DNA aptamer-based surface plasmon resonance sensing of human C-reactive protein†
Xiaohai Yang,
Yaning Wang,
Kemin Wang*,
Qing Wang,
Pei Wang,
Min Lin,
Nandi Chen and
Yuyu Tan
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Key Laboratory for Bio-Nanotechnology and Molecular Engineering of Hunan Province, Hunan University, Changsha 410082, China. E-mail: kmwang@hnu.edu.cn; Fax: +86 731 88821566; Tel: +86 731 88821566
First published on 4th July 2014
Abstract
Human C-reactive protein (CRP) is a powerful predictor and risk factor of inflammation and cardiovascular disease. We have obtained high-affinity DNA aptamers selected by an immobilization-free screening method, and the lowest Kd value of the aptamers is 3.9 nM. The affinity and specificity of the aptamers have been further investigated using aptamer-based SPR analysis, which will be of benefit for constructing DNA aptamer-based sensors for CRP detection.
Human C-reactive protein (CRP) is a major acute-phase reactant protein produced in the liver, and is composed of five monomeric subunits.1 CRP is an inflammatory marker that is currently used in clinical assays to predict the risk for cardiovascular disease (CVD), and has also been implicated in the pathogenesis of CVD.2,3 The most commonly used diagnostic methods are the antibody-based assays.4,5 Nowadays, some researchers try to isolate aptamers and develop the aptamer-based CRP sensors for diagnostics.6 Compared with antibodies, aptamers exhibit advantageous features:7 comparable affinity, easy in vitro synthesis, better stability and easy tagging with other functionalities. These advantages make them very promising in analytical and diagnostic applications.
Conventional methods for aptamer selection mainly rely on immobilizing targets or ssDNA library on solid supports to isolate target-binding aptamers from the random library.8–11 However, the immobilization of ssDNA library may cause steric hindrance to affect the binding to targets, and immobilization of targets may induce their conformational change. It was reported that the two aptamers,12,13 a 44-base RNA and a 40-base ssDNA selected by surface immobilization of CRP, specifically bound to monomeric but not pentameric form of CRP.14 Hence, there is an urgent need to isolate aptamers specific to CRP by adopting immobilization-free selection methods.
Some immobilization-free selection methods were developed to avoid the conformational change of targets, such as membrane filtration and capillary electrophoresis.15,16 However, filtration based methods suffer strong nonspecific binding toward targets and oligonucleic acids, and are limited to large target molecules,17 while capillary electrophoresis based methods usually need specialized instruments. Nevertheless, Park reported an immobilization-free selection method assisted by graphene oxide (GO-SELEX).18 It allowed the binding between target and ssDNA library in the homogeneous solution, and separated unbound DNA by the non-specific adsorption of ssDNA library on GO. The method not only avoids the immobilization of target or ssDNA library, but also overcomes the above described drawbacks of membrane filtration and capillary electrophoresis.
Here, a modified immobilization-free GO-SELEX strategy was applied to select aptamers target to CRP. The affinity and specificity of the aptamers were characterized by surface plasmon resonance (SPR). Our work not only isolated CRP-specific aptamers for constructing aptamer-based sensors of CRP detection, but also verified that the GO-SELEX strategy could act as a flexible aptamer selection platform against other targets.
The individual steps involved in one modified GO-SELEX round are illustrated in Scheme 1. In the first step of positive selection, the DNA library was firstly mixed with GO. This step would allow the library molecules to be absorbed on GO via π–π stacking. Then the mixture was incubated with the target protein (CRP). The potential aptamers for CRP are released from GO while unbound DNA and weakly bound DNA remain on GO. Subsequently, the released DNA/CRP complex was isolated by centrifugation, and the precipitate with DNA bound GO was discarded. The collected DNA/CRP complex was desalted and then amplified by PCR, and the ssDNA was generated from dsDNA PCR product by biotin–streptavidin separations for the next round of selection.19 In addition, counter selection was simultaneously performed from the 7th round to eliminate false positive binding of targets which are more likely to be present in serum. The counter-SELEX step was conducted by mixing the counter targets, including bovine serum albumin (BSA), human serum albumin (HSA) and myoglobin, with DNA library from the previous round and then incubating the mixture with GO solution. Any DNAs absorbed on GO during this process are likely to be specific aptamers for CRP. The potential aptamers absorbed on GO were collected by centrifugation and directly used to the positive selection. The modification for GO-SELEX method was aimed at improving the affinity of the selected aptamers, because the weakly bound DNA was firstly absorbed on GO and hard to form the DNA/CRP complex. However, for this method, isoelectric point (PI) of target proteins must be neutral or closer to the pH of binding buffer in order to minimize polar and charged interaction between protein and GO, since the GO is negatively charged due to the presence of the carboxylic acid and phenolic hydroxyl groups.
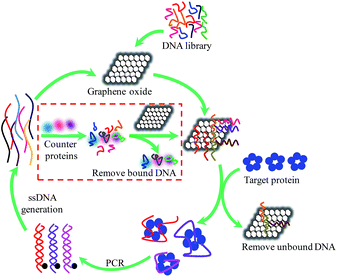 | ||
| Scheme 1 Schematic illustration of the steps involved in the modified GO-SELEX procedure of aptamer identification. | ||
In this process of selection, the DNA library was composed of 40 bp random region in the central and two 20 bp constant primer regions at both ends. Two short 20 bp DNA strands were designed to block the primer-binding sites and limit their involvement in the large conformational change in specific response to CRP binding. The sequences of DNA library and primers used in this selection were listed in Table S1 of ESI.† In order to enhance the stringency of the selection, the adsorption ratio of DNA library on GO was investigated as shown in Fig. S1.† The adsorption ratio was found to be 0.25 nmol mg−1 of GO, which was saturated to absorb any ssDNAs in the DNA library.
For monitoring the progress of selection, enrichment assays from the 3rd round to the 10th round were conducted by SPR as shown in Fig. S2a of ESI.† In brief, adding the prepared ssDNA library of each round to the CRP-immobilized Au film induced the change of resonance angle (Δθ). Δθ was significantly increased with increasing of the SELEX round (Fig. S2b†). In the 7th round of selection, the change of resonance angle attained the biggest value. The result indicated that the DNA specific to CRP was dominated in the DNA library. A further three rounds of selection could not increase the change of resonance angle. Hence, the recovered and purified ssDNAs from the 7th round selection were subjected to high-throughput sequencing.
According to the result of high-throughput sequencing, the top 20 highest repeated sequences were chosen and classified into nine groups based on their similarity of secondary structure as shown in Fig. S3 of ESI.† One sequence from each group was chosen as the candidate aptamer according to the repetitions of sequences for affinity determination. Finally, 9 full-length candidate aptamers were identified for affinity by SPR. As shown in Table 1, four aptamers CRP-80-2, CRP-80-5, CRP-80-13, and CRP-80-17 exhibited the apparent affinity for CRP, and their dissociation constants (Kd) were 1.3 μM, 24.8 nM, 40.7 nM and 3.9 nM, respectively. Aptamer CRP-80-17 was chosen as a representative of the selected aptamers for further study due to its lowest Kd. The full-length CRP-80-17 has 80 bp, but not all nucleotides are necessary for direct interaction with the target or for folding into the structure that facilitates target binding.20 In consideration of blocking the primer-binding sites in the process of selection, CRP-80-17 was optimized by cutting nucleotides of the two constant regions. Therefore, a shorter aptamer sequence with a Kd value of 16.2 nM was obtained, which contained only 40 bp and named as CRP-40-17. These results indicated that both the full-length aptamer CRP-80-17 and the shorter aptamer CRP-40-17 could bind to CRP with high affinity.
| Name | Sequence of random region (5′ to 3′)a | Kd |
|---|---|---|
| a The total sequence of candidate aptamers was 5′-AGC AGC ACA GAG GTC AGA TG-N40-CCT ATG CGT GCT ACC GTG AA-3′. Only the random regions were listed. | ||
| CRP-80-1 | TTTTAGATTTAGCTCTTATTTGTTCGAGCAATCCCAAAGA | — |
| CRP-80-2 | TACGAGCGGTGGTTTTACCCTGCAATACTTTTGGCTGTTA | 1.3 μM |
| CRP-80-3 | TCCATTATCAGGTTCTTTATTCTGTTGTTCAACTTATTAA | — |
| CRP-80-4 | GGATCTTCCCTCAATGTTTATTGTATATCTGTACTCGTAA | — |
| CRP-80-5 | TACTTATGCATTTCCTCCCACGATCTTATTTGAGAGTGAC | 24.8 nM |
| CRP-80-6 | TCCAATTCAATTCATTTCTGAACTTAGTCGGCACTTTGAC | — |
| CRP-80-7 | TATACTTCTAAAATTTGTTTGTATCTACGATGTTCTTCGT | — |
| CRP-80-13 | GCCGGATCGCGCCCCCCGTGTAAGAGGCACCCCCGGTCCC | 40.7 nM |
| CRP-80-17 | CCCCCGCGGGTCGGCTTGCCGTTCCGTTCGGCGCTTCCCC | 3.9 nM |
Aptamers are usually immobilized on the surface of supports during the construction of sensors. Thus, we investigated the ability of the immobilized aptamers to bind CRP using aptamer-based SPR analysis. Four biotinylated aptamers (Table S2†), CRP-80-17-5′ biotin, CRP-80-17-3′ biotin, CRP-40-17-5′ biotin and CRP-40-17-3′ biotin, were designed for this examination. Firstly, biotinylated aptamers were immobilized on the avidin coated Au film surface through biotin at theirs 3′ or 5′ terminus. Then, different concentrations of CRP flowed over the aptamer-immobilized Au film surface. The aptamer specifically bound to CRP and caused a change of resonance angle. All the four aptamers showed apparently binding to CRP no matter which terminus was immobilized on the Au film (Fig. 1). These results indicated that the immobilized CRP-80-17 and CRP-40-17 still preserved the affinity to CRP. Among the four immobilized aptamers, CRP-40-17-3′ biotin showed the best response to CRP with a linear range of 0–12.5 nmol L−1 (y = 0.0106 + 0.0044x), and a detection limit of 0.35 nmol L−1 (S/N = 3).
 | ||
| Fig. 1 The change of resonance angle of the four immobilized aptamers at various concentrations of CRP. | ||
Furthermore, the change of resonance angle was more significant when immobilizing the two aptamers through 3′ terminus than 5′ terminus. It may be attributed to the steric hindrance. The secondary structure of CRP-80-17 and CRP-40-17 has the apparent difference (Fig. S3 Group 9, and Fig. S4 of ESI†), and the residual ssDNA length at 3′ terminus was longer than 5′ terminus. Thus the aptamers immobilized through 3′ terminus were easier to form 3D structures for binding CRP.
Since the change of resonance angle was more apparent when immobilizing aptamers through 3′ terminus, the specificity of aptamers CRP-80-17 and CRP-40-17 was examined by immobilizing aptamer through 3′ terminus on the Au film. Five control proteins including human immunoglobulin G (IgG), HSA, haemoglobin, BSA and myoglobin were chosen, which based on the possible interference to CRP in practical usage. As shown in Fig. 2, it is clear that the aptamers CRP-80-17 and CRP-40-17 specifically binds to CRP against other control proteins, therefore, can serve as candidates for sensor applications.
Conclusions
In conclusion, we successfully selected and identificated the ssDNA aptamers for CRP using a modified immobilization-free GO-SELEX. This method avoids the conformational change induced by immobilizing CRP on a solid surface and improves the affinity of the selected aptamers. The lowest Kd value of the aptamers is 3.9 nM. Moreover, a shorter aptamer CRP-40-17 was obtained and exhibited good affinity and specificity toward CRP. The immobilized aptamer CRP-80-17 and CRP-40-17 also showed apparently binding to CRP. Thus, the results will be helpful to construct aptamer-based sensors for CRP detection.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21190040, 21375034, 21175035), National Basic Research Program (2011CB911002), International Science & Technology Cooperation Program of China (2010DFB30300), and Program of Young Teacher Grown of Hunan University.Notes and references
- M. B. Pepys and G. M. Hirschfield, J. Clin. Invest., 2003, 111, 1805–1812 CrossRef CAS.
- S. Black, I. Kushner and D. Samols, J. Biol. Chem., 2004, 279, 48487–48490 CrossRef CAS PubMed.
- S. R. Ji, Y. Wu, L. Zhu, L. A. Potempa, F. L. Sheng, W. Lu and J. Zhao, FASEB J., 2007, 21, 284–294 CrossRef CAS PubMed.
- S. K. Vashist, G. Czilwik, T. van Oordt, F. von Stetten, R. Zengerle, E. Marion Schneider and J. H. T. Luong, Anal. Biochem., 2014, 456, 32–37 CrossRef CAS PubMed.
- R. K. Gupta, A. Periyakaruppan, M. Meyyappan and J. E. Koehne, Biosens. Bioelectron., 2014, 59, 112–119 CrossRef CAS PubMed.
- M. V. Miramontes-Espino and M. M. Romero-Prado, Recent Pat. DNA Gene Sequences, 2013, 7, 195–206 CrossRef.
- A. D. Keefe, S. Pai and A. Ellington, Nat. Rev. Drug Discovery, 2010, 9, 537–550 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- R. Nutiu and Y. Li, Angew. Chem., Int. Ed., 2005, 117, 1085–1089 CrossRef.
- Y. Wu, S. Zhan, L. Wang and P. Zhou, Analyst, 2014, 139, 1550–1561 RSC.
- S. D. Kim, J. S. Ryu, H. K. Yi, S. C. Kim and B. T. Zhang, in Preliminary proceedings of the tenth international meeting on DNA computing, 2004, pp. 334–343 Search PubMed.
- C. J. Huang, H. I. Lin, S. C. Shiesh and G. B. Lee, Biosens. Bioelectron., 2010, 25, 1761–1766 CrossRef CAS PubMed.
- M. S. Wang, J. C. Black, M. K. Knowles and S. M. Reed, Anal. Bioanal. Chem., 2011, 401, 1309–1318 CrossRef CAS PubMed.
- T. Lakshmipriya, M. Fujimaki, S. C. Gopinath and K. Awazu, Langmuir, 2013, 29, 15107–15115 CrossRef CAS PubMed.
- P. Mallikaratchy, R. V. Stahelin, Z. Cao, W. Cho and W. Tan, Chem. Commun., 2006, 3229–3231 RSC.
- S. C. B. Gopinath, Anal. Bioanal. Chem., 2007, 387, 171–182 CrossRef CAS PubMed.
- J. W. Park, R. Tatavarty, D. W. Kim, H. T. Jung and M. B. Gu, Chem. Commun., 2012, 48, 2071–2073 RSC.
- C. Marimuthu, T. H. Tang, J. Tominaga, S. C. Tan and S. C. Gopinath, Analyst, 2012, 137, 1307–1315 RSC.
- S. D. Jayasena, Clin. Chem., 1999, 45, 1628–1650 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, including adsorption ratio of DNA library on GO, selection procedures, analysis of the result of high-throughput sequencing. See DOI: 10.1039/c4ra05011h |
| This journal is © The Royal Society of Chemistry 2014 |

