Understanding the fate of an anesthetic, nalorphine upon interaction with human serum albumin: a photophysical and mass-spectroscopy approach†
Abstract
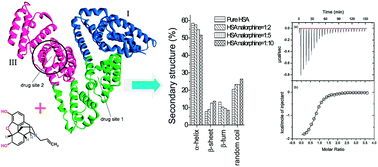
Nalorphine is an injectable opioid agonist–antagonist. For understanding its pharmacology, the binding mechanism of nalorphine to a model protein, human serum albumin (HSA), was probed by fluorescence and circular dichroism (CD) Fourier transform infrared spectroscopy (FTIR) and isothermal titration calorimetry (ITC) approaches. The binding affinity of the drug with the native HSA was 7.94 × 105 M−1 at 298 K. Meanwhile, the number of binding site was found to be approximately 1 from fluorescence, ITC and microTOF-Q data. Furthermore, the alterations of protein secondary structure in the presence of nalorphine were assessed by CD, 3D fluorescence and FTIR spectroscopy. Fluorescence resonance energy transfer (FRET) analysis proved the high probability of energy transfer from Trp residue to the drug molecule. The binding site of the drug on HSA was located on the motional restricted hydrophobic pocket of subdomain IIIA, namely site II, and the drug–protein complex was mainly stabilized by electrostatic forces.

 Please wait while we load your content...
Please wait while we load your content...