Probing structural and catalytic characteristics of galactose oxidase confined in nanoscale chemical environments†
Abstract
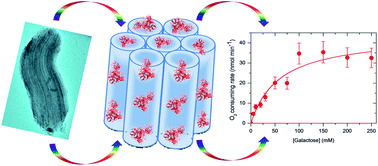
Galactose oxidase (GAOX) is a special metalloenzyme in terms of its active site structure and catalytic mechanisms. This work reports a study where the enzyme confined in a nanoscale chemical environment provided by mesoporous silicas (MPS) is probed. Two types of MPS, i.e. SBA-15 and MCF, were synthesized and used to accommodate GAOX. SBA-15-ROD is rod-shaped particles with periodically ordered nanopores (9.5 nm), while MCF has a mesocellular foam-like structure with randomly distributed pores (23 nm) interconnected by smaller windows (8.8 nm). GAOX is non-covalently confined in SBA-15-ROD, while it is covalently immobilized in MCF. Relatively high loadings in the range of 50–60 mg g−1 are achieved. Electron spin resonance (ESR) spectroscopy is used to probe the active site structures of the enzyme. The similar ESR spectra observed for GAOX in the free and immobilized states support that the electronic structure, particularly the copper catalytic centre of confined GAOX is well retained. The catalytic activity of confined enzyme is high, although the catalytic kinetics is slowed down, mainly attributed to the diffusion limitation of substrate and product in the nanoscale channels. The apparent Michaelis constant (KM) of the enzyme is largely unchanged upon immobilization, while the turnover number (kcat) is slightly reduced. The overall catalytic efficiency, represented by the ratio of kcat/KM, is retained around 70% and 60% for SBA-15 and MCF immobilization, respectively. The thermal resistance is enhanced up to 60 °C, but with no further enhancement above 60 °C.

 Please wait while we load your content...
Please wait while we load your content...