Synthesis, molecular modeling and biological evaluation of cinnamic acid derivatives with pyrazole moieties as novel anticancer agents
Wei-Ming Zhang†
ab,
Man Xing†ab,
Ting-Ting Zhaoab,
Yu-Jia Renab,
Xian-Hui Yangab,
Yu-Shun Yangab,
Peng-Cheng Lv*ab and
Hai-Liang Zhu*ab
aNanjing Institute for the Comprehensive Utilization of Wild Plant, Nanjing 210042, People's Republic of China. E-mail: zhuhl@nju.edu.cn; Fax: +86-25-8359 2672; Tel: +86-25-8359 2572
bState Key Laboratory of Pharmaceutical Biotechnology, Nanjing University, Nanjing 210093, People's Republic of China. E-mail: pclv@nju.edu.cn
First published on 5th August 2014
Abstract
A series of pyrazole derivatives (1e–30e) has been designed and synthesized, and their biological activities were evaluated for EGFR and HER-2 inhibition and tumor cell antiproliferation. Among the compounds synthesized, compound 30e exhibited excellent enzyme inhibitory activity (IC50 = 0.21 ± 0.05 μM for EGFR and IC50 = 1.08 ± 0.15 μM for HER-2). Compound 30e also showed the most potent antiproliferative activity, which inhibited the growth of MCF-7 and B16-F10 cell lines with IC50 values of 0.30 ± 0.04 and 0.44 ± 0.05 μM, respectively. The molecular docking study was performed to analyze the probable binding models and the 3D-QSAR models were built for the rational design of EGFR/HER-2 inhibitors. Based on the results obtained, compound 30e with potent EGFR and HER-2 inhibitory activity may be a potential anticancer agent.
Introduction
Although extensive efforts have been applied to co-deliver anticancer therapeutics by nanoparticles to induce synergistic anticancer effects, conventional anticancer therapy using cytotoxic drugs lacks selectivity and is prone to toxicity and drug resistance.1,2 Therefore, targeted therapies to minimize toxicity to healthy tissues have been attracting extensive attention, such as using small molecules, peptides or aptamers.3,4 Actually, epidermal growth factor receptor (EGFR) kinase, playing a key role in signal transduction pathways that regulate cell division, differentiation and migration, has been proposed as a promising target for drug design. Thus, EGFR inhibitors were widely used to treat human cancers.5–7 The EGFR family comprises the four members of EGFR/erbB-1/HER-1, erbB-2/Neu/HER-2, erbB-3/HER-3, and erbB-4/HER-4, which belong to the type I receptor tyrosine kinase (RTK) class. They play an important role in the mediating growth factor signaling.8,9 The abnormal signaling of these pathways results in disregulated cell proliferation, evasion from apoptosis, angiogenesis, migration, and metastasis. Especially, the overexpression of EGFR and HER-2 was observed in many human cancers including bladder, breast, colon, and lung cancers.10,11 As reported, using small molecules to inhibit kinase activity of EGFR and HER-2 by binding the ATP site of the intracellular tyrosine kinase domain to block the auto-phosphorylation of tyrosine kinase (TK), disrupt the activation of signal-transduction cascades, and then interfere cell proliferation, angiogenesis, invasion and metastasis, has accordingly and historically been a successful area of research for the advancement of cancer drugs.12,13 Gefitinib and erlotinib (Fig. 1) are the representative drugs for this kind of inhibitors and have been approved by US-FDA in these years.14,15Apart from the above two agents, several other compounds (Fig. 1 and 2), compounds containing pyrazole and cinnamyl moiety, for example, compound A and compound E, were found to possess high inhibitory activity against EGFR and HER-2.16–21 Pyrazole motif displays a variety of pharmacological activities, such as anti-inflammatory,22 antibacterial–antifungal,23 inhibition against cyclooxigenase-2,24 p38 MAP kinase25 and CDK2/Cyclin A.26,27 Therefore, the pyrazole ring is an advantageous choice for the synthesis of pharmaceutical compounds with different biological activities and good safety profiles.28–30 Cinnamoyl moiety was also demonstrated to possess a wide range of pharmacological activities. Particularly, cinnamic acid ester derivatives showed potential antitumor activity.31,32 For instance, Qian et al. described the SAR of the novel series of cinnamic acid metronidazole ester derivatives, and the biological activity evaluation indicated that cinnamic acid ester derivatives are potent inhibitors of EGFR and HER-2.20 Moreover, other studies also demonstrated that the incorporation of cinnamoyl moiety could contribute to the binding of target compounds with EGFR and HER-2 active site.33
Encouraged by the above mentioned research, herein we performed the synthesis and biological evaluation of a series of novel pyrzole derivatives possessing pyrazole–cinnamyl moiety. Enzyme inhibitory results show that some of them are potential EGFR and HER-2 inhibitors. Molecular modeling studies were also performed to understand the binding mode between this kind of inhibitor and the active site of EGFR/HER-2 crystal structures.
Chemistry
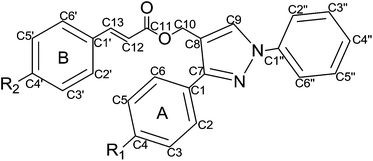
The synthetic route for pyzazole (1e–30e) is outlined in Scheme 1. Firstly, 1-phenyl-2-(1-phenylethylidene)hydrazine (b) was prepared by treating different substituted acetophenone (a) with phenylhydrazine hydrochloride in the presence of sodium acetate. Secondly, 1,3-diphenyl-1H-pyrazole-4-carbaldehyde (c) was afforded by treating compound b with POCl3. Thirdly, different substituted (1,3-diphenyl-1H-pyrazol-4-yl)methanol (d) was gotten by the reaction of compound c with sodium borohydride in DCM. Finally, compound d was coupled with different substituted cinnamic acid in the presence of DMAP and K2CO3. Twenty-seven out of the thirty target compounds (1e–30e) were reported for the first time. All of the synthetic compounds gave satisfactory elementary analytical and spectroscopic data. 1H NMR and ESI-MS spectra were consistent with the assigned structures.Results and discussion
The synthesized compounds were evaluated for their antiproliferative activities against MCF-7 and B16-F10 cells. The results were summarized in Table 2. As shown in Table 2, pyrazole derivatives bearing the pyrazole–cinnamyl moiety exhibited remarkable antiproliferative effects. Among them, compound 30e displayed the most potent antiproliferative activity (IC50 = 0.30 ± 0.04 μM for MCF-7 and IC50 = 0.44 ± 0.05 μM for B16-F10). Subsequently, structure-activity relationships (SAR) studies were performed by modification of the parent compound to determine how the substituents of the subunits affect the antiproliferative activities. Inspection of the chemical structures of the compounds 1e–30e suggested that they could be divided into two subunits: A-ring and B-ring (Table 1).As shown in Table 2, it was revealed that different substitutes on the A-ring led to different antiproliferative activities. A comparison of the para substituents on the A-ring demonstrated that an electron-donating group could improve their antiproliferative activity, and the potency order was OMe > Me. Similarly, change of substituents on the B-ring could also affect the antiproliferative activities. Among them, compounds with electron-donating substituent at para position showed higher antiproliferative activity and the potency order was similar with A-ring: OMe > Me, while compounds with electron-withdrawing substituents led to a drop for the antiproliferative activity. The potency order of para-substituents compounds is that: OMe > Me > H > Br > Cl > F. Among all the compounds tested, compound 30e that has para-OMe group on both A-ring and B-ring showed the best antiproliferative activity.
| Compound | IC50 ± SD (μM) | |||
|---|---|---|---|---|
| MCF-7a | B16-F10a | EGFRb | HER-2b | |
| a Inhibition of the growth of tumor cell lines.b Inhibition of EGFR and HER-2. | ||||
| 1e | 4.05 ± 0.37 | 3.51 ± 0.17 | 5.45 ± 0.47 | 6.33 ± 0.53 |
| 2e | 7.45 ± 0.67 | 7.51 ± 0.45 | 9.05 ± 0.68 | 16.85 ± 1.42 |
| 3e | 5.14 ± 0.51 | 5.34 ± 0.26 | 6.47 ± 0.72 | 9.26 ± 0.89 |
| 4e | 4.74 ± 0.41 | 4.84 ± 0.32 | 6.68 ± 0.53 | 9.97 ± 0.78 |
| 5e | 3.51 ± 0.31 | 2.98 ± 0.17 | 4.26 ± 0.48 | 6.24 ± 0.72 |
| 6e | 2.11 ± 0.18 | 1.82 ± 0.12 | 3.24 ± 0.49 | 4.63 ± 0.28 |
| 7e | 6.34 ± 0.47 | 6.46 ± 0.42 | 8.15 ± 0.58 | 10.52 ± 0.94 |
| 8e | 10.37 ± 0.87 | 11.47 ± 1.27 | 14.22 ± 1.16 | 19.13 ± 1.62 |
| 9e | 8.57 ± 0.79 | 9.63 ± 0.89 | 9.82 ± 0.74 | 18.04 ± 1.36 |
| 10e | 8.22 ± 0.65 | 8.21 ± 0.44 | 10.68 ± 0.86 | 18.55 ± 1.27 |
| 11e | 4.64 ± 0.30 | 4.51 ± 0.23 | 5.94 ± 0.37 | 8.12 ± 0.75 |
| 12e | 3.87 ± 0.18 | 3.27 ± 0.22 | 4.87 ± 0.53 | 6.23 ± 0.79 |
| 13e | 6.17 ± 0.46 | 5.86 ± 0.25 | 7.95 ± 0.68 | 10.35 ± 1.03 |
| 14e | 9.05 ± 0.61 | 10.59 ± 0.92 | 12.46 ± 0.92 | 19.01 ± 0.16 |
| 15e | 7.96 ± 0.69 | 7.93 ± 0.76 | 8.35 ± 0.78 | 16.41 ± 1.73 |
| 16e | 7.03 ± 0.51 | 7.31 ± 0.58 | 8.59 ± 0.62 | 12.44 ± 0.97 |
| 17e | 4.35 ± 0.44 | 4.48 ± 0.24 | 5.33 ± 0.47 | 7.060.84 |
| 18e | 2.75 ± 0.25 | 2.35 ± 0.13 | 3.58 ± 0.28 | 5.12 ± 0.62 |
| 19e | 1.27 ± 0.12 | 1.13 ± 0.09 | 1.68 ± 0.23 | 3.52 ± 0.27 |
| 20e | 2.81 ± 0.21 | 2.75 ± 0.22 | 4.78 ± 0.31 | 5.86 ± 0.53 |
| 21e | 2.35 ± 0.14 | 2.16 ± 0.11 | 3.37 ± 0.25 | 4.86 ± 0.59 |
| 22e | 1.50 ± 0.16 | 1.27 ± 0.13 | 2.05 ± 0.18 | 3.64 ± 0.37 |
| 23e | 0.67 ± 0.04 | 0.74 ± 0.08 | 0.98 ± 0.10 | 2.53 ± 0.31 |
| 24e | 0.32 ± 0.05 | 0.53 ± 0.07 | 0.44 ± 0.09 | 1.85 ± 0.24 |
| 25e | 0.92 ± 0.09 | 0.85 ± 0.11 | 1.39 ± 0.21 | 2.74 ± 0.21 |
| 26e | 2.02 ± 0.17 | 1.55 ± 0.19 | 2.29 ± 0.19 | 4.13 ± 0.35 |
| 27e | 1.97 ± 0.11 | 1.42 ± 0.14 | 2.35 ± 0.31 | 3.85 ± 0.38 |
| 28e | 1.15 ± 0.10 | 1.03 ± 0.09 | 1.37 ± 0.20 | 3.12 ± 0.25 |
| 29e | 0.56 ± 0.04 | 0.66 ± 0.05 | 0.62 ± 0.08 | 2.23 ± 0.19 |
| 30e | 0.30 ± 0.04 | 0.44 ± 0.05 | 0.21 ± 0.05 | 1.08 ± 0.15 |
| Erlotinib | 0.08 ± 0.006 | 0.12 ± 0.011 | 0.03 ± 0.002 | 0.14 ± 0.02 |
A solid-phase ELISA assay was performed with the aim to examine whether this kind of compounds could inhibit the autophosphorylation of EGFR and HER-2 kinases. As shown in Table 2, it was observed that pyrazolyl–cinnamyl derivatives showed good inhibitory activities against EGFR and HER-2. Among them, compound 30e which has the best antiproliferative activity, also displayed the most potent inhibitory activity (IC50 = 0.21 ± 0.05 μM for EGFR and IC50 = 1.08 ± 0.15 μM for HER-2). Subsequently, using Discovery Studio 3.1 (Discovery Studio 3.1, Accelrys, Inc. San Diego, CA), a molecular modeling study was performed to explore the interaction between the synthesized compounds and the active site of EGFR/HER-2. In the present study, the crystal structures of EGFR (PDB Code: 1M17.pdb) and HER-2 (PDB Code: 3PP0.pdb) were chosen, which were obtained from the RCSB protein data bank (http://www.pdb.org). After preparing the receptor and ligands, the site sphere was selected based on the ligand binding location. The binding models of the most potent compounds with corresponding target were depicted in Fig. 3 (30e with 1M17) and Fig. 4 (30e with 3PP0), respectively.
 | ||
| Fig. 3 The molecular docking of compound 30e into EGFR. Compound 30e (colored by atom: carbons: gray; nitrogen: blue; oxygen: red) is nicely bound into EGFR (entry 1M17 in the Protein Data Bank). The dotted lines show the hydrogen bond and the yellow line show the Pi-Sigma interactions. | ||
 | ||
| Fig. 4 The molecular docking of compound 30e into HER-2. Compound 30e (colored by atom: carbons: gray; nitrogen: blue; oxygen: red) is nicely bound into EGFR (entry 3PP0 in the Protein Data Bank). | ||
As shown in Fig. 3, compound 30e was nicely bound to the ATP binding site of EGFR via hydrophobic interactions and binding is stabilized by a hydrogen bond and a Pi-Sigma interaction. The oxygen atom of the OMe group in A-ring formed one hydrogen bond with the amino hydrogen of Lys A: 721 (N–H⋯O = 2.477 Å; 130.6°). A Pi-Sigma interaction was also formed between Leu694 and the benzene ring of compound 30e (bond length: 2.48 Å), which could enhance the binding action between receptor EGFR and compound 30e. Similarly, it can be seen from Fig. 4 that compound 30e can bind to the active site of HER2 via a hydrogen bond. The oxygen atom of the OMe group in A-ring formed one hydrogen bond with the amino hydrogen of Met A: 801 (N–H⋯O = 2.278 Å; 145.8°).
Finally, in order to give a systematic evaluation on pyrazole derivatives as antitumor agents and to discover more potent and selective dual EGFR/HER-2 inhibitors, a 3D-QSAR model was initiated by using a novel 3D-QSAR protocol of Discovery Studio 3.1. Thirty compounds with definite IC50 values against EGFR and HER-2 inhibitors were selected as the model dataset. By convention, we used the pIC50 scale (−lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50), in which higher value indicates exponentially greater potency, to measure the inhibitory activity. The training set and test set were chosen by the Diverse Molecules method in Discovery Studio 3.1. To ensure a good alignment, we chose the alignment conformation of each molecule with lowest energy in the docked results of CDOCKER protocol. Before building the QSAR model, we applied the alignment by the substructure (E)-(1,3-diphenyl-1H-pyrazol-4-yl)methyl 3-phenylacrylate. The correlation coefficient r2 between observed activity of testing set and training set was found to be 0.819 for the EGFR model and 0.861 for the HER-2 model, which confirm that the QSAR models built are both acceptable. The molecules aligned with the iso-surfaces of the 3D QSAR model coefficients on electrostatic potential grids and van der Waals grids are shown in Fig. 5. Electrostatic map indicates red contours around regions where high electron density (negative charge) is expected to increase activity, and blue contours represent areas where low electron density (partial positive charge) is expected to increase activity. Similarly, steric map indicates areas where steric bulk is predicted to increase (green) or decrease (yellow) activity. As for EGFR, on the ring A, a slightly bulk and low negative charged group on the para-position would facilitate the inhibitory activity, which is in good agreement with the fact that OMe-substituent compounds display more potent activity than that of F- or Cl-substituent. Meanwhile, on ring B, it is suggested that compounds with low negative charged and slightly bulk R2 group on the para-position might make the efficiency better. Similar trends were observed for HER-2 model. The 3D-QSAR models correlate well with the experimental data and could provide insights on further discovery of more potent EGFR and HER-2 inhibitors.
IC50), in which higher value indicates exponentially greater potency, to measure the inhibitory activity. The training set and test set were chosen by the Diverse Molecules method in Discovery Studio 3.1. To ensure a good alignment, we chose the alignment conformation of each molecule with lowest energy in the docked results of CDOCKER protocol. Before building the QSAR model, we applied the alignment by the substructure (E)-(1,3-diphenyl-1H-pyrazol-4-yl)methyl 3-phenylacrylate. The correlation coefficient r2 between observed activity of testing set and training set was found to be 0.819 for the EGFR model and 0.861 for the HER-2 model, which confirm that the QSAR models built are both acceptable. The molecules aligned with the iso-surfaces of the 3D QSAR model coefficients on electrostatic potential grids and van der Waals grids are shown in Fig. 5. Electrostatic map indicates red contours around regions where high electron density (negative charge) is expected to increase activity, and blue contours represent areas where low electron density (partial positive charge) is expected to increase activity. Similarly, steric map indicates areas where steric bulk is predicted to increase (green) or decrease (yellow) activity. As for EGFR, on the ring A, a slightly bulk and low negative charged group on the para-position would facilitate the inhibitory activity, which is in good agreement with the fact that OMe-substituent compounds display more potent activity than that of F- or Cl-substituent. Meanwhile, on ring B, it is suggested that compounds with low negative charged and slightly bulk R2 group on the para-position might make the efficiency better. Similar trends were observed for HER-2 model. The 3D-QSAR models correlate well with the experimental data and could provide insights on further discovery of more potent EGFR and HER-2 inhibitors.
Conclusion
In the present work, a series of novel potential anticancer agents (1e–30e) was synthesized and evaluated for their inhibitory activity against EGFR and HER-2 and antiproliferative activities against MCF-7 cell lines and B16-F10 cell lines. Compound 30e demonstrated the most potent inhibitory activity against EGFR and HER-2 with IC50 of 0.21 ± 0.05 μM and 1.08 ± 0.15 μM, which can also inhibit the growth of MCF-7 and B16-F10 cells with IC50 values of 0.30 ± 0.04 and 0.44 ± 0.05 μM respectively. Molecular docking showed that compound 30e bound nicely to the ATP binding site by hydrogen bonds which might play a crucial role in its EGFR/HER-2 inhibition and antiproliferative activities. 3D-QSAR models were also built to provide more pharmacophore understanding that could be useful for the design and discovery of novel anticancer agents with dual EGFR and HER-2 inhibitory activity.Experimental part
Chemistry
All chemicals and reagents used in the current study were of analytical grade. Melting points (uncorrected) were determined on an XT4 MP apparatus (Taike Corp., Beijing, China). All the 1H NMR spectra were recorded on a Bruker DPX300 model Spectrometer in DMSO-d6 and chemical shifts were reported in ppm (δ). ESI-MS spectra were recorded on a Mariner System 5304 Mass spectrometer. Elemental analyses were performed on a CHN–O-Rapid instrument. TLC was performed on the glass backed silica gel sheets (Silica Gel 60 GF254) and visualized in UV light (254 nm).General synthetic procedure of (1,3-diphenyl-1H-pyrazol-4-yl)methanol
A mixture of para-substituted acetophenone (a) (20 mmol) and phenylhydrazine hydrochloride (25 mmol) coupled with sodium acetate (40 mmol) was dissolved in anhydrous ethanol to yield compounds b. Then compounds b and POCl3 (8 mL) was added to a cold solution of DMF (10 mL), respectively. The mixture was stirred at 50–60 °C for 6 h. After cooling, the mixture was basified to pH 8 by the dropwise addition of 50% NaOH solution under ice bath, the precipitate was filtered and recrystallized from ethanol to form compounds c. Then, compounds c and 1.5 eq. sodium borohydride were dissolved in anhydrous CH2Cl2 solution and refluxed for 8 h to gain the desired compound d.General synthetic procedure of target compounds 1e–30e
Compounds 1e–30e were synthesized by coupling compounds d with cinnamic acids (1 eq.), with the help of DMAP (1.2 eq.) and K2CO3 (3 eq.). The mixture was refluxed in anhydrous CH2Cl2 for 8–10 h. The products were extracted with ethyl acetate. The extract was washed successively with 10% HCl, saturated NaHCO3 and water, respectively, then dried over anhydrous Na2SO4, filtered and evaporated. The residue was recrystallized from ethanol to gain the desired compounds.Spectral properties of pyrazole derivatives
Biological evaluation
Antiproliferation assay
The antiproliferative activities of the prepared compounds against MCF-7 and B16-F10 cells were evaluated as described in the literature34,35 with some modifications. Target tumor cell lines were grown to log phase in RPMI 1640 medium supplemented with 10% fetal bovine serum. After diluting to 2 × 104 cells per mL with the complete medium, 100 μL of the obtained cell suspension was added to each well of 96-well culture plates. The subsequent incubation was permitted at 37 °C, 5% CO2 atmosphere for 24 h before the cytotoxicity assessments. Tested samples at pre-set concentrations were added to six wells with Erlotinib co-assayed as positive references. After 48 h exposure period, 40 μL of PBS containing 2.5 mg mL−1 of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added to each well. Four hours later, 100 μL extraction solution (10% SDS–5% isobutyl alcohol–0.01 M HCl) was added. After an overnight incubation at 37 °C, the optical density was measured at a wavelength of 570 nm on an ELISA microplate reader. In all experiments three replicate wells were used for each drug concentration. Each assay was carried out for at least three times. The results were summarized in Table 2. All data are presented as mean ± SD unless otherwise noted.Enzyme inhibitory assay
A 1.6 kb cDNA encoded for the EGFR cytoplasmic domain (EGFR-CD, amino acids 645–1186) and 1.7 kb cDNA encoded for human HER-2 cytoplasmic domain (HER-2-CD, amino acids 676–1245) were cloned into baculoviral expression vectors pBlueBacHis2B and pFASTBacHTc (Huakang Company, China), respectively. A sequence that encodes (His)6 was located at the 50 upstream to the EGFR and HER-2 sequences. Sf-9 cells were infected for three days for protein expression. Sf-9 cell pellets were solubilized at 0 °C in a buffer at pH 7.4 containing 50 mM HEPES, 10 mM NaCl, 1% Triton, 10 μM ammonium molybdate, 100 μM sodium vanadate, 10 μg mL−1 aprotinin, 10 μg mL−1 leupeptin, 10 μg mL−1 pepstatin, and 16 μg mL−1 benzamidine HCl for 20 min followed by 20 min centrifugation. Crude extract supernatant was passed through an equilibrated Ni-NTA superflow packed column and washed with 10 mM and then 100 mM imidazole to remove nonspecifically bound material. Histidine tagged proteins were eluted with 250 and 500 mM imidazole and dialyzed against 50 mM NaCl, 20 mM HEPES, 10% glycerol, and 1 μg mL−1 each of aprotinin, leupeptin, and pepstatin for 2 h. The entire purification procedure was performed at 4 °C or on ice.Both EGFR and HER-2 kinase assays were set up to assess the level of autophosphorylation based on DELFIA/Time-Resolved Fluorometry. Compounds 1e–30e were dissolved in 100% DMSO and diluted to the appropriate concentrations with 25 mM HEPES at pH 7.4. In each well, 10 μL compound was incubated with 10 μL (5 ng for EGFR or 12.5 ng for HER-2) recombinant enzyme (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 dilution in 100 mM HEPES) for 10 min at room temperature. Then, 10 μL of 5 mM buffer (containing 20 mM HEPES, 2 mM MnCl2, 100 μM Na3VO4, and 1 mM DTT) and 20 μL of 0.1 mM ATP–50 mM MgCl2 were added for 1 h. Positive and negative controls were included in each plate by incubation of enzyme with or without ATP-MgCl2. At the end of incubation, liquid was aspirated, and plates were washed three times with wash buffer. A 75 μL (400 ng) sample of europium labeled anti-phosphotyrosine antibody was added to each well for another 1 h of incubation. After washing, enhancement solution was added and the signal was detected by Victor (Wallac Inc.) with excitation at 340 nm and emission at 615 nm. The percentage of autophosphorylation inhibition by the compounds was calculated using the following formula: 100% − [(negative control)/(positive control − negative control)]. The IC50 was obtained from curves of percentage inhibition with eight concentrations of compound. As the contaminants in the enzyme preparation are fairly low, the majority of the signal detected by the anti-phosphotyrosine antibody is from EGFR or HER-2. All data are presented as mean ± SD unless otherwise noted.
80 dilution in 100 mM HEPES) for 10 min at room temperature. Then, 10 μL of 5 mM buffer (containing 20 mM HEPES, 2 mM MnCl2, 100 μM Na3VO4, and 1 mM DTT) and 20 μL of 0.1 mM ATP–50 mM MgCl2 were added for 1 h. Positive and negative controls were included in each plate by incubation of enzyme with or without ATP-MgCl2. At the end of incubation, liquid was aspirated, and plates were washed three times with wash buffer. A 75 μL (400 ng) sample of europium labeled anti-phosphotyrosine antibody was added to each well for another 1 h of incubation. After washing, enhancement solution was added and the signal was detected by Victor (Wallac Inc.) with excitation at 340 nm and emission at 615 nm. The percentage of autophosphorylation inhibition by the compounds was calculated using the following formula: 100% − [(negative control)/(positive control − negative control)]. The IC50 was obtained from curves of percentage inhibition with eight concentrations of compound. As the contaminants in the enzyme preparation are fairly low, the majority of the signal detected by the anti-phosphotyrosine antibody is from EGFR or HER-2. All data are presented as mean ± SD unless otherwise noted.
Docking simulations
The three-dimensional structures of the aforementioned compounds were constructed using Chem. 3D ultra 12.0 software [Chemical Structure Drawing Standard; Cambridge Soft corporation, USA (2010)], then they were energetically minimized by using MMFF94 with 5000 iterations and minimum RMS gradient of 0.10. The crystal structures of EGFR domain bound to Erlotinib (PDB Code: 1M17.pdb) and HER-2 domain bound to 03Q (PDB Code: 3PP0.pdb) complexes were retrieved from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). All bound waters and ligands were eliminated from the protein and the polar hydrogen was added to the proteins. Molecular docking of all thirty compounds was then carried out using the Discovery Stutio (version 3.1) as implemented through the graphical user interface CDocker protocol. The whole EGFR/HER-2 complex was defined as a receptor and the site sphere was selected based on the ligand binding location of Erlotinib/03Q, then the Erlotinib/03Q molecule was removed and 30e was placed during the molecular docking procedure. Types of interactions of the docked protein with ligand were analyzed after the end of molecular docking.QSAR model
Among all the 30 compounds, 80% (i.e., 24) were utilized as a training set for QSAR (quantitative structure–activity relationship) modeling. The remaining 20% (i.e., 6) were chosen as an external test subset for validating the reliability of the QSAR model by the Diverse Molecules protocol in Discovery Studio 3.1. The selected test compounds were: 5e, 8e, 10e, 15e, 28e and 29e. The inhibitory activity of the compounds in literature [IC50 (mol L−1)] was initially changed into the minus logarithmic scale [pIC50 (mol L−1)] and then used for subsequent QSAR analysis as the response variable. In Discovery Studio, the CHARMm force field is applied and the electrostatic potential together with the van der Waals potential are treated as separate terms. As the electrostatic potential probe, A + le point change is used while distance-dependent dielectric constant is used to mimic the solvent effect. As for the van der Waals potential, a carbon atom with a radius of 1.73 Å is used as a probe. A partial least-squares (PLS) model is built using energy grids as descriptors. QSAR models were built by using the Create 3D QSAR Model protocol in Discovery Studio 3.1.Acknowledgements
This work was supported by Natural Science Foundation of Jiangsu Province (no. BK20130554).References
- K. Shantaram and K. B. John, Med. Res. Rev., 2006, 26, 569 CrossRef PubMed.
- Y. Liu, J. Fang, Y. J. Kim, M. K. Wong and P. Wang, Mol. Pharmaceutics, 2014, 11, 1651 CAS.
- Y. Liu, J. Man, M. K. Wong, K. I. Joo and P. Wang, BioMed Res. Int., 2013, 2013, 378380 Search PubMed.
- H. Gao, Q. Zhang, Z. Yu and Q. He, Curr. Pharm. Biotechnol., 2014 DOI:10.2174/1389201015666140617092552.
- L. Yi and S. G. Nathanael, Nat. Chem. Biol., 2006, 2, 7 CrossRef PubMed.
- R. P. Eric, B. A. Jeffrey, O. S. Harriet, I. O. Tudor, A. S. Larry and J. H. Helen, Annu. Rev. Physiol., 2008, 70, 165 CrossRef PubMed.
- S. Ayana, C. Sarat and G. Heidi, Cancer Res., 2008, 68, 589 CrossRef PubMed.
- Y. Yarden and M. X. Sliwkowski, Nat. Rev. Mol. Cell Biol., 2001, 2, 127 CrossRef CAS PubMed.
- T. Holbro and N. E. Hynes, Annu. Rev. Pharmacol. Toxicol., 2004, 44, 195 CrossRef CAS PubMed.
- J. R. Woodburn, Pharmacol. Ther., 1999, 82, 241 CrossRef CAS.
- A. Wells, Adv. Cancer Res., 2000, 78, 31 CrossRef CAS.
- A. J. Bridges, Curr. Med. Chem., 1999, 6, 825 CAS.
- D. H. Boschelli, Drugs Future, 1999, 24, 515 CrossRef CAS.
- J. Anido, P. Matar, J. Albanell, M. Guzman, F. Rojo, J. Arribas, S. Averbuch and J. Baselga, Clin. Cancer Res., 2003, 9, 1274 CAS.
- V. Chandregowda, G. V. Rao and G. C. Reddy, Org. Process Res. Dev., 2007, 11, 813 CrossRef CAS.
- F. Wei, B. X. Zhao, B. Huang, L. Zhang, C. H. Sun and W. L. Dong, Bioorg. Med. Chem. Lett., 2006, 16, 6342 CrossRef CAS PubMed.
- Y. S. Xie, X. H. Pan, B. X. Zhao, J. T. Liu, D. S. Shin and J. H. Zhang, J. Organomet. Chem., 2008, 693, 1367 CrossRef CAS PubMed.
- J. Das, S. Pany, S. Panchal, A. Majhi and G. M. Rahman, Bioorg. Med. Chem., 2011, 19, 6196 CrossRef CAS PubMed.
- B. Insuasty, A. Tigreros, F. Orozco, J. Quiroga, R. Abonia and M. Nogueras, Bioorg. Med. Chem., 2010, 18, 4965 CrossRef CAS PubMed.
- Y. Qian, H. J. Zhang, H. Zhang and H. L. Zhu, Bioorg. Med. Chem., 2010, 18, 4991 CrossRef CAS PubMed.
- D. D. Li, P. C. Lv, H. Zhang, H. J. Zhang, Y. P. Hou, K. Liu, Y. H. Ye and H. L. Zhu, Bioorg. Med. Chem., 2011, 19, 5012 CrossRef CAS PubMed.
- G. Szabo, J. Fischer, A. K. Varga and K. Gyires, J. Med. Chem., 2008, 51, 142 CrossRef CAS PubMed.
- A. Tanitame, Y. Oyamada, K. Ofuji, H. Terauchi, M. Kawasaki, M. Wachi and J. Yamagishi, Bioorg. Med. Chem. Lett., 2005, 15, 4299 CrossRef CAS PubMed.
- S. M. Rida, M. N. S. Saudi, A. M. Youssef and M. A. Halim, Lett. Org. Chem., 2009, 6, 282 CrossRef CAS.
- J. Regan, S. Breitfelder, P. Cirillo, T. Gilmore, A. G. Graham, E. Hickey, B. Klaus, J. Madwed, M. Moriak, N. Moss, C. Pargellis, S. Pav, A. Proto, A. Swinamer, L. Tong and C. Torcellini, J. Med. Chem., 2002, 45, 2994 CrossRef CAS PubMed.
- M. G. Brasca, C. Albanese, R. Amici, D. Ballinari, L. Corti, V. Croci, D. Fancelli, F. Fiorentini, M. Nesi, P. Orsini, F. Orzi, W. Pastori, E. Perrone, E. Pesenti, P. Pevarello, S. F. Riccardi, F. Roletto, P. Roussel, M. Varasi, A. Vulpetti and C. Mercurio, ChemMedChem, 2007, 2, 841 CrossRef CAS PubMed.
- P. Pevarello, D. Fancelli, A. Vulpetti, R. M. Amici, V. Villa, P. Pittalà, A. Vianello, M. Cameron, C. Ciomei, J. R. Mercurio, F. Bischoff, M. Roletto and M. G. Varasi, Bioorg. Med. Chem. Lett., 2006, 16, 1084 CrossRef CAS PubMed.
- M. R. Michaelides, PCT Int. Appl., 2010, 6, 5825 Search PubMed.
- B. Insuasty, A. Tigreros, F. Orozco, J. Quiroga, R. Abonia, M. Nogueras, A. Sanchez and J. Cobo, Bioorg. Med. Chem., 2010, 18, 4965 CrossRef CAS PubMed.
- M. Labbozzetta, R. Baruchello, P. Marchetti, M. C. Gueli, P. Poma, M. Notarbartolo, D. Simoni and N. D. Alessandro, Chem.-Biol. Interact., 2009, 181, 29 CrossRef CAS PubMed.
- C. Mariano, M. Marder, V. C. Blank and L. P. Roguin, Bioorg. Med. Chem., 2006, 14, 2966 CrossRef PubMed.
- S. Dallavalle, R. Cincinelli, R. Nannei, L. Merlini, G. Morini, S. Penco, C. Pisano, L. Vesci, M. Barbarino, V. Zuco, M. D. Cesare and F. Zunino, Eur. J. Med. Chem., 2009, 44, 1900 CrossRef CAS PubMed.
- Y. Luo, K. M. Qiu, X. Lu, K. Liu, J. Fu and H. L. Zhu, Bioorg. Med. Chem., 2011, 19, 4730 CrossRef CAS PubMed.
- C. Bhattacharya, Z. Yu, M. J. Rishel and S. M. Hecht, Biochemistry, 2014, 53, 3264 CrossRef CAS PubMed.
- Z. Yu, R. M. Schmaltz, T. C. Bozeman, R. Paul, M. J. Rishel, K. S. Tsosie and S. M. Hecht, J. Am. Chem. Soc., 2013, 135, 2883 CrossRef CAS PubMed.
Footnote |
| † These two authors equally contributed to this paper. |
| This journal is © The Royal Society of Chemistry 2014 |