Integrated bioprinting and imaging for scalable, networkable desktop experimentation†
Abstract
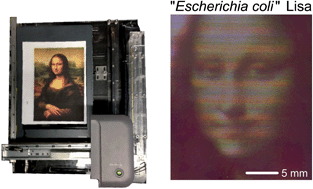
Adaptations of mass-market consumer electronics are increasingly used to aid experimentation in engineering, life sciences, and education. Inspired by recent bioprinting and imaging research, we have developed a low-cost, networkable, scalable bioprinter with integrated imaging that enables automated fluid deposition with monitoring biological materials over a large area that can interface with standard plasticware. We re-engineered a desktop printer and scanner with mechanical workarounds (rather than software) to leverage the unmodified software, creating a complete system for ∼$700. We found that the resulting print accuracy and precision of this system were close to those of the unmodified printer and scanner. We demonstrate the reach of this system by printing multiple strains of Escherichia coli, performing quantitative time-lapse recordings of bacterial growth, and then separating different fluorescent strains according to color. A fluid-deposition and imaging platform, like the one developed here, could be integrated for do-it-yourself research, remote experimentation, and (on-line) education.

 Please wait while we load your content...
Please wait while we load your content...