using acridine orange/ethidium bromide (AO/EB) by treating the cancer cell line with the biopolymer (IC50–187.5 μg mL−1) at different time intervals (12, 24 & 36 h) (Fig. 3A). Acridine orange stains both live and dead cells, and emit green fluorescence, whereas ethidium bromide stains the cells whose membrane integrity is lost, emitting red fluorescence.10 Hence in the present study, distinct cytological changes within the cells have been characterized according to the fluorescence emission and morphological features, and are shown in Fig. 3A. This figure shows (a) well-organized, uniform live cells stained green; (b) early apoptotic cells with condensed chromatin at the perinuclear region in red; (c) preliminary late apoptotic cells emitting bright red fluorescence; and (d) occurrence of prominent cell death by apoptosis and swollen necrotic bodies.
 |
| | Fig. 3 Morphological assessment of MDA-MB-231 cancer cells using fluorescence staining (A) AO/EB staining; (B) Hoechst 33259 staining; (C) JC-1 at different time intervals with the IC50 concentration of the biopolymer – (a) untreated as control; (b) 24 h treatment; (c) 36 h treatment; (d) 48 h treatment. | |
Hoechst staining
Hoechst staining was performed to differentiate dead from live cells by examining their nuclear morphology under a fluorescence microscope.11 Hoechst 33258 is a cationic lipophilic dye that permeates the cell membrane and effectively binds the AT-rich region of double-stranded DNA.12 A time-dependent study was carried out with the IC50 concentration of the biopolymer (IC50–187.5 μg mL−1), and showed typical apoptotic nuclear morphology, namely nuclear shrinkage, DNA condensation and fragmentation (Fig. 3B). However, the untreated cells retained an intact nuclear morphology. The drastic change occurring within the cells could be due to the controlled release of phloroglucinol. It has also been previously observed that change in nuclear morphology of cancer cells is directly proportional to controlled release of phloroglucinol.6 Altogether, the loss of cell volume over time is attributed to cytoskeleton breakdown and plasma membrane blebbing.13
Mitochondria membrane potential (Δψm)
Mitochondria membrane potential (Δψm) is a valuable indicator to assess the induction of cellular apoptosis.14 As mitochondria are considered to be the “power house” of the cell, several notable anticancer drugs target them to induce apoptosis without affecting normal cells. Keeping this in mind, to study the effect of biopolymer on the mitochondrial Δψm, JC-1 staining was performed. JC-1 is a cationic lipophilic dye that selectively enters the mitochondria and emits red (for untreated cells) and green (for treated cells) fluorescence depending upon the mitochondrial Δψm.15 As expected, in the present study, the time-dependent treatment confirms the loss of mitochondrial Δψm against the MDA-MB-231 breast cancer cell line as shown in Fig. 3C. In normal/untreated cells, JC-1 enters the mitochondria forming J-aggregates enabling red fluorescence. However, in the case of apoptotic (or) unhealthy cells with low mitochondria Δψm, JC-1 remains in a monomeric form and emits green fluorescence.16 Further, this result confirms the release of apoptogenic proteins such as cytochrome c and apoptosis inducing factor (AIF) into the cytoplasm leading to the activation of the caspase cascade.17
DNA fragmentation
DNA fragmentation is considered to be a hallmark event during the progression of apoptosis. However, apoptosis can also occur without DNA fragmentation. In the present study, the biopolymer-treated cells showed apoptotic DNA fragmentation whereas the untreated cells showed no such event (Fig. 4). Several in vitro studies pertain to explain the participation of caspase-9-dependent activation of caspase-3 in the development of DNA fragmentation.18 The caspase-3 enzyme readily cleaves the inhibitor of caspase-activated DNase (ICAD) and activates the caspase-activated DNase (CAD) enzyme. On the other hand CAD cleaves the internucleosomal DNA, leading to apoptotic DNA fragmentation.
 |
| | Fig. 4 DNA fragmentation of MDA-MB-231 breast cancer cells – (1) ladder; (2) untreated cells as control; (3) IC50 treated cells at 24 h; (4) IC50 treated cells at 48 h. | |
Cell cycle arrest
The effect of the biopolymer on cell cycle progression was also examined by treatment with IC50 concentration after 24 & 48 h incubation (ESI Table 2†). After 24 h, the biopolymer caused an increase of cells in S phase as compared to G2 phase and untreated cells. This may be due to controlled release of phloroglucinol that affects replication fork formation by inhibiting topoisomerase I, leading to the occurrence of double-stranded DNA breaks.19 However, there is a steady increase in G2 phase as compared to S phase after 48 h, which indicates the DNA damage from S phase has prevented G2 phase from entering the process of mitosis.20
The role of caspases in apoptosis induced by biopolymer
Caspases are aspartate-specific cysteine protease enzymes that play a vital role in the induction, transduction and amplification of intracellular apoptotic signals in eukaryotes.21 Based on the amino acid composition, caspases are divided into three sub-families, namely activator, executioner and mediator caspases. Most importantly, the initiator and executioner caspases get involved in the apoptotic signalling cascade. For the first time, the molecular mechanism was studied for the effect of the phloroglucinol-encapsulated starch biopolymer on the MDA-MB-231 breast cancer cell line. The results from the study suggest the involvement of both extrinsic and intrinsic pathways towards the induction of apoptotic cell death. To ensure this, caspases were quantified based upon a colorimetric assay kit on treatment with the IC50 concentration of the biopolymer at different time intervals (24, 36 and 48 h). Caspase-3 and 8 showed a time-dependent increase in activity as compared to untreated cells (Fig. 5). The results taken together also confirmed the involvement of caspases-3 and 9 in the apoptotic event.
 |
| | Fig. 5 Phloroglucinol-encapsulated starch biopolymer-induced caspase-3 and 8 activity assays at different time intervals. | |
These results have led us to study the expression pattern of caspases at the gene and protein level during the induction of apoptosis. During internalization of the biopolymer, the modified form of the starch matrix interacts with Fas (or) TNF receptor in the plasma membrane to recruit procaspase-8. Meanwhile, the autoactivation of procaspase-8 into caspase-8 leads to the activation of downstream caspases.22 This has been confirmed by the upregulation of caspase-8 through differential gene expression (Fig. 6a) and western blotting analysis (Fig. 7c). An increase in the expression pattern of pro-caspase-8 and active caspase-8 at the IC50 concentration was also observed within 24 h. This suggests that a minimum time interval is sufficient for the modified form of starch to interact with receptors in the plasma membrane. In addition, the cleavage of pro-caspase-8 to active caspase-8 was time-dependent. Hence, the modified form of starch matrix plays an important role in the diffusion and internalization. After the internalization, the weak hydrogen bonds between the soluble starch and phloroglucinol disrupt the controlled release.6 This paves the way for phloroglucinol to attack mitochondria and increase its mitochondrial membrane potential towards the activation of caspase-9, according to JC-1 staining and DNA fragmentation.23 The expression pattern of caspase-9 was found to depend on the expression time, as shown by semi-quantitative RT-PCR (Fig. 6b). This results in an increase in the mitochondrial membrane potential, which causes a release of cytochrome c into the cytosol, which in turn binds pro-caspase-9 leading to formation of the apoptosome.24 Meanwhile, these changes activate caspase-3 to play a crucial role in apoptosis via the intrinsic cell death pathway (Fig. 6c and 7b). It has also been reported that executionary caspases (caspase-3) may cleave cytokeratins, PARP, the plasma membrane cytoskeletal protein alpha fodrin, the nuclear protein NuMA and other related proteins by activating cytoplasmic endonuclease.25 In our study, it was also found that the expression patterns of the gene and protein for β-actin (internal control) were the same (Fig. 6d and 7a). Meanwhile, the expression fold ratio of western blot analysis for caspase-3 and 8 at different time intervals during the treatment with IC50 concentration biopolymer is significantly higher as compared to control (Fig. 8). The overall findings suggest that caspase-mediated apoptosis is provoked by phloroglucinol-encapsulated starch biopolymer in MDA-MB-231 cancer cells (Fig. 9). Hence, the phloroglucinol-encapsulated starch biopolymer may merit serving as an anticancer or chemotherapeutic agent for treating breast cancer in the near future.
 |
| | Fig. 6 Semi-quantitative RT-PCR – Effect of the biopolymer on expression of (a) caspase-8; (b) caspase-9; (c) caspase-3; and (d) β-actin (control) at different time intervals. | |
 |
| | Fig. 7 A time-dependent treatment of MDA-MB-231 breast cancer cells with the IC50 concentration of the phloroglucinol-encapsulated starch biopolymer. SDS-PAGE was carried out and proteins were detected by Western blotting – (a) β-actin; (b) caspase-3; (c) caspase-8. | |
 |
| | Fig. 8 Changes in pro-apoptotic protein (caspase-3 & caspase-8) induced by the phloroglucinol-encapsulated starch biopolymer at different time intervals assessed by Western blot analysis in MDA-MB-231 breast cancer cells. Significant differences from the control (untreated) are indicated by ***p = 0.001 and ###p = 0.002. | |
 |
| | Fig. 9 Possible overall mechanism involved in caspase-mediated apoptosis of MDA-MB-231 breast cancer cells induced by the phloroglucinol-encapsulated starch biopolymer. | |
Conclusion
In conclusion, phloroglucinol-encapsulated starch biopolymer induces apoptosis in MDA-MB-231 breast cancer cells via extrinsic and intrinsic pathways. These findings suggest that the encapsulation of the phenolic compound will pave the way for a new approach to prevent and treat cancer.
Acknowledgements
The authors are grateful to Natural Resource Data Management System (DST-NRDMS), Government of India, New Delhi for their financial assistance through major research project. The authors acknowledge Dr Hairul Islam, Pondicherry Centre for Biological Sciences, Puducherry for his help in cell culture facilities.
References
- G. Vilar, J. Tulla-Puche and F. Albericio, Curr. Drug Delivery, 2012, 9, 1 CrossRef.
- K. E. Uhrich, Chem. Rev., 1999, 99, 3181 CrossRef CAS PubMed.
- A. Rodrigues and M. Emeje, Carbohydr. Polym., 2012, 87, 987 CrossRef CAS PubMed.
- A. Munin and F. Edwards-Levy, Pharmaceutics, 2012, 3, 793 CrossRef PubMed.
- K. B. Pandey and S. I. Rizvi, Oxid. Med. Cell. Longevity, 2009, 2, 270 CrossRef PubMed.
- P. Kumar, S. Senthamilselvi and M. Govindaraju, RSC Adv., 2014, 4, 26787 RSC.
- S. H. Eom, Y. M. Kim and S. W. Kim, Food Chem. Toxicol., 2012, 50, 3251 CrossRef CAS PubMed.
- P. Singh, J. Sidana, S. B. Bharate and W. J. Foley, Nat. Prod. Rep., 2010, 27, 393 RSC.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55 CrossRef CAS.
- T. J. Liegler, W. Hyun, T. S. Yen and D. P. Stites, Clin. Diagn. Lab. Immunol., 1995, 2, 369 CAS.
- S. Yamakawa, A. Demizu, Y. Kawaratani, Y. Nagaoka, Y. Terada, S. Maruyama and S. Uesato, Biol. Pharm. Bull., 2005, 31, 916 Search PubMed.
- R. K. Schlicher, J. D. Hutcheson, H. Radhakrishna, R. P. Apkarian and M. R. Prausnitz, Ultrasound Med. Biol., 2011, 36, 677 CrossRef PubMed.
- D. Ly, D. R. Grubb and A. Lawen, Apoptosis, 2003, 8, 115 CrossRef.
- A. Perelman, C. Wachtel, M. Cohen, S. Haupt, H. Shapiro and A. Tzur, Cell Death Dis., 2012, 3, 430 CrossRef PubMed.
- A. Mathur, Y. Hong, B. K. Kemp, A. A. Barrientos and J. D. Erusalimsky, Cardiovasc. Res., 2000, 46, 126 CrossRef CAS.
- T. Hishita, S. Tada-Oikawa, K. Tohyama, Y. Miura, T. Nishihara, Y. Tohyama, Y. Yoshida, T. Uchiyama and S. Kawanishi, Cancer Res., 2001, 61, 2878 CAS.
- H. Zhang and M. Xu, Cell Res., 2000, 10, 205 CrossRef CAS PubMed.
- W. H. Zhang, A. Poh, A. A. Fanous and A. Eastman, Cell Cycle, 2008, 7, 1668 CrossRef CAS.
- Y. Hu, Y. Yang, Q. D. You, W. Liu, H. Y. Gu, L. Zhao, K. Zhang, W. Wang, X. T. Wang and Q. L. Guo, Biochem. Biophys. Res. Commun., 2006, 351, 521 CrossRef CAS PubMed.
- T. J. Fan, L. H. Han, R. S. Cong and J. Liang, Acta Biochim. Biophys. Sin., 2005, 37, 719 CrossRef CAS PubMed.
- M. Kikuchi, S. Kuroki, M. Kayama, S. Sakaguchi, K. K. Lee and S. Yonehara, J. Biol. Chem., 2012, 287, 41165 CrossRef CAS PubMed.
- M. Brentnall, L. Rodriguez-Menocal, R. B. De Guevara, E. Cepero and L. H. Boise, BMC Cell Biol., 2013, 14, 32 CrossRef CAS PubMed.
- S. W. G. Tait and D. R. Green, Nat. Rev., 2010, 11, 621 CrossRef CAS PubMed.
- S. Elmore, Toxicol. Pathol., 2007, 35, 495 CrossRef CAS PubMed.
- S. M. Huang, C. W. Cheung, C. S. Chang, C.
H. Tang, J. F. Liu, Y. H. Lin, J. H. Chen, S. H. Ko, K. L. Wong and D. Y. Lu, J. Cell. Biochem., 2011, 112, 643 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra06664b |
|
| This journal is © The Royal Society of Chemistry 2014 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) and Hoechst 33258 (5 mg mL−1). The stained cells were visualized under a fluorescence microscope (Olympus BX-51, Tokyo, Japan) and photographed. The untreated cells were used as control.
1 ratio) and Hoechst 33258 (5 mg mL−1). The stained cells were visualized under a fluorescence microscope (Olympus BX-51, Tokyo, Japan) and photographed. The untreated cells were used as control.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C) and the pellet was maintained in lysis buffer (10 mM Tris–Cl, pH 7.5, 1 mM EDTA and 0.2% Triton X-1000) for 30 min. The supernatant was collected and incubated with DNA extraction buffer (0.1 mg mL−1 proteinase K & 0.2 mg mL−1 RNase) for 1 h, at 60 °C. The resulting mixture was extracted with phenol/chloroform (1
000 rpm for 10 min at 4 °C) and the pellet was maintained in lysis buffer (10 mM Tris–Cl, pH 7.5, 1 mM EDTA and 0.2% Triton X-1000) for 30 min. The supernatant was collected and incubated with DNA extraction buffer (0.1 mg mL−1 proteinase K & 0.2 mg mL−1 RNase) for 1 h, at 60 °C. The resulting mixture was extracted with phenol/chloroform (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and run on an agarose gel containing 2% ethidium bromide. The DNA fragments were visualized under a UV-transilluminator and photographed.
1) and run on an agarose gel containing 2% ethidium bromide. The DNA fragments were visualized under a UV-transilluminator and photographed.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min) and resuspended in 1 mL of ice-cold phosphate buffer solution (free from Mg & Ca). The cells were then fixed with ethanol at 4 °C for 12 h, and stained with propidium iodide before analysis by flow cytometry (FACSCalibur, BD Biosciences).
000 rpm for 10 min) and resuspended in 1 mL of ice-cold phosphate buffer solution (free from Mg & Ca). The cells were then fixed with ethanol at 4 °C for 12 h, and stained with propidium iodide before analysis by flow cytometry (FACSCalibur, BD Biosciences).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 min. About 50 μl of supernatant was incubated with specific substrate (at 37 °C) for 2 h in a water bath. The absorbance of the cleaved substrate was measured at 405 nm using a microtiter plate reader (BioRad, UK).
000 rpm for 1 min. About 50 μl of supernatant was incubated with specific substrate (at 37 °C) for 2 h in a water bath. The absorbance of the cleaved substrate was measured at 405 nm using a microtiter plate reader (BioRad, UK).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C. The total protein content present in the sample was determined by using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). The obtained protein (30 μg) was subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinyl difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was then blocked and probed with primary antibodies (anti-caspase-3 & 8) overnight at 4 °C. The membrane was again incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Finally, the blots were visualized using enhanced chemiluminescence blotting detection kit (Perkin Elmer Life Sciences, Inc., Boston, MA, USA). β-actin was used as an internal control.
000 rpm for 10 min at 4 °C. The total protein content present in the sample was determined by using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). The obtained protein (30 μg) was subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinyl difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was then blocked and probed with primary antibodies (anti-caspase-3 & 8) overnight at 4 °C. The membrane was again incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Finally, the blots were visualized using enhanced chemiluminescence blotting detection kit (Perkin Elmer Life Sciences, Inc., Boston, MA, USA). β-actin was used as an internal control.









