Design, synthesis, biological evaluation and molecular docking of novel metronidazole derivatives as selective and potent JAK3 inhibitors
Ya-Li Sang†
,
Yong-Tao Duan†,
Han-Yue Qiu,
Peng-Fei Wang,
Jigar A. Makawana,
Zhong-Chang Wang,
Hai-Liang Zhu* and
Zhen-Xiang He*
State Key Laboratory of Pharmaceutical Biotechnology, Nanjing University, Nanjing 210093, People's Republic of China. E-mail: zhuhl@nju.edu.cn; zxhe@nju.edu.cn; Fax: +86-25-83592672; Tel: +86-25-83592672
First published on 26th March 2014
Abstract
In the JAK/STAT pathway, sustainable activation of JAK with the capabilities of regulating cell growth and apoptosis can produce abnormal proliferation in tumor cells. A series of novel metronidazole derivatives containing the 1,4-benzodioxan moiety as potential inhibitors targeting JAK have been designed, synthesized and their biological activities were also evaluated. Among all synthesized compounds, compound 4t possessed the most potent antitumor activity against A549, Hela, HepG-2 and U251 in vitro, with IC50 values of 65, 21, 16 and 44 nM, respectively, which has been proved by the result of a flow cytometry (FCM) assay. Docking simulations demonstrated that compound 4t could bind tightly with the crystal structure of the JAK3 active site and act as a potential JAK3 inhibitor.
1. Introduction
The JAK/STAT (Janus associated kinase-signal transducer and activator of transcription) pathway involved in signal transduction by transmitting extracellular signals (growth promoting factors, cytokines or hormones) to the nucleus to control gene expression has a prominent part in regulating cell fates, such as cell growth, survival, differentiation, proliferation and apoptosis. Deregulated JAK/STAT signaling which has only recently begun to be explored is intimately linked to tumorigenesis,1 hematological malignancies,2 and myeloproliferative disorder.3 JAK family composed of JAK1-3 and Tyk2 is PTK (protein tyrosine kinase) binding to the juxtamembrane region of cytokine receptors.4,5 Triggered by dimerization of receptor, which is the result of binding of a ligand with its receptor, JAK will be activated. Subsequently activated JAK phosphorylates STAT, close behind is the dimerization of specific phosphorylated STAT proteins and ensuing activated STATs translocate into the nucleus, regarding as transcription factors to regulate gene expression.6,7 Following this molecular mechanism (Fig. 1), the JAK/STAT3 pathway has been found to be key events in the occurrence, growth and progression of tumors, as well as it may offer us targets for the cancer therapy.8The sustainable activation of STAT3 is required for the development of skin cancer,9 head and breast tumors10 etc. Furthermore, STAT3 plays a critical role in cancer inflammation and immunity.11 Since tumorigenesis is a multistage process, activated STAT3 will not lead to cell deformation alone, but is the key inducing factor of tumorigenesis. STAT3, activated by JAK, partners JAK in vivo.12 Consequently, JAK is actively pursued as targets in the research of therapeutic agents, not only against cancer, but also many other diseases by blocking the signal transduction of JAK/STAT3 pathway.
A new phase of targeted therapies is rapidly emerging. Meanwhile, compounds that target PTK are in preclinical and clinical research. These compounds are generally separated into two classes: monoclonal antibodies and small-molecule anti-tumor agents,13 and the current progress of the small molecules acting as TKIs (tyrosine kinases inhibitors) is promising.14 During the research of JAK inhibitors, a host of chemotypes have been designated (Fig. 2).15 Among which, Ruxolitinib (INCB18424) serving as potent and selective JAK inhibitor recently gets approval of FDA, supplying the best available therapy for myelofibrosis,16 erythrocytosis17 and other indications18 by interrupting JAK/STAT signal transduction. Equally as INCB18424, notably, AZD1480, MK0457 also have imidazole ring which has an unequalled place in the field of synthesizing medicine.19 Imidazole derivatives are active against bacteria, viruses, fungi and some imidazole drugs have anticancer properties. Particularly, imidazole ring is capable of high specificity, selectivity and affinity bonding to proteins by hydrogen bonds or metals as a ligand and drugs,20 which has been demonstrated by the following docking simulations. Meanwhile, within 1,4-benzodioxan, the oxygen at location 4 maintains the stability of an optimum conformation for drug–receptor interaction complexes. On the other hand, the oxygen at location 1 contributes to the binding by interacting with receptor polar pockets,21 which also occurs in our docking simulations. Moreover, 1,4-benzodioxan derivatives reveal powerful biological activities (anticancer,22 anti-fungal and antipsychotic activities23). For example, 1,2,4-triazoles having 1,4-benzodioxan (A) fragments was synthesized as potential MetAP2 inhibitors,22 WB 4101 (B) were regarded as subtype-selective alpha1-adrenoreceptor antagonists (Fig. 3).24
Furthermore, our research team has already made some progress in the research of imidazole derivatives as TKIs.25,26 All of these inspire us to do further research in the direction of finding new imidazole derivatives containing 1,4-benzodioxan moiety as promising TKIs. Herein we depict the synthesis, antitumor activity, succeeding structure–activity relationship (SAR) and Flow Cytometry (FCM) studies of target compounds. Docking simulations are also accomplished by binding the X-ray crystallographic structure of the JAK3 with inhibitors to predict the binding modes and affinity.
2. Result and discussion
2.1. Chemistry
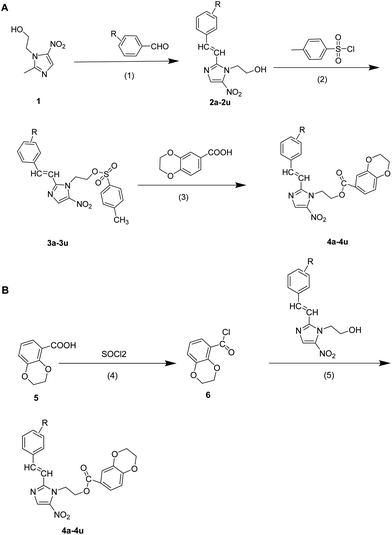
The synthetic routes of a series of novel metronidazole derivatives including 1,4-benzodioxan moiety targeting JAK (4a–4u) were outlined in Scheme 1A. First of all, different substituted (E)-2-(5-nitro-2-styryl-1H-imidazol-1-yl)ethan-1-ol (2a–2u) were synthesized by reaction of metronidazole with diverse substituted benzaldehydes in DMSO with the presence of sodium methylate as catalyst. Compounds 2a–2u were converted into different substituted (E)-2-(5-nitro-2-styryl-1H-imidazol-1-yl)ethyl 4-methylbenzenesufonate (3a–3u) by reaction with 4-toluene sulfonyl chloride using triethylamine as promoter in CH2Cl2 at room temperature in high yield. Finally, compounds 3a–3u and 1,4-benzodioxan were dissolved in DMF under a refluxing and K2CO3-base condition, stirring together the reactants to an adequate degree. The target compounds could be obtained with yields in the range of 60–80%. Meanwhile, the desired products could be produced following another pathway outlined in Scheme 1B. Initially, treat 1,4-benzodioxan in refluxing thionyl chloride with a few drops of DMF for 4 h to get 2,3-dihydrobenzo[b][1,4]dioxine-5-carbonyl chloride (6). Then, different substituted (E)-2-(5-nitro-2-styryl-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-6-carboxylate (4a–4u) were synthesized in CH2Cl2 by treating compounds 3a–3u with solution of compound 6 in CH2Cl2 dropwise using triethylamine as promoter in high yield.2.2. Bioassay
| Compounds | R | IC50 (µM) | |||
|---|---|---|---|---|---|
| A549 | Hela | U251 | HepG-2 | ||
| 3a | o-F | 3.01 | 3.35 | 5.75 | 2.74 |
| 3b | m-F | 2.87 | 0.98 | 1.66 | 2.73 |
| 3c | p-F | 3.85 | 1.20 | 2.87 | 1.80 |
| 3d | o-Cl | 5.08 | 2.17 | 2.41 | 2.17 |
| 3e | m-Cl | 2.94 | 2.41 | 1.71 | 1.24 |
| 3f | p-Cl | 4.57 | 3.43 | 2.15 | 3.67 |
| 3g | o-Br | 6.67 | 4.35 | 2.45 | 5.34 |
| 3h | m-Br | 3.94 | 1.27 | 1.19 | 1.28 |
| 3i | p-Br | 4.17 | 2.84 | 3.43 | 2.35 |
| 3j | o-CH3 | 10.18 | 5.36 | 4.93 | 7.06 |
| 3k | m-CH3 | 5.21 | 3.39 | 3.05 | 2.97 |
| 3l | p-CH3 | 9.29 | 3.07 | 2.08 | 5.13 |
| 3m | o-NO2 | 8.07 | 6.83 | 5.46 | 6.25 |
| 3n | m-NO2 | 6.88 | 4.49 | 1.11 | 4.59 |
| 3o | p-NO2 | 8.81 | 4.75 | 7.45 | 4.97 |
| 3p | o-OCH3 | 9.22 | 1.38 | 3.34 | 3.03 |
| 3q | m-OCH3 | 4.14 | 2.31 | 2.94 | 2.94 |
| 3r | p-OCH3 | 7.15 | 4.46 | 5.86 | 2.99 |
| 3s | 2-Cl-6-F | 1.74 | 0.52 | 1.69 | 1.18 |
| 3t | 2,4-2Cl | 1.68 | 0.98 | 0.87 | 0.72 |
| 3u | –H | 15.26 | 9.68 | 6.13 | 12.24 |
| Compounds | R | IC50 (nM) | ||||
|---|---|---|---|---|---|---|
| A549 | Hela | U251 | HepG-2 | JAK 3 | ||
| 4a | o-F | 108 | 33 | 132 | 161 | 15 |
| 4b | m-F | 95 | 48 | 59 | 87 | 23 |
| 4c | p-F | 161 | 53 | 48 | 118 | 46 |
| 4d | o-Cl | 266 | 76 | 65 | 232 | 49 |
| 4e | m-Cl | 101 | 47 | 74 | 94 | 31 |
| 4f | p-Cl | 383 | 53 | 193 | 133 | 33 |
| 4g | o-Br | 223 | 177 | 89 | 358 | 54 |
| 4h | m-Br | 116 | 62 | 63 | 103 | 42 |
| 4i | p-Br | 219 | 66 | 107 | 221 | 74 |
| 4j | o-CH3 | 435 | 91 | 215 | 363 | 67 |
| 4k | m-CH3 | 227 | 68 | 133 | 135 | 50 |
| 4l | p-CH3 | 309 | 71 | 162 | 106 | 46 |
| 4m | o-NO2 | 576 | 132 | 386 | 555 | 82 |
| 4n | m-NO2 | 264 | 94 | 164 | 249 | 25 |
| 4o | p-NO2 | 475 | 86 | 204 | 173 | 49 |
| 4p | o-OCH3 | 231 | 102 | 75 | 247 | 52 |
| 4q | m-OCH3 | 122 | 62 | 101 | 128 | 57 |
| 4r | p-OCH3 | 372 | 18 | 140 | 111 | 33 |
| 4s | 2-Cl-6-F | 84 | 25 | 69 | 76 | 13 |
| 4t | 2,4-2Cl | 65 | 21 | 44 | 16 | 9 |
| 4u | –H | 744 | 78 | 405 | 314 | 102 |
| Tofacitinib citrate | 114 | 5 | 27 | 12 | 1 | |
As depicted in Table 2, among these compounds, compound 4t has the most effective anti-proliferative ability (IC50 = 21 nM for Hela and IC50 = 44 nM for U251) in comparison to the positive control tofacitinib citrate (IC50 = 5 nM for Hela and IC50 = 27 nM for U251). Hela was found to display higher sensitivity toward these compounds than other cell lines. Nevertheless, A549 was observed to be relatively poor sensitive toward them with IC50 ranging from 65 to 744 nM. Regarding HepG-2, compounds 4d and 4t particularly revealed potent inhibiting effect (IC50 = 76 and 16 nM, respectively), compared to tofacitinib citrate (12 nM).
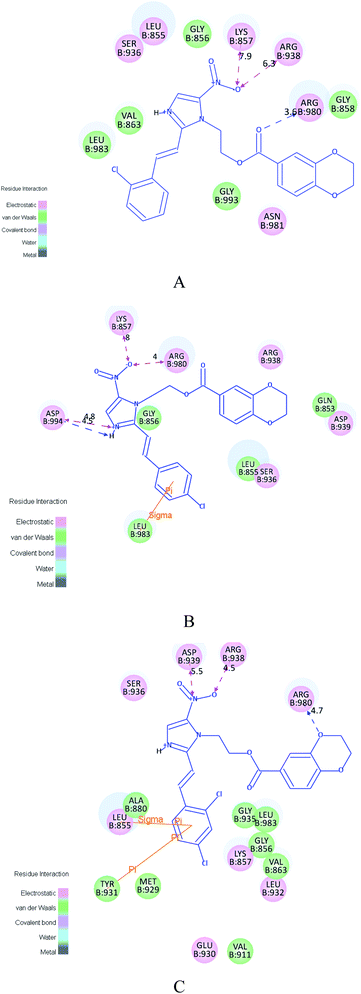
The ensuing structure–activity relationship (SAR) can be observed by the different JAK inhibitory activities of these compounds with varying substituted parts in Tables 1 and 2. By and large, the inhibitory abilities of 1,4-benzodioxan derivatives (4a–4u) became much better than 3a–3u. Primarily, to these compounds, structure–activity relationship (SAR) study showed that stronger electron-withdrawing substitute has better JAK inhibitory activities (e.g. 4b, 4e, 4h, 4k, 4n, 4q), whereas the difference was slight. Besides, we proceeded to detect the molecular docking of potent antitumor agents (4d, 4f, 4t) with the JAK3 crystal structure (PDB code: 3FUP). All docking programs were operated by Discovery Studio 3.5. The binding models of inhibitors (4d, 4f, 4t) with 3FUP were illustrated in Fig. 4. In Fig. 4C, benzene ring containing two electron-withdrawing halogen atoms tightly bound to 3FUP via one π–π interaction and one π–sigma interaction. Meanwhile, the two chlorine atoms of 4t interacted with amino acid residues of JAK3 by electrostatic interactions, van der Waals and covalent bonds. While, in Fig. 4A and B, one-substituted chlorine atom occupied smaller space and displayed weaker electronegativity compared to di-substituted moieties. The molecular docking results revealed that compound 4t bound stronger to 3FUP than 4d and 4f. The antitumor activities showed compounds with di-substituted moieties (e.g. 4t, 4s) indicated more improved antitumor activities than those with one-substituted atom (e.g. 4d, 4f). According to SAR analysis, the active gradient was 2,4-2Cl > 2-Cl-6-F > –F > –Cl > –Br > –OCH3 > –CH3 > H > –NO2. Moreover, a meta substitute (4e) had improved antitumor activity comparing to the ortho (4d) or para (4f) position. The foregoing SAR analysis showed that compounds 4t was the most effective JAK inhibitory agent, meanwhile, other compounds were also found to be potent inhibitors because of their effective antitumor activities, compared to the potent positive control tofacitinib citrate.
2.3. Molecular docking study
All our present efforts have been directed towards gaining better understanding on the effectiveness of the synthesized compounds and guiding further SAR studies. In order to achieve this goal, we proceeded to detect the molecular docking of potent antitumor agents (3t, 4t) with the JAK3 crystal structure (PDB code: 3FUP). All docking programs were operated by Discovery Studio 3.5. The binding models of inhibitors (3t, 4t) with 3FUP were illustrated in Fig. 6. The amino acid residues which bound with 3FUP were labeled. In the binding mode (Fig. 6B), compound 4t nicely bound to 3FUP via three hydrogen bonds, one π–π interaction, one π–sigma interaction and other interactions (e.g., electrostatic interaction, van der Waals). One oxygen atom on the triazole ring with ARG938 (distance N–O⋯H = 4.5 Å) and the nitrogen atom of –NO2 with ASP939 (distance O–N⋯H = 5.5 Å) made important contributions to the hydrogen bonding interaction. The third hydrogen bond was formed by oxygen atom on the 1,4-benzodioxan ring with ARG980 (distance H–O⋯H = 4.7 Å), together with the above two hydrogen bonds, being a probable crucial factors for its potent activity. In addition, the π–π interaction and π–sigma interaction were formed between the benzene ring and LEU855 (distance = 2.9 Å), TYR931 (distance = 6.3 Å), respectively. Besides, the binding model was enhanced by electrostatic interaction formed between compound 4t and residues (e.g. LYS857, LEU932, ASN981) and van der Waals formed by compound 4t and residues (e.g. GLY935, VAL863, ASP976). Meanwhile, it was observed that the potent inhibitor 4t was retained tightly by the binding pocket of 3FUP, from the receptor surface model in Fig. 6C. However, in Fig. 6A, compound 3t bound to 3FUP via two π–cation interactions and other interactions (e.g. electrostatic interaction, van der Waals). These molecular docking results showed that, compare the common structural part of 3t and 4t, the latter bound stronger to JAK3. Along with the bioassay data, the docking results revealed that compound 4t was potential JAK3 inhibitor.2.4. 3D-QSAR
To obtain a systematic SAR profile on synthesized compounds as JAK3 inhibitors and explore more powerful and selective dual antitumor agents, 21 compounds with accurate IC50 values against JAK3 were chosen as the model dataset to build 3D-QSAR model by create-in QSAR software of DS 3.5 (Discovery Studio 3.5, Accelrys, Co. Ltd). In general, the pIC50 scale (−log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50) was transformed from the acquired IC50 (µM) values of these synthesized compounds and used as a means in 3D-QSAR model to select activity conformations of the designed molecules and reasonably measure the synthesized antitumor agents. Illustrated in Table 3, the training and test set was randomly selected in due proportion that ratio of training set was 0.762, ratio of test set was 0.238 by the Diverse Molecules method of Discovery Studio 3.5. The success of this model relied on the acceptable study of docking protocol and bioassay results.
IC50) was transformed from the acquired IC50 (µM) values of these synthesized compounds and used as a means in 3D-QSAR model to select activity conformations of the designed molecules and reasonably measure the synthesized antitumor agents. Illustrated in Table 3, the training and test set was randomly selected in due proportion that ratio of training set was 0.762, ratio of test set was 0.238 by the Diverse Molecules method of Discovery Studio 3.5. The success of this model relied on the acceptable study of docking protocol and bioassay results.
| Compounds | JAK3 | Residual error | |
|---|---|---|---|
| Experiment pIC50 | Predicted pIC50 | ||
| a Compounds were selected as the test sets while the rest ones were in the training sets. | |||
| 4aa | 7.387 | 7.408 | −0.021 |
| 4ba | 7.319 | 7.427 | −0.108 |
| 4c | 7.292 | 7.239 | 0.053 |
| 4d | 7.222 | 7.312 | −0.09 |
| 4ea | 7.328 | 7.433 | −0.105 |
| 4f | 7.167 | 7.079 | 0.088 |
| 4g | 7.276 | 7.351 | −0.075 |
| 4h | 7.252 | 7.314 | −0.062 |
| 4i | 7.194 | 7.247 | −0.053 |
| 4ja | 7.018 | 7.115 | −0.097 |
| 4k | 7.207 | 7.135 | 0.072 |
| 4l | 7.148 | 7.091 | 0.057 |
| 4m | 6.991 | 6.921 | 0.07 |
| 4na | 7.027 | 7.16 | −0.133 |
| 4o | 7.065 | 7.124 | −0.059 |
| 4p | 7.284 | 7.235 | 0.049 |
| 4q | 7.42 | 7.483 | −0.063 |
| 4r | 7.201 | 7.251 | −0.05 |
| 4s | 7.602 | 7.537 | 0.065 |
| 4t | 7.678 | 7.614 | 0.064 |
| 4u | 7.108 | 7.021 | 0.087 |
Among the twenty docked poses, the alignment conformation of each molecule owning the lowest CDOCKER_INTERACTION_ENERGY was maintained in default situation. The 3D-QSAR model built by DS 3.5, clarified the crucial regions (steric or electrostatic) affecting the binding affinities. It was a PLS model set up 210 independent variables (conventional R2 = 0.8396). The experimental and predicted pIC50 values and relative residual values of the training set and test set molecules in 3D-QSAR model had been showed in Table 3. Moreover, the graphical relationship of actual and predicted values had been demonstrated in Fig. 7A, in which the plot of the actual IC50 versus the predicted values proved that this model was reliable in forecasting of activity for metronidazole derivatives containing 1,4-benzodioxan moiety.
A outline plot of the electrostatic field region favorable (in blue) or unfavorable (red) for antitumor activity based on JAK3 protein target were illustrated in Fig. 7B, while the energy grids corresponding to the favorable (in green) or unfavorable (yellow) steric effects for the JAK3 affinity were shown in Fig. 7C. It was widely acceptable that a better inhibitor based on the 3D-QSAR model should have strong van der Waals attraction in the green areas and a polar group in the blue electrostatic potential areas (which were dominant close to the skeleton). As shown in these two pictures, this promising model would provide a guideline to design and optimize more effective JAK3 inhibitors based on the metronidazole derivatives containing 1,4-benzodioxan moiety and pave the way for us to further study in future.
3. Conclusion
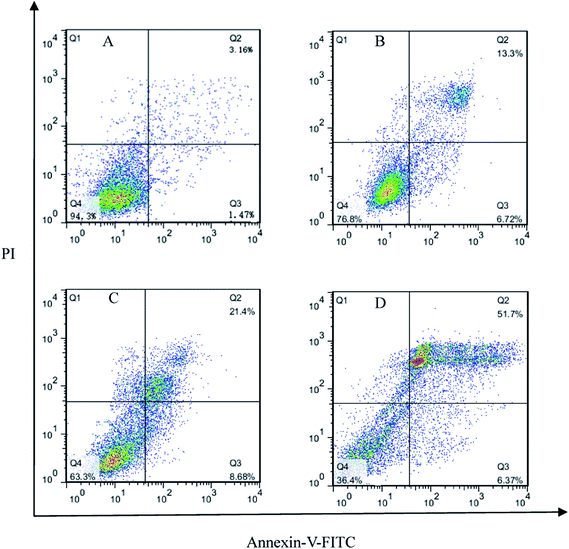
Summarily, we have designed and synthesized a novel series of metronidazole derivatives containing 1,4-benzodioxan moiety and evaluated their inhibitory activities against A549, Hela, U251 and HepG-2. Preliminary results illustrated that these compounds showed a promising prospect as antiproliferative agents. Among all compounds, compound 4t displayed potent biological activities (IC50 = 21 nM for Hela, IC50 = 44 nM for U251, IC50 = 16 nM for HepG-2 and IC50 = 65 nM for A549), which had been demonstrated by analysis of apoptosis using flow cytometry (FCM).Docking simulation provided further insight into the probable binding models and poses between the JAK3 protein and its ligand. It was observed that three hydrogen bonds, one π–π interaction, one π–sigma interaction and other interactions (e.g., electrostatic interaction, van der Waals) formed between compound 4t with the protein residues might play critical roles in its anticancer activities. The results showed that compound 4t as well as the other metronidazole derivatives has the potential to be selective and potent JAK inhibitor. Eventually, QSAR models had been created on the basis of previous antitumor assay and molecular docking study to offer a reliable tool for reasonable design of JAK inhibitors in future. The information of this QSAR study might help to search more promising anticancer agents.
4. Experiments
4.1. Materials and measurements
All chemicals utilized in present research were of analytical grade. Thin layer chromatography (TLC) was accomplished on silica gel plates (Silica Gel 60 GF254), the result could clearly be seen in UV light (254 nm). Purification of the desired products was performed by column chromatography. The amount of silica gel used in column chromatography was 50–100 times the weight charged on the column. Melting points were determined on a XT4 MP apparatus (Taike Corp, Beijing, China). All the 1H NMR spectra were recorded on a Bruker DPX 300 model Spectrometer in DMSO-d6 at 25 °C with TMS and chemical shifts (δ) were reported in parts per million (ppm). Elemental analyses were satisfactorily performed on a CHN–O-Rapid instrument within ±0.4% of the theoretical values.4.2. General procedure for the synthesis of (2-methyl-5-nitro-1H-imidazol-1-yl)methanol derivatives
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) washed by ice-water (25 mL) for three times and dried to obtain pure solid products.
5) washed by ice-water (25 mL) for three times and dried to obtain pure solid products.4.2.3.1. 2-(2-(2-Fluorostyry)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4a). Yellow powder, yield: 65.2%. m.p. 176–177 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.13 (s, 4H, O–CH2–CH2–O), 4.58 (t, J = 6.2 Hz, 2H, N–CH2–C), 5.02 (s, 2H, C–CH2–O), 6.50 (t, J = 3.6 Hz, 1H, C–CH–C), 6.91 (d, J = 8.4 Hz, 1H, C–CH–C), 7.05–7.31 (m, 5H, Ph–H), 7.79–7.95 (m, 2H, Ph–H), 8.23 (s, 1H, N–CH). MS (ESI): 440.12 ([M + H]+). Anal. calcd for C22H18FN3O6: C, 60.14; H, 4.13; N, 9.56; O, 21.85%. Found: C, 60.17; H, 4.08; N, 9.63; O, 21.78%.
4.2.3.2. 2-(2-(3-Fluorostyry)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4b). Yellow powder, yield: 58.0%. m.p. 198–200 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.12 (d, J = 5.3 Hz, 4H, O–CH2–CH2–O), 4.61 (s, 2H, N–CH2–C), 5.01 (s, 2H, C–CH2–O), 6.58 (d, J = 8.1 Hz, 1H, C–CH–C), 7.16–7.32 (m, 3H, C–CH–C, Ph–H), 7.37–7.55 (m, 3H, Ph–H), 7.59–7.75 (m, 2H, Ph–H), 8.24 (s, 1H, N–CH). MS (ESI): 440.12 ([M + H]+). Anal. calcd for C22H18FN3O6: C, 60.14; H, 4.13; N, 9.56; O, 21.85%. Found: C, 60.23; H, 4.04; N, 9.63; O, 21.73%.
4.2.3.3. 2-(2-(4-Fluorostyry)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4c). Yellow powder, yield: 64.1%. m.p. 201–203 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.09 (s, 4H, O–CH2–CH2–O), 4.60 (s, 2H, N–CH2–C), 5.00 (s, 2H, C–CH2–O), 6.57 (d, J = 7.4 Hz, 1H, C–CH–C), 7.14–7.47 (m, 4H, C–CH–C, Ph–H), 7.42–7.66 (m, 2H, Ph–H), 7.70–7.94 (m, 2H, Ph–H), 8.23 (s, 1H, N–CH). MS (ESI): 440.12 ([M + H]+). Anal. calcd for C22H18FN3O6: C, 60.14; H, 4.13; N, 9.56; O, 21.85%. Found: C, 60.05; H, 4.18; N, 9.64; O, 21.89%.
4.2.3.4. 2-(2-(2-Chlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4d). Yellow powder, yield: 65.3%. m.p. 181–182 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.07–4.18 (m, 4H, O–CH2–CH2), 4.61 (s, 2H, N–CH2–C), 5.01 (s, 2H, C–CH2–O), 6.59 (d, J = 7.5 Hz, 1H, C–CH–C), 7.16–7.32 (m, 4H, C–CH–C, Ph–H), 7.32–7.43 (m, 2H, Ph–H), 7.50 (t, J = 4.9 Hz, 2H, Ph–H), 8.24 (s, 1H, N–CH). MS (ESI): 456.09 ([M + H]+). Anal. calcd for C22H18ClN3O6: C, 57.97; H, 3.98; N, 9.22; O, 21.06%. Found: C, 57.84; H, 3.91; N, 9.32; O, 21.17%.
4.2.3.5. 2-(2-(3-Chlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4e). Yellow powder, yield: 59.2%. m.p. 190–192 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.06 (s, 2H, O–CH2–C), 4.13–4.17 (m, 2H, C–CH2–O), 4.57 (s, 2H, N–CH2–C), 5.03 (s, 2H, C–CH2–O), 6.54 (d, J = 7.3 Hz, 1H, C–CH–C), 7.19–7.35 (m, 4H, C–CH–C, Ph–H), 7.39 (d, J = 5.7 Hz, 3H, Ph–H), 7.69 (t, J = 6.7 Hz, 1H, Ph–H), 8.23 (s, 1H, N–CH). MS (ESI): 456.09 ([M + H]+). Anal. calcd for C22H18ClN3O6: C, 57.97; H, 3.98; N, 9.22; O, 21.06%. Found: C, 58.14; H, 3.83; N, 9.07; O, 21.23%.
4.2.3.6. 2-(2-(4-Chlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4f). Yellow powder, yield: 61.8%. m.p. 206–208 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.07 (s, 2H, O–CH2–C), 4.17 (s, 2H, O–CH2–C), 4.59 (s, 2H, N–CH2–C), 4.99 (s, 2H, C–CH2–O), 6.58 (d, J = 7.1 Hz, 1H, C–CH–C), 7.22 (t, J = 8.7 Hz, 2H, C–CH–C, Ph–H), 7.45 (t, J = 6.1 Hz, 3H, Ph–H), 7.70 (t, J = 6.3 Hz, 3H, Ph–H), 8.24 (s, 1H, N–CH). MS (ESI): 456.09 ([M + H]+). Anal. calcd for C22H18ClN3O6: C, 57.97; H, 3.98; N, 9.22; O, 21.06%. Found: C, 57.91; H, 4.09; N, 9.29; O, 20.92%.
4.2.3.7. 2-(2-(2-Bromostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4g). Yellow powder, yield: 65.1%. m.p. 194–196 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.09 (s, 2H, O–CH2–C), 4.18 (s, 2H, O–CH2–C), 4.59 (s, 2H, N–CH2–C), 5.01 (s, 2H, C–CH2–O), 6.67 (d, J = 7.0 Hz, 1H, C–CH–C), 7.21 (t, J = 8.7 Hz, 2H, C–CH–C, Ph–H), 7.32–7.48 (m, 3H, Ph–H), 7.64–7.85 (m, 3H, Ph–H), 8.25 (s, 1H, N–CH). MS (ESI): 500.04 ([M + H]+). Anal. calcd for C22H18BrN3O6: C, 52.82; H, 3.63; N, 8.40; O, 19.19%. Found: C, 52.74; H, 3.57; N, 8.48; O, 19.06%.
4.2.3.8. 2-(2-(3-Bromostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4h). Yellow powder, yield: 66.5%. m.p. 193–195 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.12 (s, 4H, O–CH2–CH2–O), 4.53 (s, 2H, N–CH2–C), 5.03 (s, 2H, C–CH2–O), 6.59 (d, J = 7.4 Hz, 1H, C–CH–C), 7.31–7.47 (m, 4H, C–CH–C, Ph–H), 7.52–7.64 (m, 2H, Ph–H), 7.75–7.81 (m, 2H, Ph–H), 8.25 (s, 1H, N–CH). MS (ESI): 500.04 ([M + H]+). Anal. calcd for C22H18BrN3O6: C, 52.82; H, 3.63; N, 8.40; O, 19.19%. Found: C, 52.76; H, 3.73; N, 8.51; O, 19.07%.
4.2.3.9. 2-(2-(4-Bromostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4i). Yellow powder, yield: 57.4%. m.p. 205–207 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.24 (s, 4H, O–CH2–CH2–O), 4.60 (s, 2H, N–CH2–C), 5.00 (s, 2H, C–CH2–O), 6.60 (d, J = 8.4 Hz, 1H, C–CH–C), 7.15–7.29 (m, 2H, C–CH–C, Ph–H), 7.44–7.71 (m, 6H, Ph–H), 8.23 (s, 1H, N–CH). MS (ESI): 500.04 ([M + H]+). Anal. calcd for C22H18BrN3O6: C, 52.82; H, 3.63; N, 8.40; O, 19.19%. Found: C, 52.94; H, 3.52; N, 8.48; O, 19.13%.
4.2.3.10. 2-(2-(2-Methystyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4j). Yellow powder, yield: 72.6%. m.p. 172–174 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 2.50 (s, 3H, –CH3), 4.13 (d, J = 6.0 Hz, 4H, O–CH2–CH2–O), 4.63 (s, 2H, N–CH2–C), 4.90 (s, 2H, C–CH2–O), 6.64 (d, J = 7.3 Hz, 1H, C–CH–C), 7.17–7.46 (m, 7H, C–CH–C, Ph–H), 7.84 (d, J = 5.7 Hz, 1H, Ph–H), 8.27 (s, 1H, N–CH). MS (ESI): 436.14 ([M + H]+). Anal. calcd for C23H21N3O6: C, 63.44; H, 4.86; N, 9.65; O, 22.05%. Found: C, 63.35; H, 4.89; N, 9.77; O, 21.99%.
4.2.3.11. 2-(2-(3-Methystyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylat (4k). Yellow powder, yield: 71.2%. m.p. 173–175 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 2.47 (s, 3H, –CH3), 4.15 (s, 4H, O–CH2–CH2–O), 4.61 (d, J = 4.6 Hz, 2H, N–CH2–C), 4.92 (t, J = 5.3 Hz, 2H, C–CH2–O), 6.89 (d, J = 7.3 Hz, 1H, C–CH–C), 7.05–7.34 (m, 2H, C–CH–C, Ph–H), 7.34–7.86 (m, 6H, Ph–H), 8.24 (s, 1H, N–CH). MS (ESI): 436.14 ([M + H]+). Anal. calcd for C23H21N3O6: C, 63.44; H, 4.86; N, 9.65; O, 22.05%. Found: C, 63.36; H, 4.97; N, 9.65; O, 21.98%.
4.2.3.12. 2-(2-(4-Methystyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4l). Yellow powder, yield: 71.4%. m.p. 183–185 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 2.49 (s, 3H, –CH3), 4.12 (s, 4H, O–CH2–CH2–O), 4.58 (t, J = 4.2 Hz, 2H, N–CH2–C), 4.87 (t, J = 8.4 Hz, 2H, C–CH2–O), 6.57 (d, J = 5.3 Hz, 1H, C–CH–C), 6.88–7.15 (m, 2H, C–CH–C, Ph–H), 7.34–7.48 (m, 4H, Ph–H), 7.82–7.98 (m, 2H, Ph–H), 8.27 (s, 1H, N–CH). MS (ESI): 436.14 ([M + H]+). Anal. calcd for C23H21N3O6: C, 63.44; H, 4.86; N, 9.65; O, 22.05%. Found: C, 63.33; H, 4.98; N, 9.67; O, 22.13%.
4.2.3.13. 2-(5-Nitro-2-(2-nitrostyryl)-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4m). Yellow powder, yield: 64.4%. m.p. 193–194 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 3.37 (s, 2H, O–CH2–CH2), 4.02 (m, 2H, O–CH2–CH2), 4.48 (d, J = 6.3 Hz, 2H, N–CH2–C), 5.04 (s, 2H, C–CH2–O), 6.84 (d, J = 5.4 Hz, 1H, C–CH–C), 7.27–7.49 (m, 4H, C–CH–C, Ph–H), 7.88–8.35 (m, 4H, Ph–H, N–CH), 8.47 (s, 1H, Ph–H). MS (ESI): 467.11 ([M + H]+). Anal. calcd for C22H18N4O8: C, 56.65; H, 3.89; N, 12.01; O, 27.44%. Found: C, 56.54; H, 3.94; N, 12.07; O, 27.46%.
4.2.3.14. 2-(5-Nitro-2-(3-nitrostyryl)-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4n). Yellow powder, yield: 58.2%. m.p. 195–197 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 3.96–4.15 (m, 4H, O–CH2–CH2), 4.64 (s, 2H, N–CH2–C), 5.03 (s, 2H, C–CH2–O), 6.57 (d, J = 6.5 Hz, 1H, C–CH–C), 7.17–7.28 (m, 2H, C–CH–C, Ph–H), 7.61–7.83 (m, 3H, Ph–H), 8.30–8.56 (m, 3H, Ph–H, N–CH), 8.58 (s, 1H, Ph–H). MS (ESI): 467.11 ([M + H]+). Anal. calcd for C22H18N4O8: C, 56.65; H, 3.89; N, 12.01; O, 27.44%. Found: C, 56.74; H, 3.81; N, 11.95; O, 27.53%.
4.2.3.15. 2-(5-Nitro-2-(4-nitrostyryl)-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4o). Yellow powder, yield: 60.1%. m.p. 201–203 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 3.98–4.18 (m, 4H, O–CH2–CH2), 4.57 (s, 2H, N–CH2–C), 5.01 (s, 2H, C–CH2–O), 6.63 (d, J = 5.4 Hz, 1H, C–CH–C), 7.07–7.19 (m, 2H, C–CH–C, Ph–H), 7.43–7.54 (m, 4H, Ph–H), 8.27–8.46 (m, 2H, Ph–H, N–CH), 8.42 (s, 1H, Ph–H). MS (ESI): 467.11 ([M + H]+). Anal. calcd for C22H18N4O8: C, 56.65; H, 3.89; N, 12.01; O, 27.44%. Found: C, 56.59; H, 3.94; N, 12.07; O, 27.48%.
4.2.3.16. 2-(2-(2-Methoxystyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylat (4p). Yellow powder, yield: 64.3%. m.p. 178–179 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 3.77 (s, 3H, C–OCH3), 4.07–4.09 (m, 2H, O–CH2–C), 4.17–4.20 (m, 2H, C–CH2–O), 4.60 (t, J = 7.2 Hz, 2H, N–CH2–C), 5.00 (t, J = 4.8 Hz, 2H, C–CH2–O), 6.56 (d, J = 3.9 Hz, 1H, C–CH–C), 6.94–6.97 (m, 1H, C–CH–C), 7.22–7.33 (m, 5H, Ph–H), 7.43 (d, J = 8.2 Hz, 1H, Ph–H), 7.69 (d, J = 7.4 Hz, 1H, Ph–H), 8.23 (s, 1H, N–CH). MS (ESI): 452.14 ([M + H]+). Anal. calcd for C23H21N3O7: C, 61.19; H, 4.69; N, 9.31; O, 24.81%. Found: C, 61.08; H, 4.42; N, 9.37; O, 24.93%.
4.2.3.17. 2-(2-(3-Methoxystyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4q). Yellow powder, yield: 61.3%. m.p. 176–178 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 3.76 (s, 3H, C–OCH3), 4.11 (s, 4H, O–CH2–CH2–O), 4.55 (s, 2H, N–CH2–C), 4.98 (s, 2H, C–CH2–O), 6.49 (t, J = 7.2 Hz, 1H, C–CH–C), 6.92 (t, J = 8.4 Hz, 2H, C–CH–C, Ph–H), 7.1 (d, J = 5.22 Hz, 1H, Ph–H), 7.32–7.44 (m, 4H, Ph–H), 7.70 (d, J = 4.5 Hz, 1H, Ph–H), 8.23 (s, 1H, N–CH). MS (ESI): 452.14 ([M + H]+). Anal. calcd for C23H21N3O7: C, 61.19; H, 4.69; N, 9.31; O, 24.81%. Found: C, 61.23; H, 4.51; N, 9.39; O, 24.92%.
4.2.3.18. 2-(2-(4-Methoxystyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4r). Yellow powder, yield: 65.1%. m.p. 183–185 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 3.59 (s, 3H, C–OCH3), 4.13 (s, 4H, O–CH2–CH2–O), 4.65 (s, 2H, N–CH2–C), 4.78 (s, 2H, C–CH2–O), 6.53 (d, J = 4.3 Hz, 1H, C–CH–C), 7.03 (t, J = 4.4 Hz, 2H, C–CH–C, Ph–H), 7.22–7.44 (m, 5H, Ph–H), 7.77 (d, J = 6.8 Hz, 1H, Ph–H), 8.22 (s, 1H, N–CH). MS (ESI): 452.14 ([M + H]+). Anal. calcd for C23H21N3O7: C, 61.19; H, 4.69; N, 9.31; O, 24.81%. Found: C, 61.13; H, 4.74; N, 9.36; O, 24.87%.
4.2.3.19. 2-(2-(2-Chloro-6-fluorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4s). Yellow powder, yield: 54.1%. m.p. 184–186 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.04–4.24 (m, 4H, O–CH2–CH2), 4.61 (s, 2H, N–CH2–C), 4.98 (s, 2H, C–CH2–O), 6.57 (t, J = 6.9 Hz, 1H, C–CH–C), 6.90 (d, J = 4.3 Hz, 1H, C–CH–C), 7.0 (d, J = 7.83 Hz, 1H, Ph–H), 7.26–7.46 (m, 4H, C–CH–C, Ph–H), 7.85 (d, J = 5.4 Hz, 1H, Ph–H), 8.27 (s, 1H, N–CH). MS (ESI): 474.08 ([M + H]+). Anal. calcd for C22H17ClFN3O6: C, 55.76; H, 3.62; N, 8.87; O, 20.26%. Found: C, 55.84; H, 3.51; N, 8.96; O, 20.17%.
4.2.3.20. 2-(2-(2,4-Dichlorostyryl)-5-nitro-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4t). Yellow powder, yield: 63.5%. m.p. 190–192 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.37 (s, 4H, O–CH2–CH2–O), 4.62 (d, J = 5.3 Hz, 2H, N–CH2–C), 5.02 (s, 2H, C–CH2–O), 6.58 (d, J = 7.4 Hz, 1H, C–CH–C), 7.13–7.24 (m, 2H, C–CH–C, Ph–H), 7.53–7.72 (m, 6H, Ph–H), 8.21 (s, 1H, N–CH). MS (ESI): 490.05 ([M + H]+). Anal. calcd for C22H18Cl2N3O6: C, 53.89; H, 3.49; N, 8.54; O, 19.58%. Found: C, 53.78; H, 3.56; N, 8.59; O, 19.54%.
4.2.3.21. 2-(5-Nitro-2-styryl-1H-imidazol-1-yl)ethyl 2,3-dihydrobenzo[b][1,4]dioxine-5-carboxylate (4u). Yellow powder, yield: 62.7%. m.p. 173–175 °C; 1H NMR (DMSO-d6, 300 MHz, δ ppm): 4.23 (s, 2H, O–CH2–C), 4.36 (s, 2H, C–CH2–O), 4.67 (s, 2H, N–CH2–C), 5.01 (s, 2H, C–CH2–O), 6.67 (d, J = 4.5 Hz, 1H, C–CH–C), 7.23–7.35 (m, 6H, C–CH–C, Ph–H), 7.45–7.55 (m, 3H, Ph–H), 8.21 (s, 1H, N–CH). MS (ESI): 422.13 ([M + H]+). Anal. calcd for C22H19N3O6: C, 62.70; H, 4.54; N, 9.97; O, 22.78%. Found: C, 62.78; H, 4.61; N, 9.87; O, 22.83%.
4.3. Antiproliferation assay
The antitumor activities of synthesized compounds (3a–3u and 4a–4u) against A549, Hela, U251, HepG-2 cell lines were evaluated as described elsewhere with some modifications.27 Target tumor cell lines were grown to log phase in RPMI 1640 medium supplemented with 10% fetal bovine serum. After diluting to 2 × 104 cells per mL with the complete medium, 100 µL of the obtained cell suspension was added to each well of 96-well culture plates. The subsequent incubation was permitted at 37 °C, 5% CO2 atmosphere for 24 h before the cytotoxicity assessments. Tested samples at preset concentrations were added to six wells with sorbefacient as positive references. After 48 h exposure period, 40 µL of PBS containing 2.5 mg mL−1 of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltet-razolium bromide) was added to each well. Four hours later, 100 µL extraction solution (10% SDS–5% isobutyl alcohol−0.01 M HCl) was added. After an overnight incubation at 37 °C, the optical density was measured at a wavelength of 570 nm on an ELISA microplate reader. In all experiments three replicate wells were used for each drug concentration. Each assay was carried out for at least three times.4.4. JAK3 inhibitory assay
To evaluate the effect of the compounds on JAK3 assembly in vitro, varying concentrations were pre-incubated with 10 µM JAK3 in glutamate buffer at 30 °C and then cooled to 0 °C. After addition of GTP, the mixtures were transferred to 0 °C cuvettes in a recording spectrophotometer and warmed up to 30 °C and the assembly of JAK3 was observed turbid metrically. The IC50 was defined as the compound concentration that inhibited the extent of assembly by 50% after 20 min incubation.4.5. Flow cytometry
Cells (1.3 × 105 cells per mL) were cultured in the presence or not of novobiocin analogues at 200 µM. Nvb at the same concentration served as reference inhibitor. After treatment for 48 and 72 h, cells were washed and fixed in PBS–ethanol (30/70). For cytofluorometric examination, cells (104 cells per mL) were incubated for 30 min in PBS/Triton X-100, 0.2%/EDTA (1 mM), and propidium iodide (PI) (50 µg mL−1) in PBS supplemented by RNase (0.5 mg mL−1). The number of cells in different phases of the cell cycle was determined, and the percentage of apoptotic cells was quantified. Analyses were accomplished by a FACS Calibur (Becton Dickinson, Le Pont de Claix, France). Cell Quest software was used for data acquisition and analysis.4.6. Docking simulations
For the molecular docking model, the three-dimensional X-ray structure of JAK3 (PDB code: 3FUP) acquired from the RCSB protein data bank (http://www.pdb.org) was selected as the template in which compound 4t embed. All bound waters and ligands were eliminated from the protein and the polar hydrogen was added to the proteins. The docking procedure was carried out using CDOCKER protocol for receptor–ligand interactions section of Discovery Studio (version 3.5). Initially, the three-dimensional structures of the compounds in this paper were built by Chem. 3D ultra 12.0 software [Chemical Structure Drawing Standard; Cambridge Soft corporation, USA (2012)], then they were energetically minimized by using MMFF94 with 5000 iterations and minimum RMS gradient of 0.10. Molecular docking of all compounds was then performed using the Discovery Studio (version 3.5) as implemented through the graphical user interface CDOCKER protocol. CDOCKER is an implementation of a CHARMm based molecular docking tool using a rigid receptor.4.7. QSAR model
Ligand-based 3D-QSAR approach was carried out using QSAR software of DS 3.5 (Discovery Studio 3.5, Accelrys, Co.Ltd). Among all the 21 compounds, 76.2% (that is 16) were used as a training set for QSAR modeling and the remaining 23.8% (that is 5) were selected as an external test subset for validating the reliability of the QSAR model. The corresponding pIC50 values which were converted from the acquired IC50 (µM) were used for subsequent QSAR analysis as the response variable. All the definition of the descriptors can be seen in the “Help” of DS 3.5 software and they were calculated by QSAR protocol of DS 3.5.Acknowledgements
This work was supported by the project (No. BY2012136) from the Science & Technology Agency of Jiangsu Province and by the projects (Nos. CXY1213 & CXY1222) from the Science & Technology Bureau of Lianyuangang City of Jiangsu Province.References
- O. Dreesen and A. H. Brivanlou, Stem Cell Rev., 2007, 3, 7 CrossRef CAS.
- M. Furqan, N. Mukhi, B. Lee and D. Liu, Biomarker Research, 2013, 1, 5 CrossRef.
- S. Verstovsek, R. A. Mesa, J. Gotlib, R. S. Levy, V. Gupta, J. F. DiPersio, J. V. Catalano, M. Deininger, C. Miller and R. T. Silver, et al., N. Engl. J. Med., 2012, 366, 799 CrossRef CAS.
- A. C. Ward, I. Touw and A. Yoshimura, Blood, 2000, 95, 19 CAS.
- J. A. Wells and D. A. M. Vos, Annu. Rev. Biochem., 1996, 65, 609 CrossRef CAS.
- S. C. Carter and L. S. Smit, Recent Prog. Horm. Res., 1998, 53, 61 Search PubMed.
- L. Valentino and J. Pierre, Biochem. Pharmacol., 2006, 71, 713 CrossRef CAS.
- K. S. Machado and J. Mieczkowski, et al., Cancer Biol. Ther., 2012, 13, 657 CrossRef.
- L. Pedranzini, A. Leitch and J. Bromberg, J. Clin. Invest., 2004, 114, 619 CrossRef CAS.
- J. E. Darnell, Nat. Med., 2005, 11, 595 CrossRef CAS.
- H. Yu, D. Pardoll and R. Jove, Nat. Rev. Cancer, 2009, 9, 798 CrossRef CAS.
- V. Boudyn and J. Kovarik, Europe PubMed Central, 2002, 49, 349 Search PubMed.
- S. Goel, S. Mani and R. P. Soler, Curr. Oncol. Rep., 2002, 4, 9 CrossRef.
- A. Levitzki, Annu. Rev. Pharmacol. Toxicol., 2013, 53, 161 CrossRef CAS.
- S. S. Jatiani, S. J. Baker, L. R. Silverman and P. E. Reddy, Genes Cancer, 2010, 1, 979 CrossRef CAS.
- C. Harrison, J. J. Kiladjian, H. K. Al-Ali, H. Gisslinger, R. Waltzman, V. Stalbovskaya, M. McQuitty, D. S. Hunter, R. Levy, L. Knoops, F. Cervantes, A. M. Vannucchi, T. Barbui and G. Barosi, N. Engl. J. Med., 2012, 366, 787 CrossRef CAS.
- F. Passamonti, Blood, 2012, 120, 275 CrossRef CAS.
- R. A. Mesa, IDrugs, 2010, 13, 394 CAS.
- K. Shalini, P. K. Sharma and N. Kumar, Chem. Sin., 2010, 1, 36 CAS.
- P. Molina, A. Tarraga and F. Oton, Org. Biomol. Chem., 2012, 10, 1711 CAS.
- M. Pigni, L. Brasili, M. Giannella, D. Giardina, U. Gulini, W. Quaglia and C. Melchiorre, J. Med. Chem., 1988, 31, 2300 CrossRef.
- Y. P. Hou, J. Sun, Z. H. Pang, P. C. Lv, D. D. Li, L. Yan, H. J. Zhang, E. X. Zheng, J. Zhao and H. L. Zhu, Bioorg. Med. Chem., 2011, 19, 5948 CrossRef CAS.
- Y. Aiba and D. Hasegawa, et al., Bioorg. Med. Chem. Lett., 2001, 11, 2783 CrossRef CAS.
- M. Pallavicini, R. Budriesi, L. Fumagalli, P. Ioan, A. Chiarini, C. Bolchi, M. P. Ugenti, S. Colleoni, M. Gobbi and E. Valoti, J. Med. Chem., 2006, 49, 7140 CrossRef CAS.
- J. Sun, D. D. Li, J. R. Li, F. Fang, Q. R. Du, Y. Qian and H. L. Zhu, Org. Biomol. Chem., 2013, 11, 7676 CAS.
- Y. Luo, Y. Li, K. M. Qiu, X. Lu, J. Fu and H. L. Zhu, Bioorg. Med. Chem., 2011, 19, 6069 CrossRef CAS.
- A. Boumendjel, J. Boccard, P. A. Carrupt, E. Nicolle, M. Blanc, A. Geze, L. Choisnard, D. Wouessidjewe, E. L. Matera and C. Dumontet, J. Med. Chem., 2008, 51, 2307 CrossRef CAS.
Footnote |
| † These two authors equally contributed to this paper. |
| This journal is © The Royal Society of Chemistry 2014 |