Discovery of potential biomarkers in exhaled breath for diagnosis of type 2 diabetes mellitus based on GC-MS with metabolomics†
Yanyue Yana,
Qihui Wanga,
Wenwen Lia,
Zhongjun Zhaob,
Xin Yuanb,
Yanping Huangb and
Yixiang Duan*c
aResearch Center of Analytical Instrumentation, Analytical & Testing Center, Sichuan University, Chengdu, P. R. China
bResearch Center of Analytical Instrumentation, College of Chemistry, Sichuan University, Chengdu, P. R. China
cResearch Center of Analytical Instrumentation, College of Life Sciences, Sichuan University, Chengdu, P. R. China. E-mail: yduan@scu.edu.cn; Fax: +86 028 85418180; Tel: +86 028 85418180
First published on 29th May 2014
Abstract
The aim of the study was to apply gas chromatography-mass spectrometry (GC-MS) combined with a metabolomics approach to identify distinct metabolic signatures of type 2 diabetes mellitus (T2DM) and healthy controls from exhaled breath, which are characterized by a number of differentially expressed breath metabolites. In this study, breath samples of patients with type 2 diabetes mellitus (T2DM, n = 48) and healthy subjects (n = 39) were analyzed by GC-MS. Multivariate data analysis including principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) was successfully applied to discriminate the T2DM and healthy controls. Eight specific metabolites were identified and may be used as potential biomarkers for diagnosis of T2DM. Isopropanol and 2,3,4-trimethylhexane, 2,6,8-trimethyldecane, tridecane and undecane in combination might be the best biomarkers for the clinical diagnosis of T2DM with a sensitivity of 97.9% and a specificity of 100%. The study indicated that this breath metabolite profiling approach may be a promising non-invasive diagnostic tool for T2DM.
1. Introduction
Non-invasive breath diagnosis with metabolomics and metabolic profiling add an important scientific value to established biomarkers.1,2 Furthermore, it is known that volatile breath compounds have important relationships with metabolism and pathological states, such as acetone response to glucose and fat metabolism in uncontrolled diabetes.3–6 Therefore, metabolic monitoring of volatile target compounds in exhaled breath is quite a considerable tool for the investigation of some diseases.Type 2 diabetes mellitus (T2DM) is a metabolic disorder that is characterized by high blood glucose in the context of insulin resistance and relative insulin deficiency.7 Rates of T2DM have increased markedly since 1960 in parallel with obesity: as of 2010 there are approximately 285 million people with the disease compared to around 30 million in 1985.8,9 Traditional diagnosis and management of T2DM hinges on blood tests (for plasma glucose and glycated hemoglobin), which may be expensive, unpractical, and even painful. Frequent blood testing is especially necessary for patients undergoing insulin treatment. Therefore large resources have been invested worldwide in developing non-invasive devices for diabetes diagnosis and management. Breath analysis, as is one of the most promising approaches for clinical routine, is increasingly being exploited for clinical diagnosis.10 Breath testing is non-invasive offering an attractive, inexpensive, and patient-friendly evaluation. Furthermore, sample collection is easy and can even be obtained from unconscious patients. Diabetes and its related dysmetabolic states could clearly greatly benefit from the introduction of similar non-invasive tests for diagnostic, preventive and monitoring purposes too. This study investigated breath analysis as a diagnostic tool for T2DM and identified the biomarkers in the breath volatile organic compounds (VOCs).
Due to its higher consistency, robustness, and sensitivity, gas chromatography coupled with mass spectrometry (GC-MS) is one of the most frequently used analytical techniques for profiling primary metabolites.11 Combined with public databases, the application of GC-MS for compound identification makes it of great value for metabolomics.12 It has been widely applied in disease biomarker discovery. As a metabolite profiling technique, GC-MS has been used to detect and discriminate various diseases, such as diabetic kidney disease,13 colon cancer,14 type 2 diabetes mellitus,15 cirrhosis and hepatic encephalopathy.2 For the GC-MS experiments, we performed a solid phase micro extraction (SPME) coupled with GC-MS in metabolic profiling for analysis of T2DM. SPME is a simple, rapid and solvent-free sample preparation technique that can be directly coupled to GC-MS.16 It has several advantages including, faster extraction and desorption, direct compatibility with the GC inlet, and less required sample. The breath compounds were identified as many as possible by the use of SPME.
Metabolomics is the scientific study of chemical processes involving metabolites. The metabolome represents the collection of all metabolites in a biological cell, tissue, organ or organism, which are the end products of cellular processes.17 The idea that biological fluids and tissues have important relationships with the health of an individual has existed for a long time.18 Even in ancient times, physicians recognized that certain breath odors were associated with specific pathological states, such as a ‘fishy’ smell in response to renal failure and a ‘fruity’ smell will associate with diabetes.19 Ancient Chinese doctors used ants to detect whether the urine of patients contained high levels of glucose, and hence detect diabetes.20 Recently, many scientists have studied metabolic diseases using metabolomics as a main technique. For example, the Wang-Sattler group have used statistical and bioinformatical methods for analyzing metabolite concentration profiles for the identification of candidate biomarkers of T2DM.21,22 It was demonstrated that GC-MS could be used to measure compounds present in human urine and tissue extracts by Horning et al. in 1971.23 Followed, the Horning group, along with that of Linus Pauling and Arthur B Robinson led the development of GC-MS methods to monitor the metabolites present in urine through the 1970s.24 The role of metabolomics and metabolic profiling coupled with GC-MS is a rapidly growing field in disease diagnosis,15,25 therapeutics,26,27 functional genomic28,29 and toxicology studies.30,31 In this study, the GC-MS data would then be coupled with chemometric methods to develop models to identify the biomarkers of T2DM. Data obtained from metabolomics studies are complex and diverse.32 Therefore, the common statistical approach used in metabolomics data analysis is based on multivariate statistical analysis (MVA) for biomarker discovery including principal component analysis (PCA),33,34 partial least square discriminant analysis (PLS-DA)35,36 and orthogonal partial least squares discriminant analysis (OPLS-DA).37 The method is the simplest of the true eigenvector-based multivariate analyses, which is mostly used as a tool in exploratory data analysis and for making predictive models. PCA can supply the user with an overview of the clustering trend in the data by compressing the multidimensional data into a few principal components. PLS-DA and OPLS-DA calculate principal components in cooperation with the classification information and are more powerful to deal with complex multidimensional data.32 Using this protocol, large amounts of information can be acquired, with high reproducibility, on the breath metabolome.
This article discusses the breath metabolic changes of T2DM by coupling GC-MS with multivariate data analysis that included PCA, OPLS-DA. By clearly revealing the biomarkers (retention time), OPLS-DA can be successfully applied to separate the T2DM and healthy controls groups. In addition, nonparametric testing and receiver operator characteristic curves (ROC) analysis were performed to validate the robustness of OPLS-DA model. With these analyses, we identified and selected the potential biomarkers and discussed their biological functions.
2. Materials and methods
2.1. Breath sample collection
The breath samples were collected by 3L Delin-Inert Multi-Layer Foil bag (Dalian Delin Gas Packing Company, China). After cleaning the bag four times with pure nitrogen, we collected the exhaled breath of a single exhalation from total lung capacity to residual volume at a constant flow (10–11 L min−1) over a period of 20 to 30 s against a mild resistance. It was reported that the volume of exhaled breath collected did not influence the VOCs concentration in the reservoir, and as described previously, the collected exhaled breath at a controlled flow of 10–11 L min−1 without any dead space air is good for the exhaled breath measurements.38In this study, 48 T2DM patients and 39 healthy controls were recruited for this study. All the breath samples of patients (22 males and 26 females, aged 14–85 years) were from the Second Affiliated Hospital of Jilin University, Jilin, China. The T2DM patients were diagnosed according to the criteria of WHO (World Health Organization) 1999. The inclusion criteria defined that the patients of 14–90 years old had no histories of receiving medication and none of them had been suffering from the other known chronic disease, while the patients receiving long-term medication or suffering from the other known chronic disease, or lung ventilation dysfunction were all excluded based on the exclusion criteria. Each participant wrote informed consent prior to the study. All study procedures were approved by the Ethic Committee of Chinese People's Liberation Army 208 Hospital, China. The 39 health controls (15 males and 24 females, aged 21–71 years) were from Sichuan University, Chengdu, China, without receiving long-term medication and suffering from known chronic disease. All samples were collected at a stable state without recent dietary intake and exercise.
2.2. Solid phase micro extraction GC-MS analysis
All GC-MS experiments were performed by Agilent GC (7890A)-MS (220 ion trap) (Agilent Technologies, USA). A solid phase micro extraction (SPME) fiber holder – 85 μm carboxen/polydimethylsiloxane(CAR/PDMS) (Sigma-Aldrich, USA) – was exposed in the sample bag, which was full of breath samples, at constant room temperature for breath compounds extraction. In the GC system, a 60 m (0.32 mm ID, 1.8 μm phase thickness) DB-624 polysiloxane capillary column (Agilent Technologies, USA) was used. The column temperature was programed at 40 °C for 5 min, from 40 °C to 160 °C at the rate of 10 °C min−1, 160 °C–200 °C at the rate of 5 °C min−1, then hold 15 min at 200 °C. The injection temperature was kept at 250 °C. The split ratio was 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Helium carrier gas was used at a constant flow rate of 1.0 mL min−1. The full scanning mode was used for monitoring at 20–300 m/z with a scan speed of 0.5 s/scan velocity.
1. Helium carrier gas was used at a constant flow rate of 1.0 mL min−1. The full scanning mode was used for monitoring at 20–300 m/z with a scan speed of 0.5 s/scan velocity.
2.3. Sample preparation
The bag was sealed immediately after the exhaled air was collected, then the SPME fiber was introduced and maintained into the bag during the extraction at a constant room temperature (16 °C). After a measured extraction time (35 min, as optimized), the fiber was removed and immediately introduced into the GC injection port and volatiles extracted were desorbed for 5 min at 250 °C.2.4. Standards and reagents
Acetone (>99.5% purity), ethanol (>99.7% purity), were purchased from Kelong Chemical (Chengdu, China) in analytical grade. HPLC grade isopropanol (>99.9% purity) was purchased from Tianjin Kermel Chemical (Tianjin, China). m-Xylene (>99.0% purity) was purchased from Sangon Biotech (Shanghai, China) in reagent grade.2.5. Data handling and statistics
Chromatogram acquisition, automated peak deconvolution and library searches were performed using the MS Workstation (Version 6.9.3, Varian, Inc., CA, USA) in combination with the automated mass spectrometry deconvolution and identification system (AMDIS, Version 2.66). The intensity of all peaks for all breath samples were studied by multivariate statistical methods. After acquisition of chromatographic raw data, and the baseline noise subtracted with representative MS spectra selected, we extracted components utilizing exact mass chromatograms and listed detected peaks as their mass and retention time along with their associated intensities. Each chromatogram with the total ion chromatogram (TIC) signal was exported as a CSV format to Excel. Normalization of peak intensities helps to reduce the systematic variation of GC-MS data. The peak intensities were normalized by eqn (1). Where Pin is the normalized peak intensity of metabolite (peak i) in each breath sample, Pi is the peak intensity of metabolite (peak i), is the sum of all peak intensities of metabolites in the breath sample. We provided two Excel files as an example in the supplement. One is the chromatography raw data of a breath sample, while the other one is the normalized data of the same breath sample.
is the sum of all peak intensities of metabolites in the breath sample. We provided two Excel files as an example in the supplement. One is the chromatography raw data of a breath sample, while the other one is the normalized data of the same breath sample.
 | (1) |
A matrix table consisting of the peak number (based on the retention time and m/z), sample name, and the normalized peak intensity, was produced in the batch job by a computer program which was coded in MATLAB R2013a (Mathworks, Natick, MA, USA). The program was performed as following two steps:
(1) an average retention time list (ARTL) which is associated with the metabolites in each chromatogram was established. Firstly, we choose a chromatographic peak list as a reference list (ARTL1). Then, the average retention time list was produced by comparison of the other peak lists (PL) as is referenced in eqn (2).
 | (2) |
(2) Chromatographic peaks in the raw data files are detected according to the average retention time and tracking the apex of the peaks in the chromatograms. The track peak parameters were as following retention time window of 0.05 min.
Therefore, a matrix of data with one column per sample (include all the samples of the metabolomics experiment) and one row per mass signal was generated. This process also leads to peak missing, which may be the severer drift of peak or the quantity is too low to be detected. If so, we should check the chromatogram to ensure the correct result.
Followed, the resulting multivariate dataset was imported into SIMCA-P 11.0 software (Umetrics, Umeå, Sweden) as variables for the principal components analysis (PCA), and the orthogonal projection to latent structures with discriminant analysis (OPLS-DA). In addition, nonparametric Mann–Whitney U testing was used to assess whether the potential biomarkers is significantly different between the T2DM and healthy control group. The results were considered significant if the estimated p < 0.05. Receiver operator characteristic curves (ROC) analysis was performed to validate the robustness of OPLS-DA model, and the areas under curve (AUC) value, specificity, sensitivity were calculated to evaluate the diagnostic value of the potential biomarkers from the differential metabolites of the disease. All additional statistical analysis was conducted using IBM SPSS Statistics 19.0 (SPSS Inc., Chicago, Illinois, USA).
Metabolites were identified by matching retention time as well as mass spectra compared with the National Institute of Standards and Technology (NIST) mass spectra library (NIST 08, NIST Mass Spectral Search Program, Version 2.0f, USA). Additionally, on-line mass spectra searches in the Human Metabolome Database (http://www.hmdb.ca), KEGG (http://www.genome.jp/kegg/ligand.html) and the MASS Bank (http://www.massbank.jp) were performed. Commercial standard reagents were used to support identification of metabolites.
3. Results and discussion
3.1. Results
Representative GC-MS TIC chromatograms of breath samples from the T2DM group and healthy control group were displayed in Fig. 1. A number of raw GC-MS data files were selected as representative examples by automated peak deconvolution using MS Workstation and AMDIS. Specific ion characteristics of each metabolite were selected after subtracting the baseline noise in MS Workstation. A matrix table mentioned above in Section 2.5 with the signals selected was produced by the computer program. The signals represent the concentration of each selected metabolite in each sample. We focused on 254 signals obtained using the method. The mass spectral data were then processed by multivariate statistical analysis. The data consisting of 254 variables were firstly analyzed by PCA. The score plot of PCA was shown in Fig. 2a. In PCA three principle components were calculated, R2X was 0.336. | ||
| Fig. 1 Typical GC-MS spectra obtained for breath from T2DM and healthy controls, highlighting the peaks corresponding to the 8-compounds model. The labels in part correspond to the labels in Table 1. | ||
Following PCA the data sets were processed using more powerful supervised statistical method, OPLS-DA. Firstly, we applied the permutation test with 200 iterations to assess whether the PLS-DA model is valid and well-fit. In our study, the R2 intercepts were 0.359 and the Q2 was below 0 using the supervised projection method PLS-DA (Fig. 2b). In OPLS-DA, the model resulted in one predictive and four orthogonal (1 + 4) components (R2X = 0.788, R2Y = 0.908, Q2 (cum) = 0.823), revealing the high discriminative and predictive ability, shown in Fig. 2c.
Fig. 2d showed the S-plot of OPLS-DA. The potential biomarkers from S-plot in our study were selected according to the parameters of VIP. In the S-plot, fifteen metabolites were highlighted. For each selected metabolites biomarkers, a nonparametric Mann–Whitney U test was performed to assess the univariate importance of the metabolite and a value of p < 0.05 was considered significant. A total of 15 differentially expressed metabolites were measured. Only eight metabolites most strongly influencing the differentiation were listed in Table 1 with the VIP > 1 and p < 0.05. The identification of compounds considered as potential biomarkers in the model was made using NIST Mass Spectral Search Program, the HMDB (http://www.hmdb.ca), and the MASS Bank (http://www.massbank.jp). The detailed method for the verification and validation of the potential biomarkers has been mentioned in the following work.
| No. | RT (min) | Metabolites | VIP | p-Value |
|---|---|---|---|---|
| 1 | 8.497 | Acetone | 9.30 | 0.004 |
| 2 | 8.720 | Isopropanol | 7.79 | <0.001 |
| 3 | 15.875 | Toluene | 1.66 | <0.001 |
| 4 | 18.265 | m-Xylene | 1.89 | 0.005 |
| 5 | 19.699 | 2,3,4-Trimethylhexane | 2.02 | <0.001 |
| 6 | 21.009 | 2,6,8-Trimethyldecane | 3.64 | <0.001 |
| 7 | 21.540 | Tridecane | 6.53 | <0.001 |
| 8 | 21.995 | Undecane | 3.32 | <0.001 |
Among these metabolites, three potential biomarkers (acetone, isopropanol, and m-xylene) were confirmed using standard samples. A series stock solution of acetone, isopropanol, and m-xylene with the concentration of 10 μmol mL−1 was prepared with ethanol. We injected 10 μL stock solutions into a 3 L bag, respectively, before introducing pure nitrogen into the bag. And the samples were stored at room temperature to fully evaporate in the bags for more than two hours. The fragmentations of all samples obtained were shown in Fig. 3. Three standard samples were matched with the results as mentioned previously by the comparison of retention time and the electron ionization mass spectra.
 | ||
| Fig. 3 Ion current chromatograms of acetone (a), isopropanol (b), and m-xylene (c) compared with ethanol and pure nitrogen in pairs performing the electron ionization mass spectra. | ||
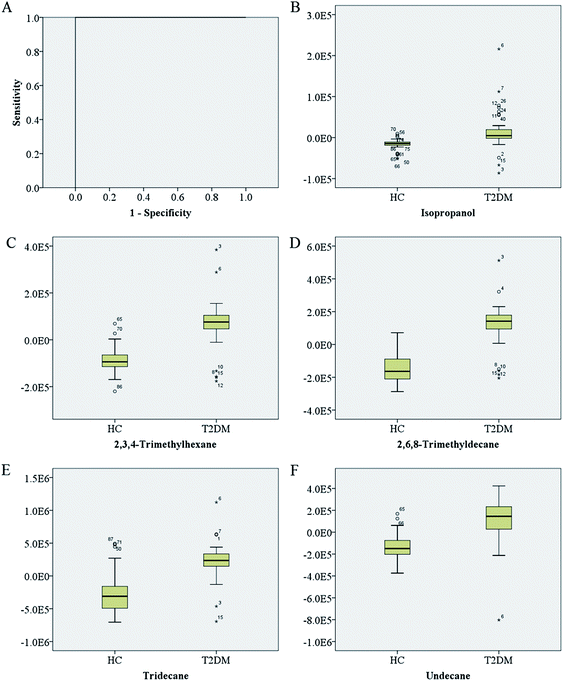
We also performed the ROC analysis to characterize these potential biomarkers of T2DM. All potential biomarkers could be divided into two groups, in which seven potential biomarkers were up-regulated in T2DM patients, and one potential biomarker was down-regulated. Fig. 4 displayed the ROC curve analysis of the eight biomarkers including acetone, isopropanol, toluene, m-xylene, 2,3,4-trimethylhexane, 2,6,8-trimethyldecane, tridecane, and undecane. Acetone, isopropanol, tridecane, 2,6,8-trimethyldecane, undecane, 2,3,4-trimethylhexane and toluene were shown higher levels in T2DM (Fig. 4a), while m-xylene were shown lower levels in T2DM (Fig. 4b). The detailed statistics of the value of area under the ROC curves (AUC), and the corresponding sensitivities and specificities for each of the potential biomarkers of T2DM were listed in Table 2. Such as the isopropanol had a sensitivity of 79.2% and a specificity of 92.3%, the calculated area under the ROC curve was 0.876 (95% confidence intervals, 0.795–0.956). To demonstrate the utility of breath metabolites for the discrimination between T2DM and healthy controls, a logistic regression model was built based on five validated biomarkers with AUC > 0.8 (isopropanol, 2,3,4-trimethylhexane, 2,6,8-trimethyldecane, tridecane, and undecane). The ROC curve was computed for the logistic regression (LR) model. As a result, we obtained a sensitivity of 97.9% and a specificity of 100% of LR model. The calculated area under the ROC curve was 1.00 (95% confidence intervals, 1.000–1.000), as shown in Fig. 5a. We also displayed the box plots of these five potential biomarkers in distinguishing T2DM from healthy controls (Fig. 5b–f).
 | ||
| Fig. 4 Receiver operating characteristic (ROC) curve analysis for the predictive power of up-regulated biomarkers (a) and down-regulated biomarkers (b) for distinguishing T2DM from healthy controls. | ||
| Metabolites | AUC (95% CIs) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| Acetone | 0.679 (0.567, 0.791) | 45.8 | 87.2 |
| Isopropanol | 0.876 (0.795, 0.956) | 79.2 | 92.3 |
| Toluene | 0.737 (0.627, 0.847) | 66.7 | 82.1 |
| m-Xylene | 0.677 (0.556, 0.798) | 69.2 | 72.9 |
| 2,3,4-Trimethylhexane | 0.910 (0.835, 0.985) | 89.6 | 94.9 |
| 2,6,8-Trimethyldecane | 0.949 (0.903, 0.995) | 89.6 | 94.9 |
| Tridecane | 0.870 (0.779, 0.962) | 89.6 | 84.6 |
| Undecane | 0.911 (0.847, 0.976) | 89.6 | 82.1 |
3.2. Discussion
PCA is a pattern recognition method that reduces the dimensionality of data to a number of summary variables, principal compounds. It can be used to describe the relationship among groups without artificial intervention. Fig. 2a showed the score plot of PCA. Each point represents a single breath sample. This score plot display any natural clustering or separation with a dataset, but the T2DM group and healthy control group in our study cannot be well separated.PCA analysis did not identify any particular similarity or large differences between sample profiles. Therefore, we used more powerful supervised statistical method, OPLS-DA. OPLS-DA is the extension orthogonal projections of the PLS-DA, while PLS-DA is a classification method based on the regression extension of PCA. In PLS-DA, generally, the original model is considered well-fit when the R2 intercepts are <0.4 and the Q2 is <0.05 in the permutation test with 200 iterations.39,40 In our study, the results (R2 = 0.359, Q2 < 0) indicated that the models were statistically valid and well-fit. The OPLS-DA method was used to test the differences in metabolite between T2DM and healthy controls, and to identify the potential biomarkers of T2DM. As shown in Fig. 2c, the results demonstrated that the OPLS-DA models were well-fit and highly discriminative and predictive, which was conclusive for supporting the presented separation between the metabolite profiles of the T2DM group and healthy controls. It was observed that healthy controls were well separated from the T2DM.
The S-plot visualizes the covariance and correlation among variables, thus it is used to identify discriminating variables.37 And the variable importance in the projection (VIP) value of OPLS-DA models is a major parameter for the detection of potential biomarkers. As shown in Fig. 2d, the 15 potential biomarkers which were highlighted from S-plot were selected with VIP > 1. Nonparametric Mann–Whitney U test was performed to assess the univariate importance of the metabolite and a value of p < 0.05 was considered significant. The results demonstrated that only eight metabolites were strongly differential with the VIP > 1 and p < 0.05 (Table 1).
All metabolites could be divided into two groups, up-regulated metabolites and down-regulated metabolites. The results indicated that acetone, isopropanol, tridecane, 2,6,8-trimethyldecane, undecane, 2,3,4-trimethylhexane and toluene were shown higher levels in T2DM, while m-xylene were shown lower levels in T2DM. Furthermore, it has been reported that ROC analysis is able to determine easily ability for identifying disease at any cutoff. AUC value from ROC is usually between 0.5 and 1.0. The AUC value is more close to 1, the higher the accuracy test is, and the bigger the diagnostic value is. Furthermore, the logistic regression model was built to demonstrate the utility of breath metabolites for the discrimination between T2DM and healthy controls. Five validated biomarkers with AUC > 0.8 were used to build the LR model. As a result, the LR model with a sensitivity of 97.9% and a specificity of 100% demonstrated that isopropanol, 2,3,4-trimethylhexane, 2,6,8-trimethyldecane, tridecane, and undecane in combination provided better prediction in T2DM.
3.3. Biological context of the new candidate biomarkers
It has been reported that more than 3000 volatile organic compounds (VOCs) had been detected in the breath of humans,41,42 including ketones, alcohols, alkanes, substituted benzenes, etc. The VOCs were derived from metabolic pathways as reported previously, such as acetone from glucose metabolism,43 alkanes from OFR-mediated lipid peroxidation of fatty acids.44,45 Since acetone was identified in human breath in 1857, it was regarded as a characteristic feature of diabetic coma.3 Acetone is one of the ketone bodies produced during the acute complication of ketoacidosis. In diabetic ketosis, ketone body production (b-hydroxy-butyrate, acetoacetate) provides fuel for vital organs (heart, brain) raising the chance of survival of the metabolic catastrophe. For this point of view, acetone metabolism is associated with diabetes mellitus, and acetone is regarded as a promising biomarker of diabetes. In our study, the actual concentration of acetone in exhaled breath is different depending on each individual. Therefore, further researches about the actual concentration in the exhaled breath to distinguish the T2DM from healthy controls should be done in the future.Isopropanol belongs to the family of alcohols and polyols compounds. The previous report indicated that isopropanol is one of the product from propanoate metabolism, and the substrate for synthesizing acetone catalyzed by the enzyme isopropanol dehydrogenase.46 It can be detected and quantified from blood, urine and cerebrospinal fluid (CSF). Isopropanol was shown higher level in T2DM patients relative to the healthy controls in our work, and we found that it followed the increased level of acetone. Moreover, as reported, Through the detection of isopropanol in a patient with diabetic ketoacidosis, it has been proved that isopropanol may be a byproduct of acetone metabolism in certain disease states.47 Thus, we suggested that the isopropanol is associated with acetone metabolism, which is believed to be a significant differential metabolite in T2DM.
2,3,4-Trimethylhexane, 2,6,8-trimethyldecane, tridecane, and undecane are the acyclic alkanes. The experimental data indicated that T2DM patients had higher levels of 2,3,4-trimethylhexane, 2,6,8-trimethyldecane, tridecane, and undecane than healthy controls. Tridecane and undecane belong to the family of fatty acyls, and might be the product of polyunsaturated fatty acids metabolism, which are considered as a group in terms of their roles in fatty acids metabolism. They are found in allspice. Tridecane is also one of the major chemicals secreted by some insects as a defense against predators, while undecane is used as a mild sex attractant for various types of moths and cockroaches, and an alert signal for a variety of ants. As reported, undecane had been detected in human urine as one of the metabolic products.48 2,3,4-Trimethylhexane and 2,6,8-trimethyldecane belong to the family of endogenous metabolite which is a metabolite that are synthesized by the enzymes encoded by the genome or the microfloral genomes in HMDB. However, their metabolic pathways and the reason with the higher level in T2DM are not yet known.
m-Xylene belonging to the family of toluenes. As reported in KEGG database, in metabolism, m-xylene is involved in redox reaction with nicotinamide adenine dinucleotide (NADH), carrying electrons from one reaction to another. This reaction forms NADH, which can then be used as a reducing agent to donate electrons. These electron transfer reactions play an important role of beta oxidation, glycolysis, and the citric acid cycle. The first step in glycolysis is phosphorylation of glucose by a family of enzymes called hexokinases to form glucose 6-phsphate (G6P). In animals, an isozyme of hexokinase called glucokinase is also used in the liver, which has a much lower affinity for glucose, and differs in regulatory properties.49 The different substrate affinity and alternate regulation of this enzyme are a reflection of the role of the liver in maintaining blood sugar levels. Glucokinase activity serves as a principal control for the secretion of insulin in response to rising levels of blood glucose.50–53 As G6P is consumed, increasing amounts of ATP initiate a series of processes that result in release of insulin. One of the immediate consequences of increased cellular respiration is a rise in the NADH concentrations. As is known, T2DM is a metabolic disorder that is characterized by high blood glucose without enough insulin, resulting in reduced glucokinase. G6P converted by the glucose decreased because of the low concentration of glucokinase. Therefore, the concentration of m-xylene in exhaled breath might be shown lower level in T2DM due to a decreased glycolysis, as demonstrated by the experimental data in our study.
Toluene is an aromatic hydrocarbon. It has been shown to exhibit beta-oxidant, depressant, hepatoprotective, anesthetic and neurotransmitter functions.54–57 Toluene can be synthesized from benzylalcohol with the oxidized ferredoxin, reported in KEGG database. Adrenal ferredoxin (adrenodoxin) is expressed in mammals including humans. The human variant of adrenodoxin is referred to as ferredoxin-1.58 Ferredoxin-1 in humans participates in the synthesis of thyroid hormones. It also transfers electrons from adrenodoxin reductase to the cholesterol side chain cleavage cytochrome P450.59,60 The reason that increased level of toluene in T2DM remains to be unknown. Nevertheless, we suggested that there is a significant involvement in adrenodoxin metabolism. Efforts should be done for the relationships of toluene and T2DM. Toluene had also been detected in the urine of breast cancer patients and normal controls.61 It was reported that toluene may influence the glutamate and taurine neurotransmitter levels to control the actions.62
T2DM is a complex disease, caused by a combination of lifestyle and genetic factors, results a number of complications, including ischemic heart disease, stroke, even non-traumatic blindness and kidney failure. Therefore, T2DM is associated with numerous metabolites, which will improve the sensitivity and specificity for T2DM detection. In our study, several potential biomarkers showed their strong predictive power for distinguishing T2DM from healthy controls. Nevertheless, the actual metabolic pathways of them are not yet known. Therefore, efforts should be made to find out their metabolic pathways and interaction with protein, enzymes, or other small molecules. It will be very helpful for pathogenesis research of T2DM. The discovered candidate biomarkers also need to be extensively validated before they can be translated into real world diagnostic and screen application.
4. Conclusions
In summary, we investigated an effective comprehensive GC-MS based metabolic profiling tool to identify distinct breath biomarkers of T2DM and healthy controls. The study demonstrates that this sufficiently robust and non-invasive profiling approach can be a promising screening tool for the clinical diagnosis of T2DM. We discovered eight potential biomarkers including acetone, isopropanol, toluene, m-xylene, 2,3,4-trimethylhexane, 2,6,8-trimethyldecane, tridecane, and undecane. These results suggested that isopropanol, 2,3,4-trimethylhexane, 2,6,8-trimethyldecane, tridecane, and undecane in combination might be the best biomarkers for the clinical diagnosis of T2DM. Moreover, the method coupled with GC-MS and all the results provide some useful information for the understanding and monitoring of T2DM.Acknowledgements
This work is financially supported from National Science Foundation of China: New Method Investigation for the Selectivity Enrichment of Trace Biomarkers in Exhaled Breath (21305095), National Recruitment Program of Global Experts (NRPGE), the Hundred Talents Program of Sichuan Province (HTPSP), and the Startup Funding of Sichuan University for setting up the Research Center of Analytical Instrumentation.References
- R. Schubert, H. Schwoebel, A. Mau-Moeller, M. Behrens, P. Fuchs, M. Sklorz, J. Schubert, S. Bruhn and W. Miekisch, Metabolomics, 2012, 8, 1069–1080 CrossRef CAS.
- T. Khalid, B. Costello, R. Ewen, P. White, S. Stevens, F. Gordon, P. Collins, A. McCune, A. Shenoy, S. Shetty, N. Ratcliffe and C. Probert, Metabolomics, 2013, 9, 938–948 CrossRef CAS.
- O. B. Crofford, R. E. Mallard, R. Winton, N. Rogers, J. Jackson and U. Keller, Trans. Am. Clin. Climatol. Assoc., 1977, 88, 128 CAS.
- S. Levey, O. J. Balchum and V. Medrano, J. Lab. Clin. Med., 1964, 63, 574–584 CAS.
- C. Tassopoulos, D. Barnett and T. Russell Fraser, The Lancet, 1969, 293, 1282–1286 CrossRef.
- O. Owen, V. Trapp, C. Skutches, M. Mozzoli, R. Hoeldtke, G. Boden and G. Reichard, Diabetes, 1982, 31, 242–248 CrossRef CAS.
- V. Kumar, A. K. Abbas, N. Fausto and J. C. Aster, Robbins & Cotran Pathologic Basis of Disease, Elsevier Health Sciences, 2009 Search PubMed.
- S. Smyth and A. Heron, Nat. Med., 2006, 12, 75–80 CrossRef CAS PubMed.
- S. Melmed, K. S. Polonsky, P. R. Larsen and H. M. Kronenberg, Williams Textbook of Endocrinology: Expert Consult, Elsevier Health Sciences, 2011 Search PubMed.
- A. Amann, Siriraj Medical Journal, 2012, 64(6), 18–19 Search PubMed.
- O. Fiehn, TrAC, Trends Anal. Chem., 2008, 27, 261–269 CrossRef CAS PubMed.
- A. Garcia and C. Barbas, in Metabolic Profiling, ed. T. O. Metz, Humana Press, 2011, vol. 708, ch. 11, pp. 191–204 Search PubMed.
- F. M. Kloet, F. W. A. Tempels, N. Ismail, R. Heijden, P. T. Kasper, M. Rojas-Cherto, R. Doorn, G. Spijksma, M. Koek, J. Greef, V. P. Mäkinen, C. Forsblom, H. Holthöfer, P. H. Groop, T. H. Reijmers and T. Hankemeier, Metabolomics, 2012, 8, 109–119 CrossRef CAS PubMed.
- D. Zimmermann, M. Hartmann, M. Moyer, J. Nolte and J. Baumbach, Metabolomics, 2007, 3, 13–17 CrossRef CAS.
- B. Tan, Y. Liang, L. Yi, H. Li, Z. Zhou, X. Ji and J. Deng, Metabolomics, 2010, 6, 219–228 CrossRef CAS.
- C. L. Arthur and J. Pawliszyn, Anal. Chem., 1990, 62, 2145–2148 CrossRef CAS.
- K. W. Jordan, J. Nordenstam, G. Y. Lauwers, D. A. Rothenberger, K. Alavi, M. Garwood and L. L. Cheng, Dis. Colon Rectum, 2009, 52, 520 CrossRef PubMed.
- J. K. Nicholson, J. C. Lindon and E. Holmes, Xenobiotica, 1999, 29, 1181–1189 CrossRef CAS PubMed.
- J. Rollo, Cases of the Diabetes Mellitus: With the Results of the Trials of Certain Acids, and Other Substances, in the Cure of the Lues Venerea, T. Gillet, London, 2nd edn, 1798 Search PubMed.
- J. van der Greef and A. K. Smilde, J. Chemom., 2005, 19, 376–386 CrossRef CAS.
- R. Wang-Sattler, Z. Yu, C. Herder, A. C. Messias, A. Floegel, Y. He, K. Heim, M. Campillos, C. Holzapfel and B. Thorand, Mol. Syst. Biol., 2012, 8 Search PubMed.
- A. Floegel, N. Stefan, Z. Yu, K. Mühlenbruch, D. Drogan, H.-G. Joost, A. Fritsche, H.-U. Häring, M. H. de Angelis and A. Peters, Diabetes, 2013, 62, 639–648 CrossRef CAS PubMed.
- E. C. Horning and M.-G. Horning, Clin. Chem., 1971, 17, 802–809 CAS.
- W. J. Griffiths and Y. Wang, Chem. Soc. Rev., 2009, 38, 1882–1896 RSC.
- E. C. Y. Chan, P. K. Koh, M. Mal, P. Y. Cheah, K. W. Eu, A. Backshall, R. Cavill, J. K. Nicholson and H. C. Keun, J. Proteome Res., 2008, 8, 352–361 CrossRef PubMed.
- Z.-D. Zeng, Y.-Z. Liang, F.-T. Chau, S. Chen, M. K.-W. Daniel and C.-O. Chan, Anal. Chim. Acta, 2007, 604, 89–98 CrossRef CAS PubMed.
- J. van der Greef, T. Hankemeier and R. N. McBurney, Pharmacogenomics, 2006, 7(7), 1087–1094 CrossRef CAS PubMed.
- E. Fischer and U. Sauer, Eur. J. Biochem., 2003, 270, 880–891 CrossRef CAS.
- O. Fiehn, J. Kopka, P. Dörmann, T. Altmann, R. N. Trethewey and L. Willmitzer, Nat. Biotechnol., 2000, 18, 1157–1161 CrossRef CAS PubMed.
- J. Aa, F. Shao, G. Wang, Q. Huang, W. Zha, B. Yan, T. Zheng, L. Liu, B. Cao, J. Shi, M. Li, C. Zhao, X. Wang and Z. Wu, Metabolomics, 2011, 7, 217–225 CrossRef CAS.
- B. Feng, S. Wu, S. Lv, F. Liu, H. Chen, X. Yan, Y. Li, F. Dong and L. Wei, J. Proteome Res., 2007, 6, 2161–2167 CrossRef CAS PubMed.
- J. Trygg, E. Holmes and T. Lundstedt, J. Proteome Res., 2006, 6, 469–479 CrossRef PubMed.
- M. Chen and R. Hofestädt, J. Biomed. Inf., 2006, 39, 147–159 CrossRef CAS PubMed.
- S. Wold, K. Esbensen and P. Geladi, Chemom. Intell. Lab. Syst., 1987, 2, 37–52 CrossRef CAS.
- P. Jonsson, J. Gullberg, A. Nordström, M. Kusano, M. Kowalczyk, M. Sjöström and T. Moritz, Anal. Chem., 2004, 76, 1738–1745 CrossRef CAS PubMed.
- Jiye A., J. Trygg, J. Gullberg, A. I. Johansson, P. Jonsson, H. Antti, S. L. Marklund and T. Moritz, Anal. Chem., 2005, 77, 8086–8094 CrossRef CAS PubMed.
- S. Wiklund, E. Johansson, L. Sjostrom, E. J. Mellerowicz, U. Edlund, J. P. Shockcor, J. Gottfries, T. Moritz and J. Trygg, Anal. Chem., 2007, 80, 115–122 CrossRef PubMed.
- P. Paredi, S. Loukides, S. Ward, D. Cramer, M. Spicer, S. A. Kharitonov and P. J. Barnes, Thorax, 1998, 53, 775–779 CrossRef CAS.
- N. Lu, D. Wei, F. Chen and S. T. Yang, Eur. J. Lipid Sci. Technol., 2012, 114, 253–265 CrossRef CAS.
- X. Yan, J. Xu, J. Chen, D. Chen, S. Xu, Q. Luo and Y. Wang, Metabolomics, 2012, 8, 299–309 CrossRef CAS.
- M. Phillips, J. Herrera, S. Krishnan, M. Zain, J. Greenberg and R. N. Cataneo, J. Chromatogr. B: Biomed. Sci. Appl., 1999, 729, 75–88 CrossRef CAS.
- T. D. C. Minh, D. R. Blake and P. R. Galassetti, Diabetes Res. Clin. Pract., 2012, 97, 195–205 CrossRef PubMed.
- R. D. Stewart and E. A. Boettner, N. Engl. J. Med., 1964, 270, 1035–1038 CrossRef CAS PubMed.
- C. Kneepkens, C. Ferreira, G. Lepage and C. Roy, Clinical and investigative medicine, Med. Clin. Exp., 1992, 15, 163 CAS.
- C. M. Frank Kneepkens, G. Lepage and C. C. Roy, Free Radicals Biol. Med., 1994, 17, 127–160 CrossRef.
- K. Hoshino and K. Udagawa, VI. Isopropanol dehydrogenase and alcohol dehydrogenase of Lactobacillus brevis var. hofuensis.[in Japanese], Nippon Nogei Kagaku Kaishi, 1960, 34, 616–619 CrossRef.
- A. E. Jones and R. L. Summers, The Journal of Emergency Medicine, 2000, 19, 165–168 CrossRef CAS.
- S. Bouatra, F. Aziat, R. Mandal, A. C. Guo, M. R. Wilson, C. Knox, T. C. Bjorndahl, R. Krishnamurthy, F. Saleem and P. Liu, PLoS One, 2013, 8, e73076 CAS.
- S. Kawai, T. Mukai, S. Mori, B. Mikami and K. Murata, J. Biosci. Bioeng., 2005, 99, 320–330 CrossRef CAS PubMed.
- I. Sweet and F. M. Matschinsky, American Journal of Physiology-Endocrinology and Metabolism, 1995, 268, E775–E788 CAS.
- H. Wang and P. B. Iynedjian, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4372–4377 CrossRef CAS.
- F. Matschinsky, B. Glaser and M. Magnuson, Diabetes, 1998, 47, 307–315 CrossRef CAS.
- P. Iynedjian, Cell. Mol. Life Sci., 2009, 66, 27–42 CrossRef CAS PubMed.
- J. Aono, E. Takimoto, T. Komatsu, A. Takeda, W. Ueda and M. Hirakawa, The Japanese journal of anesthesiology, 1990, 39, 388 CAS.
- T. Biegert, G. Fuchs and J. Heider, Eur. J. Biochem., 1996, 238, 661–668 CAS.
- K. S. Hougaard, Å. M. Hansen, U. Hass and S. P. Lund, Pharmacol. Toxicol., 2003, 92, 148–152 CAS.
- U. Tas, M. Ogeturk, S. Meydan, I. Kus, T. Kuloglu, N. Ilhan, E. Kose and M. Sarsilmaz, Toxicol. Ind. Health, 2011, 27, 465–473 CrossRef CAS PubMed.
- A. Chaikuad, C. Johansson, T. Krojer, W. W. Yue, C. Phillips, J. E. Bray, A. C. W. Pike, J. R. C. Muniz, M. Vollmar, J. Weigelt, C. H. Arrowsmith, A. M. Edwards, C. Bountra, K. Kavanagh and U. Oppermann, 2010, DOI:10.2210/pdb3p1m/pdb.
- A. V. Grinberg, F. Hannemann, B. Schiffler, J. Müller, U. Heinemann and R. Bernhardt, Proteins: Struct., Funct., Bioinf., 2000, 40, 590–612 CrossRef CAS.
- D. Beilke, R. Weiss, F. Löhr, P. Pristovšek, F. Hannemann, R. Bernhardt and H. Rüterjans, Biochemistry, 2002, 41, 7969–7978 CrossRef CAS PubMed.
- C. L. Silva, M. Passos and J. S. Câmara, Talanta, 2012, 89, 360–368 CrossRef CAS PubMed.
- T.-T. Win-Shwe, D. Mitsushima, D. Nakajima, S. Ahmed, S. Yamamoto, S. Tsukahara, M. Kakeyama, S. Goto and H. Fujimaki, Toxicol. Lett., 2007, 168, 75–82 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra01422g |
| This journal is © The Royal Society of Chemistry 2014 |