Activation of enzyme nanogel in organic solvents by PEG–substrate joint imprinting†
Abstract
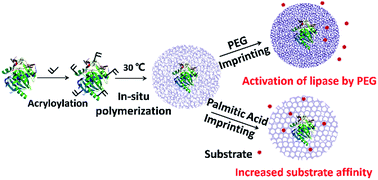
A substrate or polyethyleneglycol (PEG) imprinted lipase nanogel displayed increased apparent activity in organic solvents by 2.5–4.7 folds, compared to native lipase. It enabled a one-step synthesis of chloramphenicol palmitate with a yield of ∼99% and purity of ∼99%, indicating that the imprinted lipase nanogel is an appealing catalyst in organic media.

 Please wait while we load your content...
Please wait while we load your content...